Background: Yeast sterol regulatory element-binding protein (SREBP) proteolytic activation requires the Golgi Dsc E3 ligase and the proteasome.
Results: Genetic selection identified additional genes required for SREBP activation.
Conclusion: UBX domain protein Dsc5 and AAA ATPase Cdc48 are Dsc E3 ligase subunits required for SREBP proteolysis.
Significance: Dsc5 and Cdc48 provide a mechanistic link between the Dsc E3 ligase and proteasome in SREBP proteolysis.
Keywords: Hypoxia, Sterol, Transcription Factors, Ubiquitin Ligase, Yeast Genetics
Abstract
Schizosaccharomyces pombe Sre1 is a membrane-bound transcription factor that controls adaptation to hypoxia. Like its mammalian homolog, sterol regulatory element-binding protein (SREBP), Sre1 activation requires release from the membrane. However, in fission yeast, this release occurs through a strikingly different mechanism that requires the Golgi Dsc E3 ubiquitin ligase complex and the proteasome. The mechanistic details of Sre1 cleavage, including the link between the Dsc E3 ligase complex and proteasome, are not well understood. Here, we present results of a genetic selection designed to identify additional components required for Sre1 cleavage. From the selection, we identified two new components of the fission yeast SREBP pathway: Dsc5 and Cdc48. The AAA (ATPase associated with diverse cellular activities) ATPase Cdc48 and Dsc5, a ubiquitin regulatory X domain-containing protein, interact with known Dsc complex components and are required for SREBP cleavage. These findings provide a mechanistic link between the Dsc E3 ligase complex and the proteasome in SREBP cleavage and add to a growing list of similarities between the Dsc E3 ligase and membrane E3 ligases involved in endoplasmic reticulum-associated degradation.
Introduction
Mammalian cellular cholesterol homeostasis is maintained through the action of the well characterized SREBP4 pathway. SREBP is a membrane-bound transcription factor that is inserted into the endoplasmic reticulum (ER) membrane, with its N and C termini facing the cytosol and spaced by a short ER luminal loop (1). SREBP forms a stable complex with a second multiple transmembrane domain protein, SREBP cleavage activating protein (Scap), such that sufficient cellular cholesterol levels maintain SREBP in an inactive state through ER retention of the SREBP-Scap complex (2). When cholesterol is depleted from membranes, Scap undergoes a conformational change, and the SREBP-Scap complex traffics to the Golgi apparatus. There, two sequential proteolytic cleavage events mediated by the Golgi-resident site-1 protease (S1P) and site-2 protease (S2P) release the N-terminal transcription factor domain of SREBP from the membrane. The soluble N-terminal SREBP transcription factor travels to the nucleus and up-regulates genes involved in sterol biosynthesis and uptake from the environment (3). Consequently, cellular sterols return to sufficient levels, SREBP activity is repressed, and cholesterol homeostasis is maintained.
Fission yeast employs a conserved SREBP system for maintaining sterol homeostasis (4). However, there are several differences. For instance, Schizosaccharomyces pombe has co-opted the SREBP pathway as a principal regulator of the hypoxic response (5–7). Further, fission yeast employs a unique mechanism for SREBP processing. Surprisingly, S. pombe lacks identifiable homologs of S1P and S2P required for mammalian SREBP cleavage. Given the absence of canonical SREBP cleavage machinery, we reasoned that SREBP cleavage in S. pombe must proceed through a S1P- and S2P-independent mechanism. Through a genetic screen of the fission yeast non-essential haploid deletion collection, we identified four genes that, when deleted, confer a defect for SREBP cleavage, and we named these genes dsc1–dsc4 (8). dsc1 was the only dsc gene with a characterized homolog, Saccharomyces cerevisiae TUL1 (9). Like Tul1p, Dsc1 is an integral membrane, Golgi E3 ubiquitin ligase that has a C-terminal RING domain. Dsc2–Dsc4 are also integral membrane proteins but with largely uncharacterized functions. Dsc1–Dsc4 forms a stable Golgi-localized ubiquitin E3 ligase complex that is required for SREBP activation insomuch as the Dsc complex binds SREBP and the RING domain function of Dsc1 is essential for proper SREBP cleavage (8). In addition, Dsc-mediated SREBP cleavage required the E2 ubiquitin conjugating enzyme Ubc4 and the proteasome. Bioinformatic analysis revealed similarities between the Dsc complex and the ER-localized Hrd1 ligase complex involved in ER-associated degradation (ERAD) (8), but the mechanistic link between the Dsc E3 ligase and the proteasome is unknown.
In ERAD, the Hrd1 E3 ligase complex recognizes misfolded proteins and marks them via ubiquitination for proteasomal degradation in the cytosol (10–12). Misfolded proteins that are located in the ER lumen or integral to the ER membrane must first be extracted from the ER and released into the cytosol. The mechanical force for substrate dislocation from the ER is provided by the AAA ATPase, Cdc48, also known as mammalian p97, which forms a homohexameric complex and uses cycles of ATP hydrolysis to generate energy for a variety of cellular processes (13). The specific recruitment of Cdc48/p97 to these protein complexes often requires the activity of UBX domain-containing proteins. In the case of the Hrd1 ligase, the UBX domain-containing protein Ubx2p couples Cdc48p to the complex. Given the similarities between the Hrd1 complex and the Dsc complex, we examined whether additional components of the Dsc E3 ligase complex existed to bridge substrate recognition and substrate processing.
Here, we describe a genetic selection in S. pombe that identified new components of the Dsc E3 ligase complex required for fission yeast SREBP activation: Dsc5, a UBX domain-containing protein, and Dsc6/Cdc48. The discovery of these new members of the Dsc complex expands the similarities between the Dsc E3 ligase and the Hrd1 E3 ligase complex involved in ERAD. These findings provide a mechanism for delivery of proteins from the Dsc E3 ligase to the proteasome and establish the Dsc E3 ligase complex as an important system for understanding how cells identify and process specific membrane proteins.
EXPERIMENTAL PROCEDURES
Standard molecular biology techniques, genetic manipulations, and SREBP cleavage assays were performed as described previously (6, 14, 15). General chemicals and materials were obtained from Fisher and Sigma with other sources as indicated: Edinburgh minimal medium from MP Biomedicals; yeast extract, peptone, agar from BD Biosciences; digitonin from EMD Chemicals; oligonucleotides from Integrated DNA Technologies; alkaline phosphatase and mouse anti-GFP monoclonal antibody from Roche Applied Science; horseradish peroxidase-conjugated, affinity-purified donkey anti-rabbit and anti-mouse IgG from Jackson ImmunoResearch; anti-Myc monoclonal 9E10 IgG from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); anti-Myc polyclonal IgG (catalog no. 06-549) from Upstate Biotechnology; and prestained protein standards from Bio-Rad.
Strains and Media
Wild-type haploid S. pombe KGY425 and derived strains were grown to log phase at 30 °C in YES medium (5 g/liter yeast extract plus 30 g/liter glucose and supplements, 225 mg/liter each of uracil, adenine, leucine, histidine, and lysine) unless otherwise indicated (16). Strains used in this study are described in supplemental Table 1 (17). Strains ESY58 (h−, his3-D1::7xSRE-lacZ-kanMX6, leu1-32, ade6-M210, ura4-D18::7xSRE-ura4+-kanMX) and ESY79 (h+, his3-D1::7xSRE-lacZ-kanMX6, leu1-32, ade6-M210, ura4-D18::7xSRE-ura4+-kanMX) were generated as described previously using seven tandem copies of the Tf2–1 sterol-regulatory element to drive reporter gene activation (18). The dsc5ΔUBX::kanMX6 strain was made by deleting sequences coding for amino acids 324–427 using standard techniques (14).
