Background: To date, very few cellular factors required for secretion of flaviviruses have been described.
Results: Simultaneous depletion of class II Arf (Arf4 and Arf5) blocks dengue flavivirus secretion, without altering the constitutive secretory pathway. Dengue glycoprotein prM interacts with Arf4 and Arf5.
Conclusion: Arf4 and Arf5 play a crucial role in dengue flavivirus secretion.
Significance: Our findings reveal a molecular mechanism of dengue flavivirus secretion.
Keywords: ARF, Flaviviruses, Host-Pathogen Interactions, Positive-strand RNA Viruses, Virus Assembly, Arf4, Arf5, Dengue Virus, Recombinant Subviral Particles, Secretion
Abstract
Identification and characterization of virus-host interactions are very important steps toward a better understanding of the molecular mechanisms responsible for disease progression and pathogenesis. To date, very few cellular factors involved in the life cycle of flaviviruses, which are important human pathogens, have been described. In this study, we demonstrate a crucial role for class II Arf proteins (Arf4 and Arf5) in the dengue flavivirus life cycle. We show that simultaneous depletion of Arf4 and Arf5 blocks recombinant subviral particle secretion for all four dengue serotypes. Immunostaining analysis suggests that class II Arf proteins are required at an early pre-Golgi step for dengue virus secretion. Using a horseradish peroxidase protein fused to a signal peptide, we show that class II Arfs act specifically on dengue virus secretion without altering the secretion of proteins through the constitutive secretory pathway. Co-immunoprecipitation data demonstrate that the dengue prM glycoprotein interacts with class II Arf proteins but not through its C-terminal VXPX motif. Finally, experiments performed with replication-competent dengue and yellow fever viruses demonstrate that the depletion of class II Arfs inhibits virus secretion, thus confirming their implication in the virus life cycle, although data obtained with West Nile virus pointed out the differences in virus-host interactions among flaviviruses. Our findings shed new light on a molecular mechanism used by dengue viruses during the late stages of the life cycle and demonstrate a novel function for class II Arf proteins.
Introduction
The four serotypes of dengue virus (DENV),3 which are members of the Flavivirus genus in the Flaviridae family, are the most important vector-borne viruses, and they cause 50–100 million cases of infection per year, including 500,000 severe cases (1–3).
Assembly of DENV, like other flaviviruses, occurs at membranes of the endoplasmic reticulum (ER) (4). Virions bud into the lumen of this organelle and, before being released, traffic through the host cell secretory pathway where the cellular protease furin cleaves pre-membrane (prM) protein, resulting in the release of the pr peptide and formation of mature virions (5–7). During flavivirus infection, in addition to infectious mature virions, noninfectious subviral particles are produced and traffic along the same secretory pathway as infectious particles before being released by the host cell (8). Similar recombinant subviral particles (RSPs) can form in the absence of capsid in cells transfected solely with prM and envelope (E) glycoproteins (9–12). In a previous work, we have developed RSPs for the four dengue serotypes and have shown that they mimic budding, secretion, and maturation of DENV (12). Therefore, dengue RSP represents a safe and convenient tool for the study of virus-host interactions during DENV secretion in host cells.
Currently, the viral-host interactions during the DENV life cycle are still poorly characterized. Moreover, most studies in this field focus on the maturation process, and less research has been done to investigate the molecular mechanisms supporting secretion (13, 14). We have previously reported the development of human stable cell lines that constitutively secrete RSPs of all four dengue serotypes and their use for screening a human siRNA library targeting specifically 122 genes involved in cellular membrane trafficking (12). We noticed that two members of the ADP-ribosylation factor (Arf) family, Arf1 and Arf6, which represent the most studied Arf proteins (15), as well as an Arf-related gene, the ADP-ribosylation factor interacting protein 2 (Arfaptin 2), showed either significant reduction or enhancement of RSP release (12), strongly suggesting a role for members of the Arf family in late stages of the DENV life cycle.
