Abstract
Ample evidence suggests that almost all polypeptides can either adopt a native structure (folded or intrinsically disordered) or form misfolded amyloid fibrils. Soluble protein oligomers exist as an intermediate between these two states, and their cytotoxicity has been implicated in the pathology of multiple human diseases. However, the mechanism by which soluble protein oligomers develop into insoluble amyloid fibrils is not clear, and investigation of this important issue is hindered by the unavailability of stable protein oligomers. Here, we have obtained stabilized protein oligomers generated from common native proteins. These oligomers exert strong cytotoxicity and display a common conformational structure shared with known protein oligomers. They are soluble and remain stable in solution. Intriguingly, the stabilized protein oligomers interact preferentially with both nucleic acids and glycosaminoglycans (GAG), which facilitates their rapid conversion into insoluble amyloid. Concomitantly, binding with nucleic acids or GAG strongly diminished the cytotoxicity of the protein oligomers. EGCG, a small molecule that was previously shown to directly bind to protein oligomers, effectively inhibits the conversion to amyloid. These results indicate that stabilized oligomers of common proteins display characteristics similar to those of disease-associated protein oligomers and represent immediate precursors of less toxic amyloid fibrils. Amyloid conversion is potently expedited by certain physiological factors, such as nucleic acids and GAGs. These findings concur with reports of cofactor involvement with disease-associated amyloid and shed light on potential means to interfere with the pathogenic properties of misfolded proteins.
Keywords: Amyloid, Cell Death, Glycosaminoglycan, Nucleic Acid, Protein Folding, Oligomers Toxicity, Protein Oligomers
Introduction
Proteins are the most abundant biological macromolecules present in all types of cells. They occur in a great variety of sizes, structures, and post-translational modifications, and fulfill an enormous range of important biological functions when in their native forms. However, misfolded proteins may arise by germline mutation, erroneous transcription, or translation, failure to fold properly, spontaneous denaturation, or physical damage (1). More than two dozens aberrant polypeptides have been implicated in numerous human pathological conditions broadly referred as protein misfolding diseases (2–4). The terminal misfolded proteins accumulate as amyloid fibrils, the insoluble stable aggregates that occur extracellularly or intracellularly.
Despite the implication of specific proteins in certain diseases, increasing evidence supports the notion that all polypeptides have intrinsic properties that enable amyloid transformation. A recent genome-wide sequence survey identified the “amylome,” by which fibril-forming proteins constitutes roughly 15% of all coding polypeptides from Escherichia coli to humans (5). In fact, most proteins can be converted experimentally into amyloid under defined in vitro conditions (3, 6, 7). Bacteria assemble amyloids to form biofilm and spore structures that are critical for their survival and pathogenesis (8–11). Moreover, peptide hormones form amyloid deposits during storage within mammalian secretory granules before being released, further implying that the protein amyloid form can serve beneficial biological functions (8, 12, 13). Therefore, the two states, i.e. native (folded or intrinsically disordered) and amyloid, can in principle be adopted by almost any protein under the appropriate conditions.
The breakthrough discovery of soluble proteins oligomers provided a critical link between native proteins and their corresponding amyloid fibrils (14, 15). Soluble protein oligomers are partially misfolded intermediates that are the precursors of insoluble amyloid. Two unique features of soluble protein oligomers distinguish them from nonspecific or other types of protein aggregates: first, they display inherent cytotoxicity toward live cells (14, 15) and, second, they share a common conformational structure recognizable by a specific anti-amyloid β oligomer antibody (16, 17). In Alzheimer disease, although the extracellular accumulation of β-amyloid (Aβ) in senile plaques denotes a key pathological marker, the soluble oligomers of Aβ instead represent the primary toxic species responsible for the cognitive deficits associated with Alzheimer disease (15, 16, 18, 19). In accordance with the notion that every protein can have two states, native proteins were shown to form oligomers under specific in vitro conditions where cytotoxicity is elicited (14). Therefore, soluble protein oligomers represent an intermediate stage of protein misfolding and are critically important, both biologically and pathologically.
There is however limited information about how soluble protein oligomers participate in the process of amyloid formation. Assembly of natural soluble protein oligomers is seemingly an unfavorable and rating limiting event for eventual amyloid deposition, evidenced by high variability in the mostly late on-set protein misfolding diseases. These intermediates are believed to serve as the seed of nucleation to propagate the misfolding process of native proteins, which promotes the development of amyloid fibrils (2, 20, 21). Although natural amyloidogenic polypeptides, such as Aβ, can form oligomers in vitro, conversion of Aβ peptide to amyloid takes place spontaneously with an intrinsic kinetics (14). Soluble protein oligomers form transiently and co-exist with native and amyloid species. Therefore, it is difficult to conduct detailed mechanistic study on soluble protein oligomers without analyzing stable oligomeric species.
A series of recent investigations indicate the potential involvement of non-proteinaceous cofactors with amyloidogenic proteins and related diseases. Heparan sulfate proteoglycan (HSPG),2 a glycosaminoglycan (GAG), commonly associates with amyloid deposits, including the cerebral amyloid plaques of Alzheimer disease and the transmissible spongiform encephalopathies (22, 23). RNA molecules can potently stimulate prion protein conversion in vitro, whereas infectious prions effectively incorporate RNA, an event critical for their infectivity in vivo (24, 25). Double-stranded DNA (dsDNA) stimulates the fibrillation of α-synuclein and is associated with the mature fibrils (26, 27). Interaction between DNA and soluble aggregates of Aβ42 has also been observed (28). Whether and how these factors influence amyloid development is not clear at this time.
Here we report the generation of a stabilized oligomeric form of common proteins that displayed unexpected cytotoxicity. Further characterization revealed that these proteins had properties similar to the soluble protein oligomers described previously in the literature. Interestingly, these stabilized protein oligomers preferentially bound to polyanionic cofactors, an interaction with important implications for amyloid development.
EXPERIMENTAL PROCEDURES
Reagents and Cells
Jurkat cells and RPMI 8226 cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 50 units/ml penicillin, and 50 μg/ml streptomycin. HEK293 cells were grown in high glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and antibiotics. Pre-casted SDS-PAGE gels were purchased from Invitrogen. According to manufacturer's instructions, mammalian genomic DNA was prepared with the PureLinkTM Genomic DNA Mini kit (Invitrogen); total RNA was prepared with TRIzol Reagent (Invitrogen). CellTiter-Blue® cell viability assay kit was purchased from Promega. Aβ(1–42) peptide was purchased from EMD Biosciences; reverse Aβ peptide was from California Peptide Research. Prion aa 106–126 was obtained from Tocris Bioscience, scrambled prion (106–126) was from Sigma-Aldrich. Trypsin (from porcine pancreas) and EGCG were obtained from Sigma-Aldrich.