Yeast Mutagenesis and Selection
Fission yeast were mutagenized using methyl methanesulfonate (MMS) with minor modifications as described (15). ESY58 cells were grown overnight to a cell density of 1 × 107 cells/ml, washed in Edinburgh minimal medium plus supplements (MM) (20 g/liter glucose, 225 mg/liter each of uracil, adenine, leucine, histidine, and lysine), and resuspended at a concentration of 1 × 108 cells/ml in MM containing 0.024% (w/v) MMS. These cultures were grown for 3 h at room temperature. Cells were washed three times with sterile MilliQ water and resuspended with sterile MilliQ water at a cell density of 5 × 106 cells/ml. Mutagenized cells were plated onto selection medium (Edinburgh minimal medium, 2% (w/v) agar, 20 g/liter glucose, 225 mg/liter each of adenine, leucine, and histidine, 50.25 mg/liter uracil, 0.1% (w/v) 5-fluoroorotic acid (5-FOA), 0.2 mm CoCl2) at a cell density 5 × 105 cells/plate. Plates were wrapped in foil and incubated at 30 °C.
Colonies from the initial mutagenesis were picked and streaked onto selection medium to isolate single colonies. These cells were patched onto selection medium and then streaked onto YES medium, containing 1.6 mm CoCl2 to test for CoCl2 sensitivity. CoCl2-sensitive isolates were then streaked a second time onto selection medium to isolate single colonies. To test whether strains contained mutations in sre1+ and scp1+, CoCl2-sensitive isolates were co-transformed with plasmids expressing sre1+ and scp1+ from the cauliflower mosaic virus promoter or the corresponding empty vector control plasmids pSLF101 and pSLF102 (19). Transformants were screened for CoCl2 sensitivity. Parental isolates of strains whose growth on CoCl2 was not rescued by plasmids expressing sre1+ and scp1+ were evaluated by Western blotting for Sre1 cleavage and β-galactosidase activity. β-Galactosidase activity was determined from cultures grown for 12 h under anaerobic conditions to a cell density of 1 × 107 cells/ml as described previously (7). Isolates that demonstrated impaired Sre1 cleavage and low β-galactosidase activity were transformed with plasmid pAH05 expressing the soluble N-terminal transcription factor domain of Sre1 (amino acids 1–440) (6). Isolates whose CoCl2 sensitivity was rescued by expression of Sre1N were backcrossed to ESY79. These backcrossed strains were used to perform linkage analysis.
An additional selection was performed as described above, using 0.012% MMS. 5-FOA-resistant isolates were characterized using a combination of the above approaches leading to the identification of additional known and unknown components of the Dsc E3 ligase complex.
Linkage Analysis
To facilitate linkage analysis, both h− and h+ strains were isolated for each of the ESY58-derived mutants of interest during backcrossing to ESY79. The progeny from the backcross were mated to one another as well as to dsc1–4Δ, sre1Δ, and scp1Δ cells. All strains tested were CoCl2-sensitive. To assay linkage, a suspension of haploid spores from the mating was plated on YES or YES containing 1.6 mm CoCl2. The inability to recover CoCl2-resistant spores indicated linkage between the mutations. Positive and negative controls were included to control for variation in mating and sporulation efficiency.
Identification of cdc48 Mutant Alleles
A strain harboring dsc6-1 was backcrossed to ESY58. Two dsc6-1 isolates were selected and backcrossed for an additional 5 generations using standard mating techniques. Genomic DNA was prepared from one isolate from each backcross, and genomic DNA was sequenced using a Helicos single molecule sequencer using the manufacturer's protocol as described previously (20). Single nucleotide polymorphism (SNP) data for the two strains were filtered using several parameters to generate a list of potential genes of interest (supplemental Fig. 1). To confirm the presence of mutations in cdc48, genomic DNA was prepared from each member of the dsc6 linkage group, the cdc48 locus was PCR-amplified with primers oJB444 and oJB460, and PCR products were sequenced with primers oJB446-oJB449, oJB453, oJB460, and oJB466-oJB467.
Antibody Preparation
Antisera to Sre1(1–240), Sre2(1–426), Dsc1(20–319), Dsc2(250–372), Dsc3(1–190), and Dsc4(150–281) have been described previously (6, 8). Hexahistidine-tagged recombinant Dsc5(251–427) antigen was purified from Escherichia coli using Ni2+-NTA (Qiagen) according to the manufacturer's instructions. Dsc5 antiserum was generated by Covance based on a standard protocol. Antiserum to Cdc48 was the kind gift of R. Hartmann-Petersen (University of Copenhagen) (21).
Immunoprecipitation
Exponentially growing S. pombe cells were washed in phosphate-buffered saline (PBS; 1 mm KH2PO4, 3 mm Na2HPO4·7H2O, pH 7.4, 155 mm NaCl) and resuspended in 25 ml of PBS containing 2 mm dithiobis-succinimidyl propionate (Thermo Scientific). After a 30-min incubation at 30 °C, the reaction was quenched in 20 mm Tris-HCl, pH 7.5, for 15 min at 30 °C. Cells were then lysed with glass beads (0.5 mm; Sigma) in digitonin lysis buffer (50 mm HEPES, pH 6.8, 1% (w/v) digitonin, 50 mm KOAc, 2 mm Mg(OAc)2, 1 mm CaCl2, 200 mm sorbitol, 1 mm NaF, 0.3 mm Na3VO4) supplemented with protease inhibitors. Protease inhibitors included leupeptin (10 μg/ml), pepstatin A (5 μg/ml), PMSF (1 mm) from Sigma and the Complete EDTA-free protease inhibitor (Roche Applied Science). Lysate was rotated at 4 °C for 40 min. Insoluble cellular debris was removed by centrifugation at 100,000 × g for 10 min at 4 °C. Cell lysate (1.5 mg of protein) in 0.7 ml was incubated with 1 μg of anti-Myc 9E10 IgG for 15 min at 4 °C prior to nutation with 40 μl of protein A beads (RepliGen) for 16 h at 4 °C. The beads were washed three times with 1 ml of digitonin lysis buffer, and immunopurified protein was eluted by boiling in SDS-PAGE loading buffer (6). For Dsc2 immunoprecipitation, the cleared lysate was incubated with 5 μl of Dsc2 antiserum (8).
In Vitro Binding Assay
GST was expressed using pGEX-KG in DH5α E. coli cells (22), induced by 0.6 mm isopropyl-1-thio-β-galactopyranoside, and purified using glutathione-Sepharose beads (GE Healthcare) according to the manufacturer's protocol. The sequence for Dsc5(251–427) containing the UBX domain was cloned into pGEX-KG and named GST-UBX. To generate GST-UBX lysate, E. coli cells were induced for GST-UBX expression as above and lysed by sonication in PBS plus protease inhibitors. After the addition of 1% (w/v) Triton X-100, the GST-UBX lysate was cleared by centrifugation at 15,000 × g for 20 min at 4 °C.