Arf proteins belong to the Ras superfamily of small GTPases. They cycle between cellular membranes and the cytosol, playing critical roles in membrane trafficking via the recruitment of various coat proteins (16, 17), initiating membrane curvature (18), and by modulating the activity of several lipid-modifying enzymes (19, 20). Six Arf proteins have been identified so far, with only five expressed in humans; Arf2 has been lost (20). Based on amino acid sequence identity, the six Arfs were grouped into three classes (21) as follows: class I (Arf1–3), class II (Arf4 and -5), and class III (Arf6). Functionally, class I members are known to be involved in the assembly of different types of coat complexes onto budding vesicles along the secretory pathway (22), and class III (viz. Arf6) regulates endosome-membrane traffic and structural organization at the cell surface (23). Less is known about the function of class II proteins, although Arf4 involvement in the trafficking of rhodopsin has recently been documented (24, 25).
In this study, we investigated the potential role of all Arf family members during DENV secretion using dengue RSPs as a model system. We have identified Arf4 and Arf5 as two novel cellular factors involved in dengue virus secretion.
EXPERIMENTAL PROCEDURES
Cells, Viruses, and Antibodies
HeLa cells, human embryonic kidney cells (293T), and human hepatic cells (HepG2) were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37 °C with 5% CO2. Dengue RSP-producing cell lines (HeLa-prME-DENV1, -DENV2, -DENV3, and -DENV4), which were established using the codon optimized DENV prME gene as described previously (12), were cultured in the same medium containing 500 μg/ml hygromycin. Mosquito Aedes pseudoscutellaris (AP61) cells were grown in L-15 medium containing 10% FBS and 1% tryptophan at 28 °C.
All work with infectious flaviviruses, including Israeli WNV strain IS-98-ST1, DENV1 strain d1d FGA/NA, DENV4 strain 63632/76 (Burma), and YFV strain (Asibi), was performed in a biosafety level 3 laboratory (Institut Pasteur, Paris, France). HepG2 cells were used to study the effect of depletion of class II Arfs by siRNAs on flavivirus replication. Virus titration of DENV1 and DENV4 was performed on AP61 cells, whereas titration of YFV and WNV was performed using VeroE6 and BHK21, respectively.
The mouse anti-E monoclonal antibodies (mAb) 4E11 and 4G2 were provided by Dr. A. Amara (Institut Pasteur, Paris, France). The mouse anti-prME antibody and sera from a patient with dengue virus infection were kindly provided by Dr. Philippe Buchy (Institut Pasteur, Cambodia). The mouse anti-prM monoclonal antibody prM-6.1 (26) was kindly provided by Dr. Sittisombut Nopporn (Chiang Mai University, Chiang Mai, Thailand). Other antibodies used were anti-GAPDH mAb from Abcam (Cambridge, MA), anti-Arf4 from ProteinTech Group Inc. (Chicago, IL), and anti-Arf5 mAb from Abnova Corp. (Walnut, CA).
Constructs
For the construction of GFP fusion proteins, Arf4 (NM_001660) and Arf5 (NM_001662) cDNAs were purchased from OriGene Technologies (Rockville, MD). A GFP cDNA derived from the pGFP-N1 plasmid (Clontech) was fused to the C-terminal end of Arf4 and Arf5 and subcloned into pcDNA3.1 (Invitrogen).
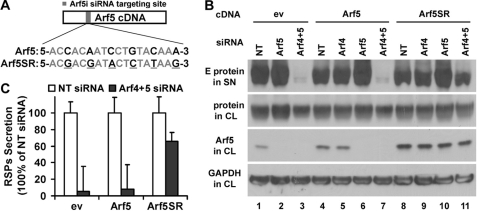
For rescue experiments, mutations were introduced into Arf5 cDNA by PCR-mediated mutagenesis at the site targeted by the Arf5i siRNA (L-011584), so that the original sequence (5-ACCACAATCCTGTACAAA-3) was changed to (5-ACGACGATACTCTATAAG-3), thus creating a siRNA-resistant Arf5 sequence (Arf5SR). Both wild-type Arf5 and Arf5SR were then subcloned into pcDNA3.1 using BamHI and XhoI restriction enzymes.