Preparation of EDC-crosslinked Proteins and Protein Aggregates
Purified HSA (Sigma-Aldrich) or human IgG (Equitech-Bio, Inc.) was dissolved in MES buffer (0.1 m 2-(N-morpholino)ethanesulfonic acid, 0.9 m NaCl, pH 4.7) at 10 mg/ml and incubated with 15 mg/ml of 1-ethyl-3-[3-dimethyl-aminopropyl] carbodiimide hydrochloride (EDC, Pierce Biotech) for 2 h at 23°C. The crosslinked samples were then dialyzed against PBS and filter sterilized. To prepare heat-aggregated samples, proteins in MES buffer were incubated at 65 °C for 2 h and dialyzed against PBS afterward. 100°C denatured proteins were boiled 5 min in PBS and then kept at 4 °C. To prepare crosslinked heat-denatured samples, EDC was added to the 65 °C denatured proteins in MES buffer using the same protocol as for native proteins. The 100 °C-denatured proteins were spun down at 5000 rpm for 5 min and resuspended in MES buffer and then EDC was added. Both samples were then dialyzed against PBS.
Size-fractionation of EDC-crosslinked Proteins
4 mg of EDC-crosslinked HSA was loaded onto a Superose 6 100/300 GL column and separated with ÄKTA FPLC system (GE Healthcare). Proteins were eluted in TBS buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl). All experiments were carried out at 4 °C and a flow rate of 0.2 ml per min. Protein content in eluted samples was measured using the absorbance reading at 280 nm. Selected fractions were separated on 8% SDS-PAGE and stained with SimplyBlueTM SafeStain buffer (Invitrogen).
Direct ELISA and Dot Blot Analysis with Anti-Aβ Oligomer Antibody
To perform dot blot analysis, 2 μl of protein samples (1 mg/ml) were spotted onto activated Immobilon-P membrane (Millipore). The blot was blocked with 5% nonfat milk in TTBS buffer prior to incubation with anti-Aβ oligomer A11 rabbit Ab (1:1000, Millipore). Anti-rabbit-HRP Ab (JacksonImm) was subsequently incubated before addition of SuperSignal West Pico Chemiluminescent Substrate (Pierce Biotech). For direct ELISA, proteins were coated at different concentrations in PBS on ELISA plate overnight in 100 μl volume per well. The plate was washed, blocked with 1% BSA in PBS, and incubated for 2 h with A11 Ab (1:1000). After three washes, the plate was incubated with anti-rabbit-HRP Ab (JacksonImm) for 2 h before addition of TMB substrate.
Measurement of Fluorescence Emission
HSA and IgG samples (0.5 mg/ml) were mixed with 50 μm of Thioflavin T (Sigma-Aldrich). The fluorescence was measured by a spectrofluorometer (Jasco FP-6500) with an excitation wavelength of 450 nm and an emission between 450–600 nm. Alternatively, proteins were mixed with 5 μm of 4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid dipotassium salt (bis-ANS; Sigma-Aldrich). The fluorescence was measured in a spectrofluorometer with an excitation wavelength of 395 nm and an emission between 420 and 580 nm. To perform the PicoGreen test, different concentrations of native and oligomeric IgG samples were mixed with 10 μg/ml salmon sperm DNA. Quant-iTTM PicoGreen® dye (Invitrogen) was added at a concentration of 0.2 mg/ml. The fluorescence of samples was measured by a spectrofluorometer with an excitation wavelength of 450 nm and an emission at 520 nm. To measure the binding of Congo Red, HSA samples (50 μg/ml) were mixed with PBS, heparin (50 μg/ml), or salmon sperm DNA (50 μg/ml) for 2 h at room temperature. Congo Red was then added (30 μg/ml, 5 mm potassium phosphate, 150 mm NaCl, pH 7.4) for 30 min. The fluorescence was measured afterward in a spectrofluorometer with an excitation wavelength of 497 nm and an emission between 600 and 700 nm.
Measurement of Cytotoxicity
Proteins were incubated with RPMI 8226 or HEK 293 cells (1 × 106/ml) at different concentrations in RPMI medium at 37 °C for 24 h. Cells were washed, resuspended in propidium iodide containing buffer (BD Bioscience) and analyzed on a FACSCaliburTM cytometer (BD Bioscience). A 10 μl aliquot of cells were stained with 10 μl of Trypan Blue dye and analyzed by microscopy. In other experiments, CellTiter-Blue® dye was added to the cultures (1/5) for 4 h and fluorescence (560Ex/590Em) was measured by a spectrofluorometer (Jasco FP-6500), using the fluorescence of non-treated cells as 100% fluorescence.
Microscopic Analysis of Amyloid Formation
Native or oligomeric HSA (500 μg/ml) were mixed with PBS, heparin (500 μg/ml) or salmon sperm DNA (500 μg/ml) for 2 h at room temperature. Samples were then deposited in wells of a positive charged Teflon printed slide, 8 well 6 mm diameter (Electron Microscopy Sciences) and air dried. Samples were fixed in 4% paraformaldehyde, washed in water and stained with 1% Congo Red in 80% ethanol, 100 mm NaOH. Slide was washed in 80% ethanol, air dried, and analyzed microscopically in bright and polarized light using Olympus BX41 microscope.
Circular Dichroism (CD) Analysis
The secondary and tertiary structures of native and oligomeric HSA and IgG were probed by the analysis of their CD spectrum (Jasco J-810). Dialyzed proteins (200 μg/ml) in PBS were analyzed using near-UV (250–350 nm) and far-UV CD (200–260 nm) in a 1 mm path length quartz cuvette. Experimental data were corrected for buffer contributions.
Trypsin Digestion of EDC-crosslinked Proteins
5 μg of native or EDC-crosslinked HSA was incubated with 1 μg of trypsin for 60 min at 50 °C in PBS buffer containing 1 mm DTT. Samples were then separated on 8% SDS-PAGE gel and stained with SimplyBlue™ SafeStain buffer. For positive control, HSA proteins were pre-boiled for 10 min in the digestion buffer prior to addition of trypsin.
Nucleic Acids Binding Gel Shift Assay
Different proteins were incubated with 0.5 μg of either circular plasmid DNA, genomic DNA or total RNA in TE buffer (10 μm Tris-Cl, pH 7.4 and 1 μm EDTA) for 60 min. The samples were then loaded onto 1% agarose gel and subjected to electrophoresis separation. To demonstrate enzymatic protection, samples were treated with DNase I (40 ng/ml, Invitrogen) and incubated at 37 °C for 10 min prior to electrophoresis.
Transmission Electron Microscopy Analysis
Protein samples (0.5 mg/ml) were incubated with either 0.2 mg/ml of DNA or 0.8 mg/ml of heparin (Lovenox, Sanofi-Aventis) in PBS for 24 h at 4 °C. The samples were placed on single-slot Formvar coated copper grids for 1 h. Excess samples were blotted with filter paper, the samples were stained with filtered 2% uranyl acetate for 1 min. Samples were allowed to dry before being examined under a JEM 1010 transmission electron microscope (JEOL USA, Inc.) at an accelerated voltage of 80 kV. Digital images were obtained using the AMT Imaging System (Advanced Microscopy Techniques).
Confocal Imaging Analysis of Protein-DNA Complex
Biotinylated IgG oligomer (5 μg/ml) was mixed with 5 μg/ml Alexa647-labeled DNA or Alexa647-labeled RNA for 30 min at 23 °C. The complexes were laid on poly-l-lysine precoated 1.5-mm round coverslips for 15 min, fixed in 5% formaldehyde for 15 min, washed three times in PBS, and then stained with Alexa488-labeled avidin diluted 1:100 in 0.1% saponin plus 10% FBS for 1 h at 23 °C. The complexes were washed three more times for 30 min with PBS, and the coverslips were mounted onto glass slides in ProLong® Gold antifade reagent. Slides were analyzed and images acquired using a confocal microscope (model TCS SP2, Leica) with a ×63 oil immersion objective.