For the binding assay, MagneGST beads (25 μl) (Promega) were blocked in PBS containing 1% bovine serum albumin and then incubated with either GST-UBX lysate (1 mg) or purified GST (50 μg) in 0.5 ml of PBS for 1 h at 4 °C. Beads were washed three times with PBS and once with binding buffer (20 mm Hepes, pH 7.2, 150 mm KOAc, 5 mm Mg(OAc)2, 250 mm sorbitol, 0.2% Nonidet P-40, protease inhibitors) and incubated with wild-type S. pombe cytosol (1.4 mg) in 0.1 ml of binding buffer for 30 min at 4 °C. After three washes with binding buffer, bound proteins were eluted by boiling in SDS-PAGE loading dye, separated by SDS-PAGE, and evaluated by Coomassie stain and Western blotting with anti-Cdc48 antibody. S. pombe cytosol was prepared by lysing exponentially growing cells with a high pressure emulsifier (EmulsiFlex-C-3, Avestin) in B88 buffer (20 mm Hepes, pH 7.2, 150 mm KOAc, 5 mm Mg(OAc)2, and 250 mm sorbitol) plus protease inhibitors and subjecting lysate to centrifugation at 100,000 × g for 10 min at 4 °C. Final concentration of protein was 14 mg/ml.
RESULTS
Mutagenesis and Selection of Genes Required for Sre1 Proteolytic Activation
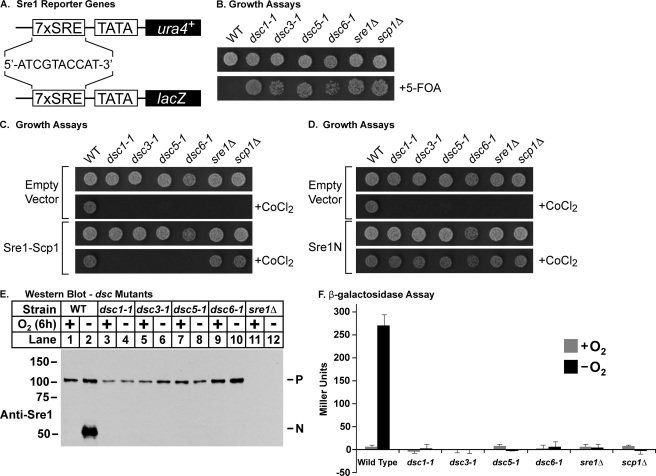
To identify genes required for Sre1 cleavage, we developed reporter systems that rely on the N-terminal transcription factor domain of Sre1 (Sre1N) for activity. We generated an Sre1N-dependent promoter by placing seven tandem copies of the sterol regulatory element from the S. pombe Tf2–1 transposon upstream of a minimal promoter from the nmt1+ gene (18, 23). This promoter directed expression of two different integrated reporter genes, ura4+ and lacZ (Fig. 1A). In this way, the activity of the reporter genes was dependent on the amount of the Sre1N in the cell.
FIGURE 1.
Identification of genes defective for Sre1 cleavage. A, diagram of integrated Sre1 reporter genes where seven tandem copies of the sterol regulatory element (sequence is shown for a single copy) are fused to the nmt1+ minimal promoter and drive expression of ura4+ or lacZ. B, wild type and the indicated mutants (1000 cells) were grown on rich medium or minimal medium containing 5-FOA. C, wild type and the indicated mutants (1000 cells) containing plasmids expressing Sre1 and Scp1 or empty vectors were grown on rich medium or rich medium containing CoCl2. D, wild type and the indicated mutants (1000 cells) containing a plasmid expressing Sre1N(1–440) or the empty vector were grown on rich medium or rich medium containing CoCl2. E, Western blot probed with anti-Sre1 IgG of phosphatase-treated whole cell lysates from wild type and the indicated mutant strains grown for 6 h in the presence or absence of oxygen. P and N, precursor and nuclear forms of Sre1, respectively. F, wild type and the indicated mutants were grown for 12 h in the presence or absence of oxygen and assayed for β-galactosidase activity. Error bars, S.D. of three biological replicates.
To select for mutants defective in Sre1 activation, we took advantage of the 7xSRE-ura4+ reporter. The ura4+ gene product, orotidine-5′-phosphate decarboxylase, initiates the conversion of 5-FOA to the toxic compound 5-fluorouracil (24). In 7xSRE-ura4+ wild-type cells, the basal amount of Sre1N generates enough Ura4 to prevent growth on minimal medium containing 5-FOA (Fig. 1B). Cells expressing 7xSRE-ura4+ were mutagenized with MMS and selected for growth on minimal medium containing 5-FOA (Fig. 1B). Cells lacking a functional Sre1 pathway fail to grow on medium containing CoCl2 (25). As a secondary screen, we tested growth of the 5-FOA-resistant mutants on rich medium containing CoCl2 and isolated 116 mutants that were both 5-FOA-resistant and CoCl2-sensitive. To eliminate strains with mutations in sre1+ and scp1+, we assayed the ability of cotransformed plasmids expressing sre1+ and scp1+ to rescue growth of mutants on CoCl2 (Fig. 1C). Finally, to identify mutants with defects upstream of Sre1N, we tested the ability of a plasmid expressing Sre1N to rescue growth on CoCl2 (Fig. 1D). Together, these assays yielded nine mutant strains with defects in Sre1N activity.
A second selection was performed with a lower concentration of MMS, producing an additional 162 5-FOA-resistant and CoCl2-sensitive mutants. To determine the number of genes identified and address whether the selection mutants represented novel components of the Sre1 pathway, we performed a linkage analysis. Each of the 5-FOA-resistant and CoCl2-sensitive mutants was mated individually to one another and to strains harboring gene deletions of the known SREBP pathway components sre1, scp1, and dsc1-4. Spores from these matings were tested for the ability to grow on rich medium containing CoCl2. Isolation of CoCl2-resistant spores indicated that the two mutations were in different genes. Conversely, failure to isolate CoCl2-resistant spores indicated genetic linkage. In total, we tested 278 mutants and identified mutations in known Sre1 pathway components, including sre1+, scp1+, dsc1+, and dsc3+. In addition, we found a number of mutants that were unlinked to known genes and thus represented potentially novel components of the Sre1 pathway. Linkage tests among these mutants revealed new linkage groups, including two (dsc5 and dsc6) that were characterized further. Sequence-verified mutants from the linkage analysis are summarized in Table 1.
TABLE 1.
Sequenced dsc mutations
| Strain | Allele | Nucleotide changea | Amino acid change |
|---|---|---|---|
| ESY58-75 | dsc1-1 | C2085T | Q673X |
| ESY58-119 | dsc1-2 | Δ857–1097 | M286T … stop |
| ESY58-120 | dsc1-3 | G1746A | W582X |
| ESY58-46 | dsc3-1 | G289T | G97X |
| ESY58-59 | dsc3-2 | Δ661 | R221G … stop |
| ESY58-121 | dsc5-1 | C518T | Q111X |
| ESY58-53 | dsc5-2 | Δ478–1422 | Δ97–411 |
| ESY58-118 | dsc5-3 | T insertion at 557 | T126Y … stop |
| ESY58-29 | cdc48-1 | A1729T | N558I |
| ESY58-17 | cdc48-2 | G2249T | E731D |
| ESY58-40 | cdc48-3 | C1813T | A586V |
| ESY58-67 | cdc48-4 | G1029A | E325K |
a Nucleotide position in unspliced mRNA relative to start codon.