To obtain separate prM and E constructs, prM or E was amplified by PCR using codon-optimized DENV1 prME as template and subcloned into pcDNA3.1 vector using BamHI and XhoI restriction enzymes. A signal sequence of vesicular stomatitis virus G envelope glycoprotein was inserted in-frame upstream of either prM or E cDNA.
For site-directed mutagenesis, the VXPX motif in the C-terminal portion of prM was either deleted (prME-ΔVXPX) or mutated to AXAX (prME-AXAX) by overlapping PCR using codon-optimized DENV1 prME as template and then subcloned into pcDNA3.1 vector using BamHI and XhoI restriction enzymes.
siRNA Experiments
All siRNAs, including nontargeting (NT) siRNA (D-001206) and transfection reagents DharmaFECT 1 (T-2001), were purchased from Dharmacon (Research Inc., Lafayette, CO). Arf1 siRNA (L-011580), Arf3 siRNA (L-011581), Arf4 siRNA (L-011582), Arf5 siRNA (L-011584), Arf6 siRNA (L-004008), and COP β (β-COP) siRNA (L-017940) were provided as SMARTpool ON-TARGET plus siRNA, which are pools of four siRNAs targeting various sites in a single gene. DENV1 E siRNA (AGATCCAGCTGACCGATT), Arf4i siRNA (AGACAACCAUUCUGUAUAA), and Arf5i siRNA (CCACAAUCCUGUACAAACU) were provided as individual siRNAs.
For siRNA experiments, reverse transfection was performed using DharmaFECT 1 reagents. Briefly, siRNAs were added to 24-well plates in DMEM without FBS and antibiotics. Twenty minutes later 0.8 ml of cells (120,000 cells/ml in DMEM supplemented with 10% FBS) were added to each well so that the final siRNA concentration was 100 nm. Cells were incubated at 37 °C for 48 h. Medium was then replaced with 0.3 ml of DMEM containing 2% FBS (without antibiotics), and 14 h later, supernatant containing secreted RSP was collected and cleared by centrifugation at 4000 rpm for 15 min. Cells were lysed in buffer containing 1% Triton X-100, 150 mm NaCl, 20 mm Tris-HCl (pH 7.5), and 1 mm EDTA for 15 min on ice, with frequent vortexing.
In rescue experiments, HeLa-prME-DENV1 cells were transfected with either Arf5SR or with an empty vector to derive three stable cell lines, following culture in medium containing 400 μg/ml G418 for 3 weeks. These cell lines were then subjected to siRNA experiments as described above.
Gel Electrophoresis, Western Blot, and RSP Quantification
4–12% NuPAGE Novex BisTris gels (Invitrogen) were used for SDS-PAGE. For Western blotting, proteins were transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were blocked overnight in 5% milk in PBS containing 0.1% Tween 20 solution and incubated with primary antibody for 1 h. After repeated washes, membranes were incubated for 1 h at room temperature with a horseradish peroxidase-labeled secondary antibody (Zymed Laboratories Inc.), and proteins were visualized using ECL Western blot detection reagent (Invitrogen) and exposure of blots to x-ray films. To quantify RSP secretion, the mean luminescence and area of supernatant and cell lysate E signals on Western blot results were measured by densitometry using the Adobe Photoshop software (Adobe Systems Incorporated, San Jose, CA). For each condition, the relative amount of RSP in supernatant was estimated by calculating the ratio E supernatant/E cell lysate. Ratios were then normalized to that of NT siRNA and expressed in percentage of reduction of secretion.