RESULTS
EDC-crosslinked Proteins Display Cytotoxicity
For a control in an independent project, we prepared bovine serum albumin (BSA) crosslinked with 1-ethyl-3-[3-dimethyl-aminopropyl]carbodiimide hydrochloride (EDC). Surprisingly, EDC-crosslinked BSA induced profound cell death when added to cultured mammalian cells (supplemental Fig. S1). After repeated confirmation and a similar observation made with EDC-crosslinked mouse immunoglobulin, we decided to investigate the underlying mechanism. To do that, we crosslinked two common and abundant human proteins, human serum albumin (HSA) and immunoglobulin G (IgG), with EDC. EDC is a zero-length crosslinking agent that effectively couples carboxyl groups to primary amines in acidic pH. After terminating the crosslinking reaction, HSA and IgG proteins were fully dialyzed against pH-neutral phosphate-buffered saline (PBS). The protein preparations were soluble and contained a mixture of multimers that can be separated by SDS-PAGE (supplemental Fig. S2). We also prepared control protein samples of HSA and IgG proteins incubated at 65 °C in the buffer used for EDC crosslinking prior to dialysis against PBS. Heat treatment resulted in the visible precipitation of proteins in solution and the formation of multimeric aggregates (supplemental Fig. S2).
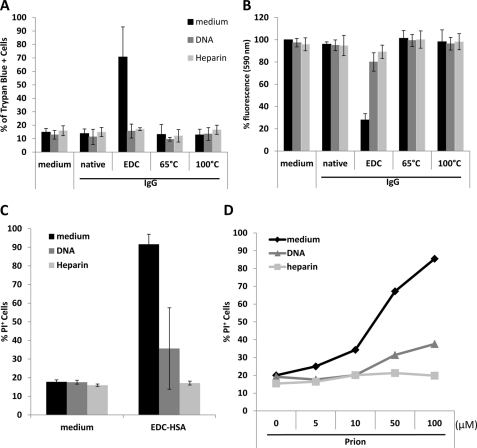
When incubated with RPMI 8226 cells, a human plasmacytoma line, EDC-crosslinked HSA, but not native or 65 °C-aggregated HSA, induced severe cell death, as determined by the positive Trypan Blue staining (Fig. 1A). We further performed a CellTiter-Blue® cell viability assay to measure the metabolic capacity of the cells (Fig. 1B), and also stained the treated cells with PI (Fig. 1C) to confirm that the cytotoxicity was exclusively associated with EDC-crosslinked HSA. This protein preparation is also toxic to HEK293 cells (Fig. 1, A and B), Jurkat cells, CHO cells, and primary human peripheral mononuclear cells (data not shown). EDC-crosslinked human IgG displayed a similar effect on the viability of cultured cells (data not shown).
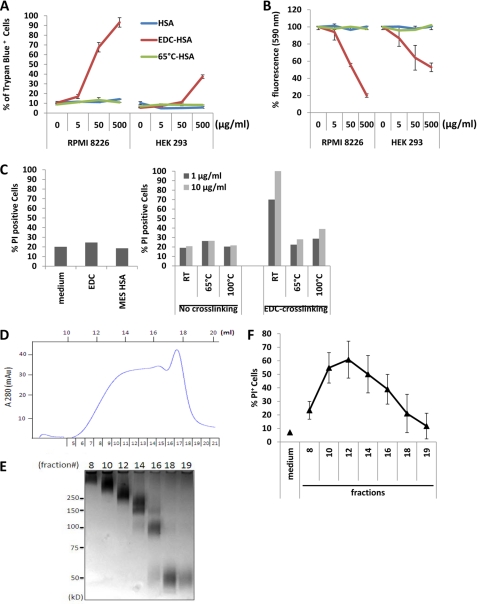
FIGURE 1.
EDC-crosslinked HSA displays cytotoxicity. A, RPMI 8226 and HEK 293 cells were cultured with different concentrations of HSA, EDC-crosslinked HSA (EDC-HSA), or 65°C-aggregated HSA. Assessment of cell death was performed by staining cells with Trypan Blue and examination under the microscope. Error bars are means ± S.D. B, assessment of cell viability with CellTiter-Blue® agent. The fluorescence at 590 nm emitted by cells cultured in medium alone with CellTiter-Blue® was used as reference (100%). Relative fluorescence emitted by cells under other conditions was calculated and plotted accordingly. C, quantification of dead cell populations in RPMI 8226 cells cultured for 24 h with medium, EDC or HSA in MES buffer was performed by PI staining (left). PI staining was also performed on RPMI 8226 cells cultured with 1 or 10 μg/ml of different forms of HSA with or without EDC crosslinking (right). D, chromatograph of size-exclusion separation of EDC-crosslinked HSA. The fraction numbers and elution volume are shown in relation to the 280 nm optical density (OD) reading. E, fractions obtained by size-exclusion chromatography were separated by SDS-PAGE. The corresponding fraction numbers are marked on the top. Also noted are the positions of molecular weight markers. F, dead cell populations in RPMI 8226 cells cultured with individual fractions from size-exclusion chromatography (1 μg/ml) was assayed by PI staining. Error bars are means ± S.D.
To test the possibility that residual EDC in the protein preparation was responsible for the observed cellular effects, we incubated RPMI cells directly with EDC or the crosslinking buffer at a dose corresponding to their levels before sample dialysis but did not observe significant cytotoxicity (Fig. 1C). To examine the effect of EDC conjugation per se on the proteins, we further crosslinked HSA proteins that had been preincubated at either 65 or 100 °C with EDC. None of these preparations displayed significant cytotoxicity (Fig. 1C).
To discern the factors in EDC-crosslinked proteins that mediate the cytotoxic effect, we performed size-fractionation of crosslinked HSA by chromatography (Fig. 1, D and E). Fractions containing monomer, dimer, trimer or higher oligomers were added to RPMI 8226 cell culture. HSA dimer (fraction 16) and a mixture of HSA trimer and tetramer (fraction 14) induced significant cytotoxicity (Fig. 1F). Fraction 12 with the highest cytotoxicity contains HSA multimers of high molecular weight (>250 kDa). In contrast, monomeric HSA isolated from EDC-crosslinked samples demonstrated limited cytotoxicity. Fractions with molecular weight significantly higher than fraction 12 somehow exerted less cell killing. These results suggest that the cytotoxic effect we observed with EDC-crosslinked samples is primarily mediated by the multimeric forms of proteins in the preparation.
EDC-crosslinked Proteins Represent Stabilized Protein Oligomers
Given the fact that EDC-crosslinked proteins are soluble mixtures of multimeric polypeptides, we hypothesized that they may mimic soluble protein oligomers involved in amyloid formation with certain structural properties. Soluble oligomers of amyloidogenic proteins, such as Aβ, prion, lysozyme, and polyglutamine, commonly express a conformational structure that differs from that of both native proteins or amyloid fibrils (16). We obtained an anti-Aβ oligomer antibody, A11, which is widely used to detect soluble protein oligomers, and found that it recognized EDC-crosslinked HSA, but not native HSA, by dot blot analysis (Fig. 2A). We further coated native, EDC-crosslinked or 65 °C-treated HSA on plastic surface in different doses and performed ELISA using the A11 antibody. Consistently, A11 dose-dependently bound to the EDC-crosslinked sample, but not to native or heat-aggregated HSA (Fig. 2B). Similar results were obtained with EDC-crosslinked human IgG (data not shown).