To confirm a defect in the production of Sre1N from the precursor, we performed Sre1 cleavage assays on these mutants. A representative Sre1 cleavage blot is shown in Fig. 1E. Wild-type cells showed robust accumulation of Sre1N when grown for 6 h under anaerobic conditions (Fig. 1E, lane 2). However, under the same conditions, the mutants failed to accumulate Sre1N (Fig. 1E, lanes 3–10). Consistent with this defect in Sre1N production, mutants failed to induce β-galactosidase expression from the 7xSRE-lacZ reporter under low oxygen (Fig. 1F). Collectively, these data demonstrate the ability of this selection scheme to isolate mutants with defects in Sre1 cleavage.
dsc5/ucp10 Is Required for Sre1 Cleavage
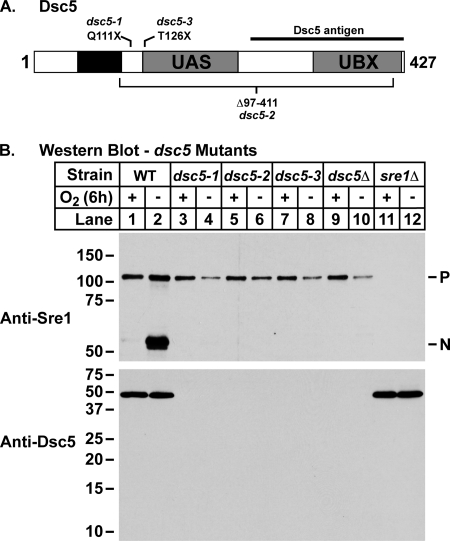
Genetic analysis revealed that one of the two new linkage groups was linked to ucp10+, which codes for a protein we identified by mass spectrometry as a candidate Dsc E3 ligase-interacting protein (Fig. 2A) (8, 26). Deletion of ucp10+ resulted in an Sre1 cleavage defect and a CoCl2-sensitive phenotype, and thus we named the gene dsc5+. dsc5+ codes for a 427-amino acid protein predicted to contain a transmembrane domain as well as UAS and UBX domains toward the C terminus (Fig. 2A). Although the function of the UAS domain remains unclear, there is significant evidence that UBX domains mediate interaction with Cdc48/p97 (13, 21, 27). Subsequently, the complete set of new mutants was tested for linkage to dsc5Δ, and this analysis identified three dsc5 mutants. DNA sequencing of the dsc5 alleles revealed mutations that result in truncations of the protein (Fig. 2A and Table 1). In each instance, Dsc5 is disrupted after its predicted transmembrane region, resulting in deletion of both the UAS and UBX domains (Fig. 2A).
FIGURE 2.
Sre1 cleavage requires dsc5. A, diagram of Dsc5, where the black box indicates a predicted hydrophobic region of the protein. Gray boxes, UAS and UBX domains identified by conserved domain prediction programs. Mutant alleles are indicated above and below the diagram. B, Western blots of phosphatase-treated whole cell lysates from wild type and the indicated mutant strains grown for 6 h in the presence or absence of oxygen probed with anti-Sre1 IgG and anti-Dsc5 serum.
To confirm that dsc5 mutants were defective for Sre1 cleavage, we grew wild-type, sre1Δ, and dsc5Δ cells along with each of the mutants for 6 h in the presence or absence of oxygen. Wild-type cells showed robust cleavage of Sre1 under low oxygen (Fig. 2B, lanes 1 and 2). However, each of the dsc5 mutants had a block in Sre1 cleavage under low oxygen that was equivalent to the defect in a dsc5Δ strain (Fig. 2B, lanes 3–10). dsc5 mutant cell extracts showed no detectable Dsc5 protein due to the deletion of the region recognized by the Dsc5 antiserum (Fig. 2B, bottom).
cdc48 Is Required for Sre1 Cleavage
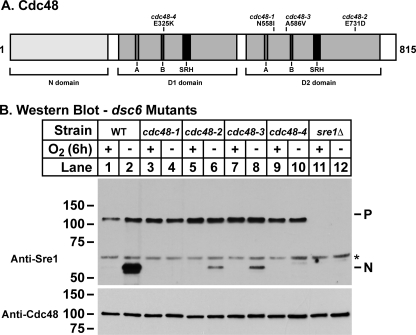
Linkage tests among the mutants identified the dsc6 linkage group that contained four independently isolated, CoCl2-sensitive mutants. Multiple attempts to clone dsc6 using plasmid library complementation of the CoCl2 growth defect failed. As an alternative approach to identify dsc6, we sequenced the genomes of two backcrossed strains harboring the dsc6-1 allele. Comparison of the two genomes with the wild-type reference genome revealed 741 SNPs present in both isolates (28). In analyzing the data, we made several assumptions to reduce the number of SNPs to a manageable number (see supplemental Fig. 1). As a result, we identified 10 candidate genes that contained open reading frame SNPs that satisfied our criteria. cdc48 was among these 10 genes. Given the central role of Cdc48 in the ubiquitin-proteasome system and the role of the Dsc E3 ubiquitin ligase complex in Sre1 processing, we considered cdc48+ a strong candidate for dsc6+. Linkage tests using tetrad analysis between an existing cdc48-353 temperature-sensitive mutant and dsc6-1 demonstrated that dsc6-1 was an allele of cdc48 (29). Next, we sequenced the cdc48 locus for each dsc6 mutant and found that each allele harbored a missense mutation in cdc48 (Fig. 3A and Table 1). cdc48 is an essential gene, possibly explaining the absence of nonsense mutations (29). Reconstitution of the individual cdc48 mutations in an otherwise wild-type background conferred a CoCl2-sensitive phenotype to the strains (data not shown). Thus, dsc6 is cdc48.
FIGURE 3.
Sre1 cleavage requires cdc48/dsc6. A, diagram of Cdc48/Dsc6 from S. pombe. The substrate and cofactor binding N domain as well as the AAA domain containing D1 and D2 domains are outlined in light gray boxes. The D1 and D2 domains each contain three important highly conserved domains. These domains, the Walker A, Walker B, and SRH, are indicated by gray, dark gray, and black boxes, respectively. These functional regions of the protein are based on alignments with the Mus musculus VCP/p97 homolog of Cdc48/Dsc6. Mutation for each allele is represented above the diagram. B, Western blot probed with anti-Sre1 IgG of phosphatase-treated whole cell lysates from wild type and the indicated mutant strains grown for 6 h in the presence or absence of oxygen. *, nonspecific, cross-reacting protein.
To confirm that each cdc48 mutation was defective for Sre1 cleavage, we grew wild-type and sre1Δ cells along with each of the cdc48 mutants for 6 h in the presence or absence of oxygen to induce Sre1 cleavage. Wild-type cells accumulated Sre1N under low oxygen (Fig. 3B, lanes 1 and 2). In contrast, each cdc48 mutant failed to accumulate Sre1N (Fig. 3B, lanes 3–10). Although each cdc48 mutant possessed a clear defect in Sre1 cleavage, variation existed among the cdc48 alleles with respect to the severity of the defect. dsc6-1 and dsc6-4 strains showed a complete block in Sre1 cleavage, whereas dsc6-2 and dsc6-3 strains showed a low level of Sre1 cleavage under low oxygen (Fig. 3B, lanes 3–10). Together, these data demonstrate that cdc48 is required for proper function of the Sre1 pathway.