Flow Cytometry Analysis
HeLa-prME-DENV1 cells were simultaneously transfected with either Arf4GFP or Arf5GFP together with siRNAs targeting Arf4 or Arf5. Medium was replaced 4 h post-transfection with DMEM containing 10% FBS. Cells were fixed in 3.2% paraformaldehyde 48 h later and analyzed using a FACSCalibur flow cytometer (BD Biosciences).
Virus Titration
DENV and WNV were titrated by focus immunodetection assay, essentially as described previously (27). Briefly, AP61 cells were seeded in 24-well plates. After 1 day, cells were washed three times, and 10-fold serial dilutions of infectious supernatants (10−2 to 10−5 for DENV and 10−3 to 10−6 for WNV) were added to cells for 90 min at 28 °C. Cells were subsequently incubated in a final volume of 800 μl of L-15 medium supplemented with 10% FBS.
Following the appropriate incubation time at 28 °C (5 days for DENV and 3 days for WNV), cells were fixed in 3% formaldehyde for 20 min, permeabilized with 0.1% Triton X-100 for 4 min at room temperature, and incubated for 30 min at 37 °C with the mouse monoclonal 4G2 antibody that can detect E protein from different flaviviruses (28). Cells were incubated with secondary anti-mouse HRP-conjugated antibodies for 1 h at 37 °C, and foci were revealed using diaminobenzidine (Sigma).
YFV was titrated by plaque assay using Vero cells seeded in 24-well plates. One day after seeding, 10-fold serial dilutions (10−3 to 10−6) of infectious supernatants were added, and cells were incubated for 90 min at 37 °C. Cells were subsequently incubated in a final volume of 800 μl of L-15 medium supplemented with 10% FBS and incubated at 37 °C for 5 days. After repeated washings, cells were stained with 3% formaldehyde crystal violet for 20 min at room temperature, and plaques were manually counted.
Fluorescence Microscopy
For fluorescence microscopy, HeLa-prME-DENV1 cells grown on glass coverslips were fixed, permeabilized, and incubated with the following antibodies: anti-E (1:1000), anti-calreticulin (1:200), anti-ERGIC53 mAb (1:1000), and anti-Golgin-97 mAb (1:50) followed by incubation with appropriate secondary antibodies conjugated with fluorescein isothiocyanate (FITC) or TRITC. Nuclei were stained with DAPI, and coverslips were mounted on glass slides for analysis. Fixed cells were visualized under an AxioObserver inverted motorized fluorescent microscope, using the ApoTome module piloted through the Axiovision 4.6 software, and images were acquired through a high resolution MRm AxioCam CCD camera (Carl Zeiss, Jena, Germany).
Co-immunoprecipitation
Subconfluent monolayers of HeLa cells, HeLa-prME-DENV1 cells, or 293T cells (107 cells per 100-mm dish) transfected with pcDNA-prM and pcDNA-E plasmids were washed twice in PBS and lysed on ice with 1 ml of RIPA buffer containing protease inhibitors (complete, EDTA-free; Roche Applied Science) and 1 mm PMSF for 30 min. Cell debris was removed by centrifugation at 13,000 rpm for 15 min at 4 °C, and lysates were precleared by incubation with protein G-Sepharose beads (Amersham Biosciences) for 1 h. Precleared lysates were then incubated for 2 h at 4 °C with protein G-Sepharose beads previously treated with either specific antibodies or control IgGs. Subsequently, beads were collected by centrifugation at 13,000 rpm for 30 s and washed four times. Bound proteins were solubilized by boiling in SDS-PAGE loading buffer, separated by SDS-PAGE, and analyzed by Western blotting.
Horseradish Peroxidase Assay and Quantification of Secretion
HeLa cells were co-transfected with NT or class II Arf siRNAs and pN1-HRP plasmid coding for a secreted form of the horseradish peroxidase (ss-HRP, a gift from Dr. Vivek Malhorta, Centre for Genomic Regulation, Barcelona, Spain). Culture medium was changed with fresh medium 48 h post-transfection and then collected 14 h later. To measure the activity of horseradish peroxidase, cleared supernatant was mixed with ECL Western blot detection reagent (Invitrogen), and luminescence intensity was then measured using the Microbeta luminometer (PerkinElmer Life Sciences).