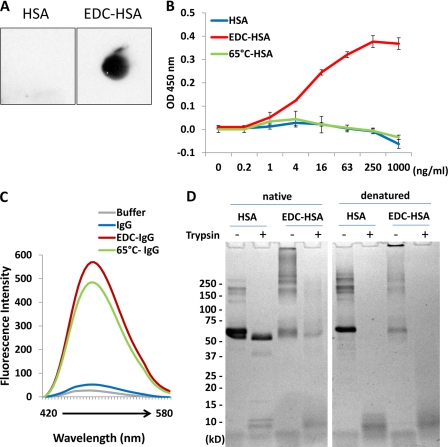
FIGURE 2.
Characterization of EDC-crosslinked proteins. A, membrane-bound native HSA and EDC-crosslinked HSA were examined with anti-Aβ-oligomer (A11) antibody by dot blot analysis. B, direct binding of A11 antibody to different amounts of native, EDC-crosslinked, or 65 °C-aggregated HSA pre-coated on an ELISA plate. Error bars are means ± S.D. of absorbance at 450 nm. C, fluorescence emission profiles of bis-ANS obtained after incubating with various IgG-derived proteins in comparison with PBS buffer. D, SDS-PAGE separation of HSA and EDC-crosslinked HSA, which had been subjected to partial trypsin digestion for 60 min (Native, left). As a control, both proteins were denatured by boiling prior to the identical trypsin digestion procedure (Denatured, right).
To further understand the structural changes in EDC-crosslinked proteins, we employed a fluorescent dye, 4,4′-bis(1-anilinonaphthalene 8-sulfonate) (bis-ANS), which preferentially binds to hydrophobic areas of a given structure. Both EDC-crosslinked and 65 °C-aggregated IgG, but not the native form, generated prominent fluorescent emission profiles with bis-ANS (Fig. 2C), indicating the presence of exposed hydrophobic regions in these protein preparations that are otherwise buried in natively folded structures. Comparison of native and EDC crosslinked proteins by far-UV (supplemental Fig. S3) and near-UV CD (supplemental Fig. S4) did not reveal significant change in the characteristic shapes of the spectra, suggesting that the proteins maintain somewhat native secondary as well as tertiary structures. Difference in the magnitude of CD signal upon crosslinking is likely due to slight differences in protein concentration. Nevertheless, when being compared with native HSA, EDC-crosslinked HSA was highly sensitive to trypsin digestion, albeit both proteins when fully denatured were equally susceptible to this protease (Fig. 2D). Together with bis-ANS binding, EDC-crosslinked proteins seemingly display features of structure misfolding.
To reveal the relevance of EDC-cross-linked proteins to amyloid fibrils, we stained different IgG preparations with ThT, a fluorescent dye specifically reactive to β-sheet-rich amyloid (29). EDC-crosslinked IgG did not bind significantly to ThT, nor did native IgG. However, 65 °C-aggregated human IgG, which forms precipitant in solution, displayed strong ThT binding, indicating the presence of β-sheet-rich structure (supplemental Fig. S5).
In short, EDC-crosslinked proteins are soluble, multimeric, have strong cytotoxicity, display a common conformational structure shared with other protein oligomers, exhibit structural alterations; but are not amyloid per se. Therefore, we consider them stabilized protein oligomers, which closely resemble the intermediate protein aggregates implicated in protein misfolding diseases.
Stabilized Protein Oligomers Preferentially Bind to Nucleic Acids
The soluble HSA oligomers we obtained by EDC crosslinking remained stable in PBS buffer, without any sign of spontaneous amyloid formation. We were intrigued by the reports that non-proteinaceous polyanionic cofactors, such as RNA and sulfated glycans, play an essential role in the infectivity of prions (30) and hypothesized that these factors may somehow interact with misfolded proteins formed along the amyloid development pathway.
As a positive control for our investigation, we obtained a prion fragment containing aa 106–126 that is both neurotoxic and amyloidogenic (31). The prion peptide readily bound to dsDNA in a gel shift assay (Fig. 3A), an interaction that conveys protection of DNA from DNase digestion (supplemental Fig. S6). However, a scrambled control peptide failed to bind or protect the DNA under the same conditions. Similarly, Aβ peptide, in contrast to a control peptide with reversed sequence, demonstrated the capacity to interact with DNA in vitro (Fig. 3B). Therefore, we confirmed the direct interaction between DNA and the prototypic amyloidogenic peptides, as previously published (32, 33).
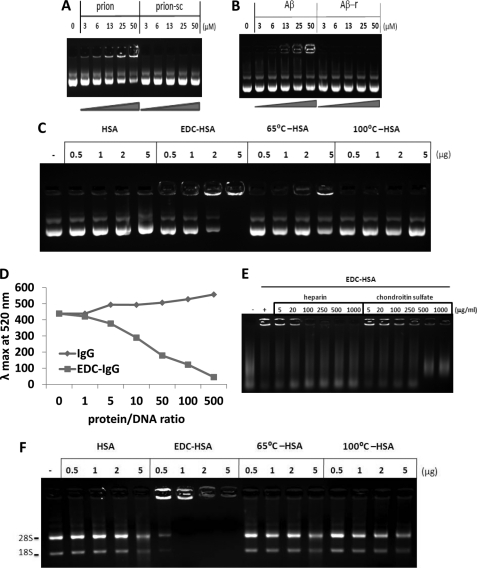
FIGURE 3.
EDC-crosslinked proteins directly bind to nucleic acids. A, different amounts of prion peptide or scrambled control peptide pre-mixed with plasmid DNA were analyzed by agarose gel electrophoresis. B, different amounts of Aβ peptide or a control peptide with reversed sequence pre-mixed with DNA were analyzed by agarose gel electrophoresis. C, different amounts of different forms of HSA-derived proteins pre-mixed with plasmid DNA were analyzed by agarose gel electrophoresis. D, 0.5 μg of plasmid DNA was incubated with increasing amounts of native or EDC-crosslinked IgG. Emission intensities at 520 nm were measured after addition of dye PicoGreen. E, EDC-crosslinked HSA pre-incubated with different amounts of heparin or chondroitin sulfate was mixed with DNA, then analyzed by agarose gel electrophoresis. F, different amounts of HSA-derived proteins were incubated with 0.5 μg of RNA then analyzed by agarose gel electrophoresis.
To test our stabilized HSA oligomers, native, EDC-crosslinked, 65 °C-aggregated, and 100 °C-denatured HSA were pre-incubated with a plasmid DNA then separated by agarose gel electrophoresis. EDC-crosslinked HSA oligomers bound to the DNA in a dose-dependent manner, which resulted in significantly delayed migration of the nucleic acids (Fig. 3C). At the 5 μg/ml dose, 65 °C-aggregated HSA weakly interacted with the DNA. Neither native nor 100°C-denatured HSA bound to the DNA. In addition to plasmid DNA, EDC-crosslinked oligomeric HSA readily bound to genomic DNA of vertebrate animal cells, circular double-stranded plasmid and single-stranded DNA (ssDNA) (supplemental Fig. S7), plus genomic DNA of bacteria and of mammalian cells (data not shown). Similar results were also obtained with IgG-derived proteins (data not shown). To further confirm the interaction between protein oligomers and DNA, we utilized PicoGreen, a fluorescent dye that emits at 520 nm when binding to the minor groove of dsDNA. A direct interaction with HSA oligomers condensed DNA and significantly reduced PicoGreen fluorescence in a dose-dependent manner (Fig. 3D).