dsc5 and cdc48 Are Required for Sre2 Cleavage
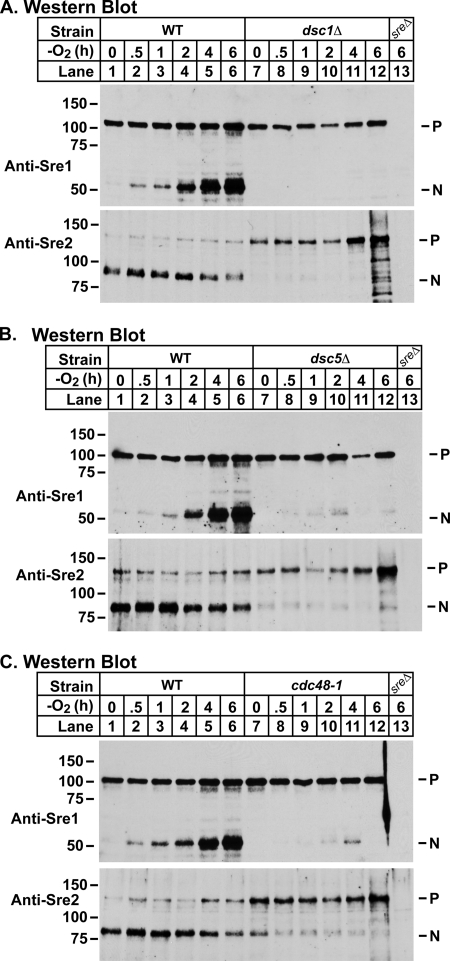
To further characterize the requirement of dsc5 and cdc48 for Sre1 cleavage, we performed a series of low oxygen time course experiments, in which we monitored Sre1 cleavage in wild-type, dsc1Δ, dsc5Δ, and cdc48-1 cells. In wild-type cells, Sre1N was produced by 30 min and accumulated throughout the course of the experiment (Fig. 4, A–C, lanes 1–6). In dsc1Δ cells, Sre1N was not detected at any time point (Fig. 4A, lanes 7–12). In contrast, in both dsc5Δ and cdc48-2 cells, a low level of Sre1N was transiently detected at around 2–4 h (Fig. 4, B and C, lanes 7–12).
FIGURE 4.
Sre2 cleavage requires dsc5 and cdc48. A, Western blot probed with anti-Sre1 IgG (top) or anti-Sre2 serum (bottom) of phosphatase-treated whole cell lysates from sre2-GFP and sre2-GFP dsc1Δ cells grown for the indicated times in the absence of oxygen. B, Western blot probed with anti-Sre1 IgG (top) or anti-Sre2 serum (bottom) of phosphatase-treated whole cell lysates from sre2-GFP and sre2-GFP dsc5Δ cells grown for the indicated times in the absence of oxygen. C, Western blot probed with anti-Sre1 IgG (top) or anti-Sre2 serum (bottom) of phosphatase-treated whole cell lysates from sre2-GFP and sre2-GFP cdc48-1 cells grown for the indicated times in the absence of oxygen. For each immunoblot, sreΔ denotes sre1Δ and sre2Δ strains for the anti-Sre1 and anti-Sre2 blots, respectively.
S. pombe has a second SREBP homolog called Sre2 that possesses a similar domain structure and membrane topology as Sre1, but Sre2 lacks the C terminus that imparts sterol regulation to Sre1 cleavage (6). As a result, Sre2 is constitutively cleaved. We previously reported that deletion of dsc1–dsc4 impaired Sre2 cleavage (5). To determine whether Dsc5 and Cdc48 are required for Sre2 cleavage, we performed an immunoblot analysis of the low oxygen time course experiments described above, using Sre2 antiserum. In wild-type cells, the nuclear form of Sre2 (Sre2N) was initially the predominant species, decreasing slightly over the course of the experiment (Fig. 4, A–C, bottom panels, lanes 1–6). dsc1Δ cells had a severe defect in Sre2 cleavage in the presence of oxygen and began to accumulate the precursor form at ∼4–6 h after shifting to low oxygen (Fig. 4A, bottom, lanes 7–12). Both dsc5Δ and cdc48-1 cells demonstrated a similar Sre2 cleavage defect as observed in dsc1Δ cells with a decrease in the production of Sre2N and an accumulation of the precursor form beginning at ∼4–6 h. Collectively, these experiments illustrate that Dsc5 and Dsc6 are required for normal cleavage of fission yeast SREBPs in the presence and absence of oxygen.
Dsc5 and Cdc48 Interact with Dsc Complex
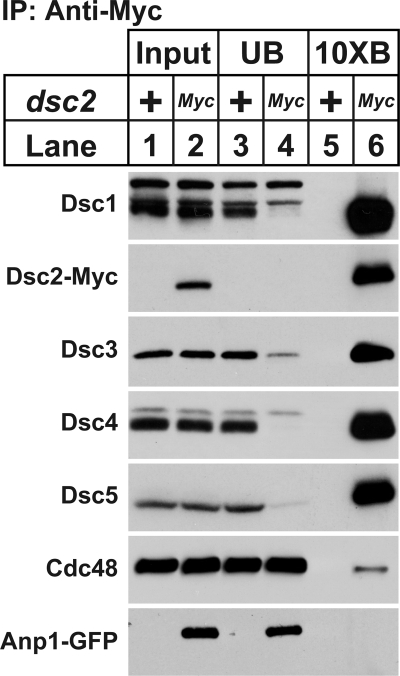
Dsc1–Dsc4 form a stable complex required for SREBP activation (8). Given the role of Dsc5 and Dsc6 in SREBP cleavage, we sought to determine whether these proteins interacted with known Dsc complex components. To this end, we grew wild-type cells or cells expressing a Myc-tagged Dsc2 and the Golgi membrane protein Anp1-GFP in the presence of oxygen. Dsc2-13xMyc-interacting proteins were purified from digitonin-solubilized cell extracts and subjected to immunoblot analysis with antibodies directed against Dsc1–Dsc5, Cdc48, and GFP. Consistent with our previous results, Dsc2 bound Dsc1, Dsc3, and Dsc4 but not the cis-Golgi membrane protein, Anp1 (Fig. 5, lanes 5 and 6). Dsc2 also bound both Dsc5 and Cdc48 (Fig. 5, lanes 5 and 6), demonstrating that Dsc5 and Dsc6 are components of the Dsc E3 ligase complex.
FIGURE 5.
Dsc5 and Cdc48 interact with Dsc2. Digitonin-solubilized extracts were prepared from cross-linker-treated, wild-type, and dsc2-13xMyc anp1-GFP cells. Dsc2-13xMyc and its interacting proteins were immunopurified (IP) with anti-Myc IgG-9E10 monoclonal antibody. Equal amounts of input (lanes 1 and 2), unbound (lanes 3 and 4), and 10-fold bound (lanes 5 and 6) fractions were analyzed by immunoblot with anti-Dsc1 IgG, anti-Myc IgG, anti-Dsc3 serum, anti-Dsc4 serum, anti-Dsc5 serum, anti-Cdc48 serum, and anti-GFP IgG.
Dsc5 UBX Domain Binds Cdc48 but Is Not Required for Sre1 Cleavage
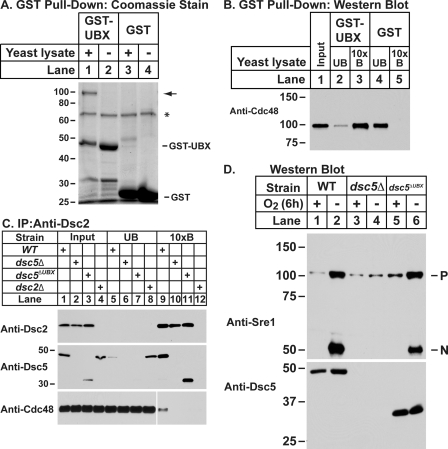
Dsc5 is predicted to contain a UBX domain, a known Cdc48-interacting motif (30). To determine whether the Dsc5 UBX domain was functional, we expressed it as a GST fusion protein (GST-UBX) and examined its ability to bind Cdc48. GST or GST-UBX was purified from E. coli, immobilized on magnetic beads, and incubated with wild-type S. pombe cytosol. Bound proteins were eluted and evaluated by SDS-PAGE with Coomassie staining. Comparison of the eluates from the GST-UBX and GST-only loaded beads showed a protein of ∼100 kDa that was present specifically in the GST-UBX eluate (Fig. 6A, lanes 1 and 3). To determine whether the 100 kDa band was Cdc48, we performed an immunoblot on the bound and unbound fractions from the experiment in Fig. 6A with anti-Cdc48 serum. Cdc48 bound specifically to the GST-UBX-loaded and not the GST only-loaded beads (Fig. 6B, lanes 3 and 5), indicating that the Dsc5 UBX domain binds Cdc48.