Because both glycosaminoglycans and nucleic acids were implicated in infectious prions, we tested the effects of two sulfated GAGs on HSA oligomers in DNA gel shift assay. Both heparin and chondroitin sulfate effectively inhibited the complex formation between HSA oligomers and DNA (Fig. 3E). Lastly, we incubated native, EDC-crosslinked and heat-aggregated HSA with total RNA isolated from mammalian cells and observed a strong interaction between HSA oligomers and RNA (Fig. 3F). Thus, the stabilized protein oligomers can invariably bind to both DNA and RNA, an interaction that is sensitive to inhibition by GAGs.
Binding with Nucleic Acids and GAGs Converts Stabilized Protein Oligomers to Amyloid
To explore the potential change in stabilized protein oligomers induced by binding to nucleic acids and GAGs, we monitored the oligomer-specific conformational structure of EDC-crosslinked HSA that had been pre-mixed with different concentrations of DNA or heparin by a dot blot analysis. The epitope recognized by the anti-Aβ oligomer antibody A11 was significantly diminished when the protein oligomer was mixed with DNA or heparin at a stoichiometric ratio (Fig. 4A), though certain levels of A11 reactivity reappeared in the presence of higher concentration of polyanions. Strikingly, within minutes after the polyanions were added to HSA oligomers, the sample solutions became turbid and insoluble matter formed in the solution (Fig. 4B). Such precipitation induced by heparin or DNA was not observed with native HSA protein.
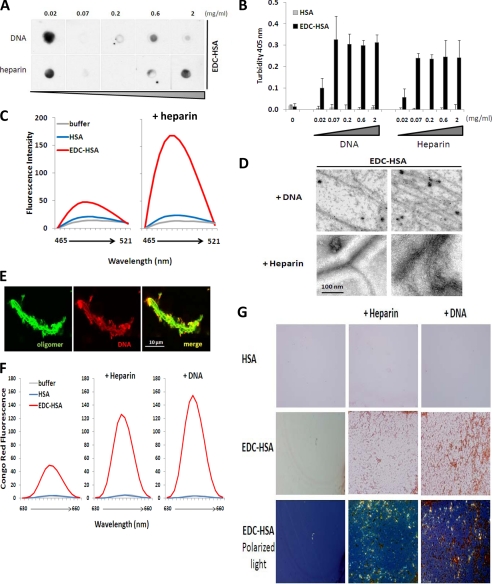
FIGURE 4.
Binding with nucleic acids and heparin converts EDC-crosslinked proteins to amyloid. EDC-crosslinked HSA (0.2 mg/ml) was incubated with different amounts of DNA or heparin. The resulting samples were analyzed by (A) dot blot with A11 antibody and (B) OD at 405 nm to measure the solution turbidity. C, native or EDC-crosslinked HSA were incubated with buffer or heparin, before mixing with dye ThT. Differential fluorescence emission profiles of the resulting samples are shown. D, transmission electron microscopy analysis of EDC-crosslinked HSA complexed with either DNA (top) or heparin (bottom). E, confocal imaging analysis of EDC-crosslinked IgG (green) complexed with fluorescently labeled DNA (red). F, and G, native or EDC-crosslinked HSA were incubated with buffer, heparin, or DNA, before mixing with dye Congo Red. Differential fluorescence emission profiles (F) and microscopic analysis (G) of the resulting samples are shown. In G top two rows: brightfield light; bottom row: transmitted polarized light (bottom row). Magnification, ×40.
To characterize the materials precipitated from the mixtures of HSA oligomers with DNA or heparin, we first tested their ability to bind to Thioflavin T. Unfortunately, significant binding with DNA masked the ThT emission profile of the HSA oligomer/DNA mixture. Nevertheless, the HSA oligomer/heparin mixture exhibited significant ThT emission compared with the oligomer alone, suggesting the formation of β-sheet-rich amyloid structures (Fig. 4C). In contrast, heparin addition failed to enhance ThT fluorescence in the native HSA protein.
To visualize amyloid formation, we performed transmission electron microscopy (TEM) analysis and confirmed the presence of fibril structures in the mixtures of HSA oligomer/DNA and HSA oligomer/heparin (Fig. 4D). The shape of the fibers ranged from single or branched rods to connected networks. By confocal microscopy, we observed the assembly of fibrous structures containing both DNA and stabilized IgG oligomers (Fig. 4E). Those insoluble aggregates have the capacity to bind to the amyloid-specific dye Congo Red, as we observed significant fluorescence emission of the dye on the complexes of oligomeric HSA-heparin or oligomeric HSA-DNA (Fig. 4F). No fluorescence was detectable when native HSA was mixed with heparin or DNA. Interestingly, soluble EDC-crosslinked HSA also bound to Congo Red, which resulted in moderate fluorescence emission, a property not associated with native HSA. We further confirmed the specific binding of Congo Red by observing microscopically the samples under transmitted polarized light. Whereas oligomeric HSA alone never precipitated on microscope slides, the oligomeric protein complexed with heparin or DNA, which led to the generation of Congo Red-positive insoluble precipitate (Fig. 4G). The binding of Congo Red is specific to the presence of β-sheet rich amyloid structures since an intense apple-green birefringence of the dye was readily visible under the transmitted polarized light (Fig. 4G, bottom panel).
When incubated with RNA, EDC-crosslinked protein oligomers lost the conformational epitope, turned insoluble and rapidly formed amyloid fibers (supplemental Fig. S8), a transformation similar to what happened when they encounter DNA. Noticeably, A11 reactivity diminishes most significantly in the presence of RNA at 3:1 and 1:1 protein:RNA ratio, but re-appeared with higher RNA concentration, a phenomenon reproducible in our hands. These results strongly suggest that the interaction between nucleic acids or GAGs and protein oligomers facilitates the rapid conversion of soluble oligomers into insoluble amyloid.
Binding with Nucleic Acids and GAGs Diminishes Cytotoxicity of Stabilized Protein Oligomers
With all the biochemical changes to stabilized protein oligomers induced by nucleic acids and GAGs, we asked whether the cellular functions of the stabilized protein oligomers are affected. Soluble protein oligomers exert strong cytotoxicity toward mammalian cells. However, in the presence of DNA or heparin, protein oligomers were no longer able to elicit a significant toxic effect on cells, as judged by Trypan Blue, CellTiter-Blue®, and propidium iodide staining (Fig. 5, A–C). In accordance with this, cell death induced by prion peptide was also significantly reduced in the presence of DNA and heparin (Fig. 5D). Therefore, nucleic acids and GAGs exert profound effects on the pathogenic function of soluble protein oligomers.
FIGURE 5.