FIGURE 6.
UBX domain of Dsc5 is required for Cdc48 binding but not for SREBP cleavage. A, the UBX domain of Dsc5 was expressed as a GST fusion protein (GST-UBX) in E. coli. Purified GST-UBX or GST was bound to magnetic beads and incubated with whole cell lysates prepared from wild-type cells. GST-UBX and GST along with their interacting proteins were eluted and analyzed by SDS-PAGE and Coomassie staining. B, equal amounts of input (lane 1), unbound (lanes 2 and 4), and 10-fold bound (lanes 3 and 5) fractions from samples in A were analyzed by immunoblot with anti-Cdc48 serum. C, digitonin-solubilized lysates from wild-type, dsc5Δ, dsc5ΔUBX, and dsc2Δ cells treated with cross-linker were prepared, and Dsc2-interacting proteins were immunopurified (IP) using anti-Dsc2 antibodies. Equal amounts of input (lanes 1–4), unbound (lanes 5–8), and 10-fold bound (lanes 9–12) fractions were analyzed by immunoblot with anti-Dsc2, anti-Dsc5, and anti-Cdc48 antibodies. An empty lane was included between lanes 8 and 9 on the Cdc48 gel to prevent contamination but was later removed. The two images are from the same exposure of a single blot. D, whole cell lysates from wild-type, dsc5Δ, and dsc5ΔUBX cells grown for 6 h in the presence or absence of oxygen were prepared and analyzed by immunoblotting with anti-Sre1 IgG or anti-Dsc5 serum.
To determine whether the UBX domain of Dsc5 is required to recruit Cdc48 to the Dsc complex in vivo, we generated a strain expressing a truncated version of Dsc5 lacking the C-terminal UBX domain (amino acids 324–427) (dsc5ΔUBX). Digitonin-solubilized lysates from wild-type, dsc5Δ, dsc5ΔUBX, and dsc2Δ cells were prepared, and Dsc2-interacting proteins were purified using anti-Dsc2 serum. In wild-type cells, both Dsc5 and Cdc48 bound Dsc2 (Fig. 6C, lane 9), whereas deletion of dsc5 resulted in a loss of Cdc48 binding to Dsc2 (Fig. 6C, lane 10). In dsc5ΔUBX cells, Dsc5ΔUBX bound to Dsc2, but binding between Dsc2 and Cdc48 was abolished (Fig. 6C, lane 11). Thus, the Dsc5 UBX domain is required for Cdc48 recruitment to Dsc2 but not Dsc5 binding to Dsc2.
Given the requirement for Cdc48 in SREBP cleavage and the function of the UBX domain in Cdc48 recruitment, we tested whether Sre1 cleavage required the Dsc5 UBX domain. Wild-type, dsc5Δ, and dsc5ΔUBX cells were assayed for Sre1 cleavage in the presence or absence of oxygen. In wild-type cells, Sre1N is generated under low oxygen (Fig. 6D, lane 2), but deletion of dsc5 blocked Sre1N cleavage (Fig. 6D, lane 4). Surprisingly, Sre1N accumulates under low oxygen in the dsc5ΔUBX strain, albeit to a somewhat diminished extent compared with wild-type cells (Fig. 6D, compare lanes 6 and 2). Collectively, these data demonstrate that the UBX domain of Dsc5 recruits Cdc48 to the Dsc complex but is not absolutely required for Sre1 cleavage.
DISCUSSION
Identification of New Dsc Complex Components
Fission yeast Sre1 is a principal regulator of the hypoxic response (6, 7). Sre1 indirectly detects oxygen through sterol levels and facilitates growth under oxygen-limiting conditions (5). Sre1 is a membrane-bound transcription factor, and Sre1 cleavage activation occurs through a mechanism that requires Dsc E3 ligase activity and the proteasome (8). We previously identified the Dsc E3 ligase complex through a genetic screen of the S. pombe non-essential haploid deletion collection. Although successful, the approach ignored essential genes, and the deletion collection screened was incomplete, containing 75% of the ∼3600 non-essential S. pombe genes (31). Here, we used a forward genetic selection to identify additional components of the fission yeast Sre1 pathway. We discovered a number of linkage groups representing both known and unknown genes that participate in Sre1 cleavage. However, the failure to identify alleles of dsc2 or dsc4 indicated that the mutagenesis was not saturating. In this study, we characterized two new linkage groups, dsc5 and dsc6, coding for a UBX domain-containing protein and Cdc48, respectively.
Several lines of evidence demonstrate that Dsc5 and Cdc48 are functional components of the Dsc E3 ligase complex required to cleave full-length Sre1. First, the ability of the active Sre1N transcription factor to rescue the CoCl2 sensitivity of dsc5 and cdc48 mutants indicated that these genes function at or prior to Sre1 cleavage (Fig. 1D). Second, mutations in either dsc5 or cdc48 blocked Sre1 and Sre2 cleavage (Figs. 2 and 3). Third, protein interaction studies demonstrated that Dsc2 binds Dsc5 and Cdc48 along with the known binding partners, Dsc1, Dsc3, and Dsc4 (Fig. 5). The majority of Dsc1, Dsc3, Dsc4, and Dsc5 exist in complexed form. However, only a small fraction of Cdc48 bound to Dsc2 (Fig. 5). This finding was expected because Cdc48 is an abundant protein and functions in numerous cellular processes (13). Thus, Dsc5 and Cdc48 are new components of the Dsc E3 ligase complex.
Function of Dsc5 and Cdc48 in SREBP Cleavage
Dsc5 is predicted to be a UAS domain- and UBX domain-containing membrane protein and is the closest S. pombe homolog of Ubx2p in S. cerevisiae. Ubx2p contains a UBX domain that recruits Cdc48 to the Hrd1 E3 ligase complex, resulting in dislocation of ERAD substrates (32, 33). UBX domains bind Cdc48 and provide spatial and temporal control over Cdc48 activity and are an important feature of many Cdc48-interacting proteins (13). Given the presence of a UBX domain in Dsc5 and the growing similarities between the Dsc and Hrd1 E3 ligase complexes, we hypothesized that Dsc5 and Ubx2p have equivalent functions with regard to Cdc48 (5). Indeed, the UBX domain of Dsc5 was capable of binding Cdc48 in vitro. Consistent with this result, deletion of the UBX domain from Dsc5 eliminated Cdc48 recruitment to the Dsc complex in vivo. Interestingly, deletion of the UBX domain from Dsc5 resulted in only a modest decrease in Sre1N accumulation under low oxygen (Fig. 6) as compared with the complete block in Sre1 cleavage in strains harboring dsc5 alleles that lack both the predicted UAS and UBX domains (Fig. 2B). This difference suggests a role for the UAS domain of Dsc5 in SREBP cleavage. The UAS domain resembles thioredoxin in structure and is also called a thioredoxin-like domain (34). Although a significant number of UAS domain-containing proteins also have UBX domains, the function of UAS domains is poorly understood. A better understanding of the UAS domain function in Dsc5 will yield insight into the broader roles of UAS domain-containing proteins.