Cytotoxicity of stabilized protein oligomers is inhibited by DNA and heparin. RPMI 8226 cells cultured with 10 μg/ml HSA-derived proteins in the presence of 10 μg/ml DNA or 10 μg/ml heparin for 24 h. The viability of the culture cells were examined by (A) Trypan Blue, (B) CellTiter-Blue®, and (C) PI staining. D, dead cell populations in RPMI 8226 cells cultured with different amounts of prion peptide in the presence of control, 10 μg/ml DNA, or 10 μg/ml heparin was assessed by PI staining.
EGCG Inhibits Amyloid Conversion of Protein Oligomers
By directly binding to the natively unfolded polypeptides, polyphenol (−)-epigallocatechin gallate (EGCG) has been shown to efficiently inhibit the fibrillogenesis of amyloidogenic proteins, such as α-synuclein, amyloid-β, and huntingtin, and prevent their conversion into toxic intermediates (34). We were curious to know whether EGCG could similarly intercept the amyloid transformation of stabilized protein oligomers.
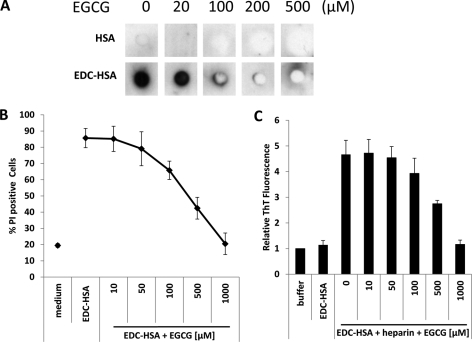
First, we incubated different amounts of EGCG with native HSA or HSA oligomers and performed dot blot analysis with A11 antibody. Similar to its effect on α-synuclein oligomers, EGCG diminished the structure on soluble HSA oligomers detectable by this antibody (Fig. 6A). Furthermore, the cytotoxicity of HSA oligomers was greatly reduced by EGCG in a dose dependent manner (Fig. 6B), suggesting a direct interaction between EGCG and HSA oligomers. Because heparin can effectively convert soluble protein oligomers to amyloid (Fig. 4), we included different doses of EGCG in the mixture of heparin and HSA oligomers and observed that EGCG was able to significantly reduce the formation of β-sheet-rich structures in the mixed sample, judged by reduced ThT fluorescence (Fig. 6C). These results are in full accordance with the previous finding that the small molecule EGCG can redirect misfolded protein intermediates to off-pathway species.
FIGURE 6.
EGCG inhibits conversion of stabilized protein oligomers to amyloid. A, HSA or EDC-crosslinked HSA were pre-incubated with increasing amounts of EGCG. Dot blot analysis was performed with A11 antibody. B, EDC-crosslinked HSA (10 μg/ml) was pre-incubated with increasing amounts of EGCG then added to the culture of RPMI 8226 cells. After 24 h, the percentage of dead cell population was measured by PI staining. C, EDC-crosslinked HSA was pre-incubated with increasing amounts of EGCG then mixed with heparin. Emission by Thioflavin T at 486 nm after adding to the resulting protein samples was measured. Fluorescence emission by the control buffer was used as a reference. Relative fluorescence emission of other samples was calculated and plotted. Error bars are means ± S.D.
DISCUSSION
Here we report an unexpected discovery that EDC-crosslinked common proteins display several key features analogous to those of soluble oligomeric amyloidogenic proteins. By preferential binding to polyanionic factors, such as nucleic acids and glycosaminoglycans, these soluble oligomers quickly convert to amyloid fibrils in vitro.
Although it has been shown that soluble oligomers of native proteins can be generated in vitro (14), it was hard for us initially to comprehend how soluble protein oligomers can be derived by crosslinking native HSA or IgG, which are long polypeptides with well-folded globular structures. Protein aggregation per se cannot explain the phenomenon because multimeric HSA crosslinked with other chemicals, such as dimethyl pimelimidate (DMP), disuccinimidyl suberate (DSS), and glutaraldehyde, failed to show cellular toxicity (data not shown). However, we did notice that, distinct from other chemical crosslinkers, which require a neutral to basic pH environment, EDC performs conjugating reactions under acidic pH conditions. Indeed, EDC is ineffective in buffer with a pH ≥ 7. EDC-crosslinked HSA prepared at pH 3∼5.5 behaved very similarly in our in vitro tests, whereas EDC-crosslinked HSA at pH 6.5 had reduced aptitude in the properties we have described here. The significance of acidic pH condition in favoring the protein misfolding pathway can be inferred from numerous published studies. An acetate buffer solution at pH 5.5 with trifluoroethanol was used to induce soluble oligomers of SH3 domain from bovine phosphatidyl-inositol-3′-kinase and the amino-terminal domain of the E. coli HypF protein (14). Human β2-microglobulin and transthyretin form amyloid fibrils efficiently under acidic conditions in vitro (35–37). For functional amyloidogenic protein Pme17, a critical pH regime between 5 ± 0.5 is required for fibril formation in a specific melanosome compartment (38).
Interestingly, BSA, which has 76% sequence homology with HSA, undergoes reversible native conformational isomerization at isoelectric point of pH 4.8 (39). It has been shown that, a combinational condition similar to EDC crosslinking with high protein concentration, low pH and salt, enables monomeric BSA to adopt a molten-globule-like state and form so-called molten oligomers (40). Therefore, in EDC-crosslinked proteins, the acidic condition under which proteins are partially denatured when the polypeptides are crosslinked may be the key to the adoption of a structure that is normally associated with transiently formed soluble protein oligomers. Apparently, proteins refold sufficiently after EDC crosslinking and exchange back to neutral pH, as they maintain both secondary and tertiary structures similar to their native counterparts. Nevertheless, EDC-stabilized proteins have exposed hydrophobic regions, are more sensitive to trypsin digestion and display a conformational epitope recognized by A11 antibody. Detailed structural characterization on these proteins is needed in the future to better understand the molecular basis of the unique properties associated with soluble protein oligomers.
It is intriguing that all EDC-crosslinked proteins we have tested demonstrate a remarkable capacity to interact with different forms of nucleic acids and sulfated GAGs. The precise molecular basis for this observation is unknown to us at this time, although it is consistent with several recent publications reporting the involvement of cofactors in amyloidogenic proteins as well as in protein misfolding diseases. In particular, non-proteinaceous cofactors in the forms of polyanions, such as RNA and phospholipids, are seemingly required to produce infectious prions, possibly by forming physical complexes with prion (25, 30). All soluble aggregates of Aβ42 interact with DNA in vitro, which reportedly induces conformational condensation of DNA (28, 41–43). DNA binding is also reported for other amyloidogenic proteins (reviewed in Ref. 44). Although the interactions between DNA and these proteins were interpreted as a potential indication of the nuclear functions of amyloidogenic proteins in some cases (44, 45), other studies suggested that they played a role in amyloid promotion. dsDNA stimulates the fibrillation of α-synuclein and is associated with the mature fibrils (26, 27). Nucleic acids facilitate polymerization of prion peptides, a process that accompanies structural changes in DNA in vitro (33, 46). Separately, cell-associated HSPG glypican-1 has been shown to facilitate prion conversion in lipid raft, and HSPG can effectively promote amyloid fibrillization in vitro (12, 23, 47, 48).