Cdc48 is a versatile, highly evolutionarily conserved molecular chaperone and the homolog of the mammalian protein p97 (35). As an AAA (ATPase associated with diverse cellular activities) domain-containing protein, Cdc48 is thought to provide the mechanical force for an array of cellular functions, including but not limited to chromosome segregation and cell cycle regulation, membrane fusion, and ubiquitin and/or proteasome-mediated protein degradation. Its functional unit is a homohexamer, comprised of six identical 90-kDa subunits (13). Each Cdc48 subunit contains three important domains; the N-terminal region of the protein is a substrate recognition domain that mediates a number of protein-protein interactions, followed by two tandem AAA ATPase domains (D1 and D2). The D1 and D2 domains are highly similar, and each contains three conserved regions: the Walker A and Walker B domains and the second region of homology (SRH) domain (36). The Walker A and Walker B domains are important for ATP binding and hydrolysis, whereas the SRH domain is thought to be important for both intersubunit and intrasubunit communication. Mutations within these domains impair Cdc48 function, leading to the accumulation of insoluble proteins, a hallmark of several human diseases (37, 38).
In this study, we discovered a role for Cdc48 in fission yeast SREBP cleavage and identified four mutations, E325K, N558I, A586V, and E731D, that abrogate its function in the SREBP pathway. Although each cdc48 allele confers an SREBP cleavage defect, variation exists in the severity of the block (Fig. 3). Direct comparison of Sre1 cleavage in strains harboring these different cdc48 alleles shows that the N558I and E325K mutations confer a complete block in Sre1 cleavage, whereas the A586V and E731D mutations are severely impaired for Sre1 cleavage but not completely defective. The disparity in SREBP cleavage among the different mutants is not due to a decrease in steady state levels of the Cdc48 protein (Fig. 3B, bottom). In the case of the E325K mutation, it is likely that the Sre1 cleavage defect can be attributed to a decrease in ATPase activity because this mutation corresponds to a previously characterized E305Q mutation in p97 (39). Given the proximity of the N558I and A586V mutations to the D2 domain Walker A and Walker B motifs, respectively, it is possible that these mutations affect ATP binding or hydrolysis. The E731D mutation is within the D2 domain but lies outside any subdomain of known importance. Given that ATP hydrolysis creates a conformational change that can drive cellular processes, this mutation could prevent productive translation of the conformational change within the Cdc48 subunit. With any of these mutants, it is possible that the observed Sre1 cleavage defects are due to differences in catalytic activity or binding to substrates and cofactors. The mechanistic details of how these mutations impair Cdc48 function remain to be determined.
Previous structure-function studies with Cdc48/p97 have largely relied on site-directed mutagenesis based on known biochemical or structural data (36, 39). Although practical, these studies limit themselves to interrogating regions of the protein already recognized as important. Furthermore, cdc48+ is an essential gene, and complete loss-of-function mutants would be inviable (29). The success of our unbiased approach makes it an attractive platform for identifying important functional residues of Cdc48. A collection of cdc48 mutants would be a valuable resource and yield information on the function of Cdc48 within a wide variety of cellular functions.
Parallels between Dsc E3 Ligase Complex and Hrd1 E3 Ligase Complex
During the initial characterization of the Dsc E3 ligase, we found that the Dsc1–Dsc4 subunits shared organizational and structural similarities with subunits of the Hrd1 E3 ligase involved in ERAD and that Sre1 cleavage required the proteasome (8). However, it was unclear how the Dsc complex delivered SREBP to its candidate protease, the proteasome. Dsc5 and Cdc48 may link these two Sre1 pathway components. In ERAD, the UBX-domain protein Ubx2p recruits Cdc48 to the Hrd1 E3 ligase complex to facilitate membrane dislocation of ubiquitinated substrates for delivery to the proteasome (32, 33). In the Sre1 pathway, Dsc5 may serve an analogous role to Ubx2p, facilitating the recruitment of Cdc48 to the Dsc E3 ligase to promote Sre1 delivery to the proteasome. However, unlike ERAD substrates, the Sre1 precursor must be incompletely processed by the proteasome to produce the functional Sre1N transcription factor. Nonetheless, Dsc5 and Cdc48 collectively provide a potential mechanistic link between the Dsc E3 ligase complex and the proteasome. In addition, Dsc5 and Cdc48 may provide a means for regulating SREBP activation through the differential binding of Dsc5 to the Dsc E3 ligase or through control of Dsc5-Cdc48 binding. Given the emerging parallels between the Dsc and Hrd1 E3 ligase complexes, the future characterization of additional linkage groups from this genetic selection and a clear understanding of Dsc complex function will advance our understanding of the ERAD pathway and protein quality control.
Supplementary Material
Acknowledgments
We thank Nick Rhind (University of Massachusetts Medical School), Phung Trang and James Bochicchio (Broad Institute Genome Sequencing Platform), Patrice Milos (Helicos BioSciences), and Avak Kahvejian (Helicos BioSciences) for advice and sequencing support. We are grateful to the following investigators for sharing strains and reagents: Mitsuhiro Yanagida (Okinawa Institute of Science and Technology), Dieter Wolf (Sanford-Burnham Medical Research Institute), Colin Gordon (Medical Research Council, UK), and Rasmus Hartmann-Petersen (University of Copenhagen).
This work was supported, in whole or in part, by National Institutes of Health Grant HL077588 (to P. J. E.).

This article contains supplemental Table 1 and Fig. 1.
- SREBP
- sterol regulatory element-binding protein
- AAA
- ATPase associated with diverse cellular activities
- UBX
- ubiquitin regulatory X
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- MMS
- methyl methanesulfonate
- 5-FOA
- 5-fluoroorotic acid
- SRH
- second region of homology
- S1P and S2P
- site-1 and -2 protease, respectively.
REFERENCES
- 1. Goldstein J. L., DeBose-Boyd R. A., Brown M. S. (2006) Cell 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 2. Nohturfft A., Yabe D., Goldstein J. L., Brown M. S., Espenshade P. J. (2000) Cell 102, 315–323 [DOI] [PubMed] [Google Scholar]
- 3. Osborne T. F., Espenshade P. J. (2009) Genes Dev. 23, 2578–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Espenshade P. J., Hughes A. L. (2007) Annu. Rev. Genet. 41, 401–427 [DOI] [PubMed] [Google Scholar]
- 5. Bien C. M., Espenshade P. J. (2010) Eukaryot. Cell 9, 352–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hughes A. L., Todd B. L., Espenshade P. J. (2005) Cell 120, 831–842 [DOI] [PubMed] [Google Scholar]
- 7. Todd B. L., Stewart E. V., Burg J. S., Hughes A. L., Espenshade P. J. (2006) Mol. Cell. Biol. 26, 2817–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stewart E. V., Nwosu C. C., Tong Z., Roguev A., Cummins T. D., Kim D. U., Hayles J., Park H. O., Hoe K. L., Powell D. W., Krogan N. J., Espenshade P. J. (2011) Mol. Cell 42, 160–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reggiori F., Pelham H. R. (2002) Nat. Cell Biol. 4, 117–123 [DOI] [PubMed] [Google Scholar]
- 10. Carvalho P., Goder V., Rapoport T. A. (2006) Cell 126, 361–373 [DOI] [PubMed] [Google Scholar]
- 11. Denic V., Quan E. M., Weissman J. S. (2006) Cell 126, 349–359 [DOI] [PubMed] [Google Scholar]
- 12. Gauss R., Sommer T., Jarosch E. (2006) EMBO J. 25, 1827–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stolz A., Hilt W., Buchberger A., Wolf D. H. (2011) Trends Biochem. Sci. 36, 515–523 [DOI] [PubMed] [Google Scholar]
- 14. Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998) Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- 15. Alfa C., Fantes P., Hyams J., McLeod M., Warbrick E. (1993) Experiments with Fission Yeast: A Laboratory Course Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 16. Burke J. D., Gould K. L. (1994) Mol. Gen. Genet. 242, 169–176 [DOI] [PubMed] [Google Scholar]
- 17. Bimbó A., Jia Y., Poh S. L., Karuturi R. K., den Elzen N., Peng X., Zheng L., O'Connell M., Liu E. T., Balasubramanian M. K., Liu J. (2005) Eukaryot. Cell 4, 799–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hughes A. L., Stewart E. V., Espenshade P. J. (2008) J. Lipid Res. 49, 2001–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Forsburg S. L. (1993) Nucleic Acids Res. 21, 2955–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thompson J. F., Steinmann K. E. (2010) Current Protocols in Molecular Biology, Chapter 7, Unit 7.10, John Wiley & Sons, Inc., New York [Google Scholar]
- 21. Hartmann-Petersen R., Wallace M., Hofmann K., Koch G., Johnsen A. H., Hendil K. B., Gordon C. (2004) Curr. Biol. 14, 824–828 [DOI] [PubMed] [Google Scholar]
- 22. Guan K. L., Dixon J. E. (1991) Anal. Biochem. 192, 262–267 [DOI] [PubMed] [Google Scholar]
- 23. Lee C. Y., Stewart E. V., Hughes B. T., Espenshade P. J. (2009) EMBO J. 28, 135–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. (1987) Methods Enzymol. 154, 164–175 [DOI] [PubMed] [Google Scholar]
- 25. Lee H., Bien C. M., Hughes A. L., Espenshade P. J., Kwon-Chung K. J., Chang Y. C. (2007) Mol. Microbiol. 65, 1018–1033 [DOI] [PubMed] [Google Scholar]
- 26. Hartmann-Petersen R., Semple C. A., Ponting C. P., Hendil K. B., Gordon C. (2003) Int. J. Biochem. Cell Biol. 35, 629–636 [DOI] [PubMed] [Google Scholar]
- 27. Alexandru G., Graumann J., Smith G. T., Kolawa N. J., Fang R., Deshaies R. J. (2008) Cell 134, 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wood V., Gwilliam R., Rajandream M. A., Lyne M., Lyne R., Stewart A., Sgouros J., Peat N., Hayles J., Baker S., Basham D., Bowman S., Brooks K., Brown D., Brown S., Chillingworth T., Churcher C., Collins M., Connor R., Cronin A., Davis P., Feltwell T., Fraser A., Gentles S., Goble A., Hamlin N., Harris D., Hidalgo J., Hodgson G., Holroyd S., Hornsby T., Howarth S., Huckle E. J., Hunt S., Jagels K., James K., Jones L., Jones M., Leather S., McDonald S., McLean J., Mooney P., Moule S., Mungall K., Murphy L., Niblett D., Odell C., Oliver K., O'Neil S., Pearson D., Quail M. A., Rabbinowitsch E., Rutherford K., Rutter S., Saunders D., Seeger K., Sharp S., Skelton J., Simmonds M., Squares R., Squares S., Stevens K., Taylor K., Taylor R. G., Tivey A., Walsh S., Warren T., Whitehead S., Woodward J., Volckaert G., Aert R., Robben J., Grymonprez B., Weltjens I., Vanstreels E., Rieger M., Schäfer M., Müller-Auer S., Gabel C., Fuchs M., Düsterhöft A., Fritzc C., Holzer E., Moestl D., Hilbert H., Borzym K., Langer I., Beck A., Lehrach H., Reinhardt R., Pohl T. M., Eger P., Zimmermann W., Wedler H., Wambutt R., Purnelle B., Goffeau A., Cadieu E., Dréano S., Gloux S., Lelaure V., Mottier S., Galibert F., Aves S. J., Xiang Z., Hunt C., Moore K., Hurst S. M., Lucas M., Rochet M., Gaillardin C., Tallada V. A., Garzon A., Thode G., Daga R. R., Cruzado L., Jimenez J., Sánchez M., del Rey F., Benito J., Domínguez A., Revuelta J. L., Moreno S., Armstrong J., Forsburg S. L., Cerutti L., Lowe T., McCombie W. R., Paulsen I., Potashkin J., Shpakovski G. V., Ussery D., Barrell B. G., Nurse P., Cerrutti L. (2002) Nature 415, 871–880 [DOI] [PubMed] [Google Scholar]
- 29. Yuasa T., Hayashi T., Ikai N., Katayama T., Aoki K., Obara T., Toyoda Y., Maruyama T., Kitagawa D., Takahashi K., Nagao K., Nakaseko Y., Yanagida M. (2004) Genes Cells 9, 1069–1082 [DOI] [PubMed] [Google Scholar]
- 30. Schuberth C., Buchberger A. (2008) Cell Mol. Life Sci. 65, 2360–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim D. U., Hayles J., Kim D., Wood V., Park H. O., Won M., Yoo H. S., Duhig T., Nam M., Palmer G., Han S., Jeffery L., Baek S. T., Lee H., Shim Y. S., Lee M., Kim L., Heo K. S., Noh E. J., Lee A. R., Jang Y. J., Chung K. S., Choi S. J., Park J. Y., Park Y., Kim H. M., Park S. K., Park H. J., Kang E. J., Kim H. B., Kang H. S., Park H. M., Kim K., Song K., Song K. B., Nurse P., Hoe K. L. (2010) Nat. Biotechnol. 28, 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Neuber O., Jarosch E., Volkwein C., Walter J., Sommer T. (2005) Nat. Cell Biol. 7, 993–998 [DOI] [PubMed] [Google Scholar]
- 33. Schuberth C., Buchberger A. (2005) Nat. Cell Biol. 7, 999–1006 [DOI] [PubMed] [Google Scholar]
- 34. Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Lu F., Marchler G. H., Mullokandov M., Omelchenko M. V., Robertson C. L., Song J. S., Thanki N., Yamashita R. A., Zhang D., Zhang N., Zheng C., Bryant S. H. (2011) Nucleic Acids Res. 39, D225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Q., Song C., Li C. C. (2004) J. Struct. Biol. 146, 44–57 [DOI] [PubMed] [Google Scholar]
- 36. Wang Q., Song C., Irizarry L., Dai R., Zhang X., Li C. C. (2005) J. Biol. Chem. 280, 40515–40523 [DOI] [PubMed] [Google Scholar]
- 37. Petrucelli L., Dawson T. M. (2004) Ann. Med. 36, 315–320 [DOI] [PubMed] [Google Scholar]
- 38. Halawani D., LeBlanc A. C., Rouiller I., Michnick S. W., Servant M. J., Latterich M. (2009) Mol. Cell. Biol. 29, 4484–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. DeLaBarre B., Christianson J. C., Kopito R. R., Brunger A. T. (2006) Mol. Cell 22, 451–462 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.