It is well described that intrinsically disordered proteins, which constitute a significant fraction of eukaryotic proteome, undergo significant structural rearrangement, in whole or partly, upon binding to their physiological targets (50–53). Since a number of amyloidogenic polypeptides contain intrinsically disordered regions (50, 53), it is plausible that polyanionic cofactors play a somewhat similar and critical role in selectively inducing β-sheet formation. Our results with stabilized protein oligomers strongly imply that polyanionic molecules function as a key contributor to the direct and rapid conversion of soluble oligomers to insoluble amyloid. Though stable by themselves in solution, the protein oligomers preferentially complex with nucleic acids or GAGs to form amyloid fibrils, which represent a more stable state for the polypeptides. This in vitro phenomenon is in line with the fact that nucleic acids and GAGs are commonly found in most amyloid deposits from patients (22, 23, 44).
Although the pathophysiological role of soluble protein oligomers is becoming increasingly appreciated in various human diseases, the biochemical and biophysical features of these toxic species remain obscure. Natural Aβ oligomers in a wide range of molecular weights have been found in the brains of patients with Alzheimer disease (18). Synthetic Aβ, like many other amyloidogenic polypeptides, was also shown to assemble into toxic oligomers in vitro, albeit with remarkable heterogeneity in terms of size, morphology, structure and toxic functions (7, 17, 49). It is generally recognized that these oligomeric assemblies are highly unstable, transient, flexible and have dynamic structures with exposed hydrophobic patches on their surface (6, 7, 17). Based on the presence of common structures recognized by polyreactive antibodies, soluble protein oligomers generally assimilate in two groups: the prefibrilar oligomers identifiable by A11 reactivity and other fibrilar oligomers lacking an A11-specific epitope (17). The fibrilar oligomers display the capacity to incorporate monomers to enlongate the fibril, whereas the prefibrilar oligomers convert to amyloid fibril by “en bloc” conformational change (17). In lieu of this classification, we regard the stabilized protein oligomers that we obtained as more likely being the prefibrilar type, as they positively expressed the A11-specific epitope and, in the presence of polyanionic cofactors, immediately adopted rapid conformational change to form amyloid. This classification is also consistent with the fact that we did not detect any spontaneous development of amyloid from these oligomeric proteins in the presence of additional protein monomers, although there is a possibility that intermolecular crosslinking imposes certain structural restraints that may interfere with dynamic fiber growth. Our finding that prefibrilar oligomers require cofactors for de novo conversion is important for a comprehensive understanding of the basic principles governing the development of amyloid in general.
The conversion of soluble oligomers of most common proteins into amyloid is in agreement with the now prevailing consensus that every protein might have a native state as well as a fibrous state (54) and that most human proteins contain segments capable to mediate the formation of amyloid fibrillization (3). Our results also support the view that amyloid fibril formation occurs as a hierarchical process starting with misfolded/unfolded monomers and going through oligomeric intermediates. Furthermore, we have identified non-proteinaceous physiological factors that likely participate in the end-stage amyloid conversion after the formation of prefibrilar oligomers (Fig. 7). On the basis of the experimental data presented here, one can postulate that the cytotoxic function of soluble protein oligomers, presumably the principle pathophysiological aspect of these amyloidogenic species, can be limited by biomolecules that directly bind to them, e.g. extrageneous HSPG, nucleic acids or EGCG. Further studies are necessary to fully understand the mechanism responsible for cofactor binding, the cytotoxic effects and conformational features of soluble protein oligomers.
FIGURE 7.
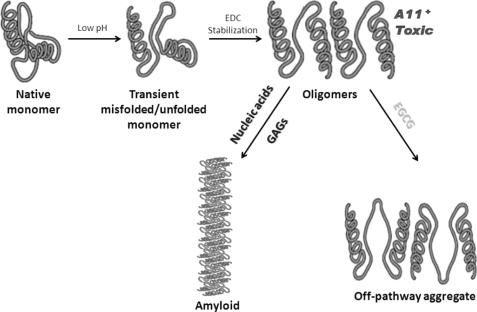
Schematic model of amyloid development from native proteins. Native proteins undergo transient misfolding under low pH condition. The misfolded or partially unfolded monomers aggregate, which is stabilized by EDC crosslinking in vitro. The resulting oligomers exhibit cytotoxic effects and adopt a conformational structure (A11+) that is commonly found in prefibrillar oligomers. These oligomers can proceed rapidly to form fibrous amyloid in the presence of nucleic acids or GAGs or develop into off-pathway aggregates by binding with EGCG.
Supplementary Material
Acknowledgments
We thank the professional assistance of Kenneth Dunner, Jr., at the M. D. Anderson High Resolution Electron Microscopy Facility. We appreciate valuable discussions with and suggestions by Drs. Michel Gilliet, Shao-Cong Sun, and Stephanie Watowich. We are grateful for helpful discussions with and professional assistance by Dr. David Hawke at The Proteomics Facility at The University of Texas M. D. Anderson Cancer Center.
This work is supported, in whole or in part, by National Institutes of Health Grant AI074809 (to W. C.) and grants from The University of Texas M. D. Anderson Cancer Center Institutional Research Grant Program (to W. C.). This work is also supported, in part, by the National Institutes of Health through M. D. Anderson's Cancer Center Support Grant (CA016672).

This article contains supplemental Figs. S1–S8.
- HSPG
- heparan sulfate proteoglycan
- bis-ANS
- 4,4′-bis(1-anilinonaphthalene 8-sulfonate)
- EDC
- 1-ethyl-3-[3-dimethyl-aminopropyl] carbodiimide hydrochloride
- EGCG
- polyphenol (−)-epigallocatechin gallate
- GAG
- glycosaminoglycan
- HSA
- human serum albumin
- IgG
- immunoglobulin G
- ThT
- Thioflavin T.
REFERENCES
- 1. Dobson C. M. (2003) Nature 426, 884–890 [DOI] [PubMed] [Google Scholar]
- 2. Selkoe D. J. (2003) Nature 426, 900–904 [DOI] [PubMed] [Google Scholar]
- 3. Schnabel J. (2010) Nature 464,, 828–829 [DOI] [PubMed] [Google Scholar]
- 4. Goldberg A. L. (2003) Nature 426, 895–899 [DOI] [PubMed] [Google Scholar]
- 5. Goldschmidt L., Teng P. K., Riek R., Eisenberg D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3487–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stefani M. (2010) FEBS J. 277, 4602–4613 [DOI] [PubMed] [Google Scholar]
- 7. Sakono M., Zako T. (2010) FEBS J. 277, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 8. Fowler D. M., Koulov A. V., Balch W. E., Kelly J. W. (2007) Trends Biochem. Sci. 32, 217–224 [DOI] [PubMed] [Google Scholar]
- 9. Chapman M. R., Robinson L. S., Pinkner J. S., Roth R., Heuser J., Hammar M., Normark S., Hultgren S. J. (2002) Science 295, 851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barnhart M. M., Chapman M. R. (2006) Annu. Rev. Microbiol. 60, 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Claessen D., Rink R., de Jong W., Siebring J., de Vreugd P., Boersma F. G. H., Dijkhuizen L., Wösten H. A. B. (2003) Genes Dev. 17, 1714–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maji S. K., Perrin M. H., Sawaya M. R., Jessberger S., Vadodaria K., Rissman R. A., Singru P. S., Nilsson K. P., Simon R., Schubert D., Eisenberg D., Rivier J., Sawchenko P., Vale W., Riek R. (2009) Science 325, 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Badtke M. P., Hammer N. D., Chapman M. R. (2009) Sci. Signal. 2, pe43- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bucciantini M., Giannoni E., Chiti F., Baroni F., Formigli L., Zurdo J., Taddei N., Ramponi G., Dobson C. M., Stefani M. (2002) Nature 416, 507–511 [DOI] [PubMed] [Google Scholar]
- 15. Walsh D. M., Klyubin I., Fadeeva J. V., Cullen W. K., Anwyl R., Wolfe M. S., Rowan M. J., Selkoe D. J. (2002) Nature 416, 535–539 [DOI] [PubMed] [Google Scholar]
- 16. Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., Glabe C. G. (2003) Science 300, 486–489 [DOI] [PubMed] [Google Scholar]
- 17. Glabe C. G. (2008) J. Biol. Chem. 283, 29639–29643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lesné S., Koh M. T., Kotilinek L., Kayed R., Glabe C. G., Yang A., Gallagher M., Ashe K. H. (2006) Nature 440, 352–357 [DOI] [PubMed] [Google Scholar]
- 19. Ellis R. J., Pinheiro T. J. T. (2002) Nature 416, 483–484 [DOI] [PubMed] [Google Scholar]
- 20. Jarrett J. T., Lansbury P. T., Jr. (1993) Cell 73, 1055–1058 [DOI] [PubMed] [Google Scholar]
- 21. Stefani M. (2004) Biochim. Biophys. Acta 1739, 5–25 [DOI] [PubMed] [Google Scholar]
- 22. McBride P. A., Wilson M. I., Eikelenboom P., Tunstall A., Bruce M. E. (1998) Exp. Neurol. 149, 447–454 [DOI] [PubMed] [Google Scholar]
- 23. Zhang X., Li J.-P., Lijuan Z. (2010) Progress in Molecular Biology and Translational Science, pp. 309–334, Academic Press [Google Scholar]
- 24. Deleault N. R., Lucassen R. W., Supattapone S. (2003) Nature 425, 717–720 [DOI] [PubMed] [Google Scholar]
- 25. Wang F., Wang X., Yuan C. G., Ma J. (2010) Science 327, 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rysavá R. (2007) Kidney Blood Press Res. 30, 359–364 [DOI] [PubMed] [Google Scholar]
- 27. Hegde M. L., Rao K. S. J. (2007) Arch Biochem. Biophys. 464, 57–69 [DOI] [PubMed] [Google Scholar]
- 28. Barrantes A., Rejas M. T., Benítez M. J., Jiménez J. S. (2007) J. Alzheimer Dis. 12, 345–355 [DOI] [PubMed] [Google Scholar]
- 29. LeVine H., 3rd (1993) Protein Sci. 2, 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rönnblom L. E., Alm G. V., Oberg K. E. (1990) J. Intern. Med. 227, 207–210 [DOI] [PubMed] [Google Scholar]
- 31. De Gioia L., Selvaggini C., Ghibaudi E., Diomede L., Bugiani O., Forloni G., Tagliavini F., Salmona M. (1994) J. Biol. Chem. 269, 7859–7862 [PubMed] [Google Scholar]
- 32. Silva J. L., Lima L. M., Foguel D., Cordeiro Y. (2008) Trends Biochem. Sci. 33, 132–140 [DOI] [PubMed] [Google Scholar]
- 33. Nandi P. K. (1998) Arch. Virol. 143, 1251–1263 [DOI] [PubMed] [Google Scholar]
- 34. Ehrnhoefer D. E., Bieschke J., Boeddrich A., Herbst M., Masino L., Lurz R., Engemann S., Pastore A., Wanker E. E. (2008) Nat. Struct. Mol. Biol. 15, 558–566 [DOI] [PubMed] [Google Scholar]
- 35. McParland V. J., Kalverda A. P., Homans S. W., Radford S. E. (2002) Nat. Struct. Mol. Biol. 9, 326–331 [DOI] [PubMed] [Google Scholar]
- 36. McParland V. J., Kad N. M., Kalverda A. P., Brown A., Kirwin-Jones P., Hunter M. G., Sunde M., Radford S. E. (2000) Biochemistry 39, 8735–8746 [DOI] [PubMed] [Google Scholar]
- 37. Liu K., Cho H. S., Lashuel H. A., Kelly J. W., Wemmer D. E. (2000) Nat. Struct. Mol. Biol. 7, 754–757 [DOI] [PubMed] [Google Scholar]
- 38. Pfefferkorn C. M., McGlinchey R. P., Lee J. C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 21447–21452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vetri V., D'Amico M., Foderà V., Leone M., Ponzoni A., Sberveglieri G., Militello V. (2011) Arch. Biochem. Biophys. 508, 13–24 [DOI] [PubMed] [Google Scholar]
- 40. Bhattacharya M., Jain N., Mukhopadhyay S. (2011) J. Phys. Chem. B 115, 4195–4205 [DOI] [PubMed] [Google Scholar]
- 41. Yu H., Ren J., Qu X. (2007) Biophys. J. 92, 185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hegde M. L., Anitha S., Latha K. S., Mustak M. S., Stein R., Ravid R., Rao K. S. (2004) J. Mol. Neurosci. 22, 19–31 [DOI] [PubMed] [Google Scholar]
- 43. Ahn B. W., Song D. U., Jung Y. D., Chay K. O., Chung M. A., Yang S. Y., Shin B. A. (2000) Anal. Biochem. 284, 401–405 [DOI] [PubMed] [Google Scholar]
- 44. Jiménez J. S. (2010) J. Alzheim. Dis. 22, 375–391 [DOI] [PubMed] [Google Scholar]
- 45. Kegel K. B., Meloni A. R., Yi Y., Kim Y. J., Doyle E., Cuiffo B. G., Sapp E., Wang Y., Qin Z. H., Chen J. D., Nevins J. R., Aronin N., DiFiglia M. (2002) J. Biol. Chem. 277, 7466–7476 [DOI] [PubMed] [Google Scholar]
- 46. Nandi P. K., Leclerc E. (1999) Arch. Virol. 144, 1751–1763 [DOI] [PubMed] [Google Scholar]
- 47. Taylor D. R., Whitehouse I. J., Hooper N. M. (2009) PLoS Pathog 5, e1000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hooper N. M. (2011) J. Neurochem. 116, 721–725 [DOI] [PubMed] [Google Scholar]
- 49. Haass C., Selkoe D. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 101–112 [DOI] [PubMed] [Google Scholar]
- 50. Fink A. L. (2005) Curr. Opin. Struct. Biol. 15, 35–41 [DOI] [PubMed] [Google Scholar]
- 51. Wright P. E., Dyson H. J. (2009) Curr. Opin. Struct. Biol. 19, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Babu M. M., van der Lee R., de Groot N. S., Gsponer (2011) Curr. Opin. Struct. Biol. 21, 432–440 [DOI] [PubMed] [Google Scholar]
- 53. Uversky V. N., Oldfield C. J., Dunker A. K. (2008) Annu. Rev. Biophys. 37, 215–246 [DOI] [PubMed] [Google Scholar]
- 54. Astbury W. T., Dickinson S., Bailey K. (1935) Biochem. J. 29, 2351–2360.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.