RESULTS
Silencing of Class II Arf Expression Inhibits Secretion of Dengue Recombinant Subviral Particles
To investigate the effect of all Arf family members on the secretion of DENV1 RSP, we transfected the HeLa-prME-DENV1 stable cell line (which constitutively secretes RSPs for DENV1) with siRNAs targeting each Arf gene. Cells treated with either NT siRNA or siRNA targeting the E glycoprotein of DENV1 were used as negative and positive controls, respectively. RSP secretion was assessed by visualizing dengue E protein in the supernatant by Western blotting. As expected, E protein expression was abolished and could not be detected in both supernatant and cell lysate after specific DENV1 E siRNA treatment (Fig. 1A, lane 9). Targeting of single Arfs did not induce any significant variation in the secretion of RSPs (Fig. 1A, upper panel, lanes 4–8), with the exception of Arf1, whose depletion partially reduced RSP secretion (Fig. 1A, upper panel, lane 4). This result is consistent with our previous data generated by screening a library of 122 siRNAs targeting genes involved in membrane traffic (12). In contrast, no significant change in intracellular expression levels of E was observed after knockdown of any of the Arfs (Fig. 1A, middle panel, lanes 4–8). It has been reported previously that Arfs could play redundant roles and that cooperation of two Arfs at the same site might be a general mechanism of Arf activity (29). Therefore, we decided to investigate the effect of all possible combinations of double Arf knockdowns on the secretion of DENV1 RSPs (Fig. 1B). For all conditions, no significant variations in expression of E protein and GAPDH were observed in cell lysates (Fig. 1B, middle and lower panels). In contrast, combinations of siRNAs targeting Arf1+4, Arf1+5, and Arf4+5 induced a clear decrease of secretion of RSPs (Fig. 1B, lanes 3, 4, and 9, respectively). Interestingly, the most dramatic reduction of DENV1 RSP secretion was observed after down-regulation of both members of class II Arfs (Arf4 and Arf5), suggesting a crucial role for these proteins in the release of RSPs (Fig. 1B, lane 9). Moreover, as the depletion of Arf4 or Arf5 alone did not decrease RSP secretion (Fig. 1A, lanes 6 and 7), our results also suggest that class II Arf proteins could compensate for each other during RSP secretion.
FIGURE 1.
Combined down-regulation of both class II Arfs inhibits dengue RSP secretion. HeLa-prME-DENV1 cells were transfected with siRNA targeting human Arf proteins. Culture medium was replaced with fresh medium 48 h post-transfection, and supernatants (SN) and cell lysates (CL) were collected 14 h later. RSP production was assessed by Western blot, visualizing dengue E protein expression in supernatant and cell lysate with the 4E11 monoclonal antibody. Cells without any treatment (Cells), nontargeting (NT) siRNA, DENV1 E siRNA, or transfection reagents alone (Mock) were used as controls. GAPDH expression level served as the loading control. A and B, HeLa-prME-DENV1 cells were transfected with siRNA targeting Arf proteins individually (A) or in combination (B). Results shown are representative of two independent experiments. C, HeLa-prME-DENV1 cells were transfected with either individual (Arf4i and Arf5i) or pooled (Arf4 and Arf5) siRNAs for class II Arfs. The efficiency of Arf siRNA was tested by Western blot using specific antibodies as described under “Experimental Procedures.” D, HeLa-prME-DENV1 cells were transiently transfected with Arf4GFP or Arf5GFP in combination with Arf4 or Arf5 siRNAs as in C. The efficiency of knockdown was assessed by calculating the percentage of GFP-positive cells by flow cytometry.
The use of pooled siRNAs, targeting four different sites in the same gene, ensured an efficient knockdown of targeted mRNA but could have resulted in an enhancement of off-targeting effect. To verify that the inhibition of DENV1 RSP secretion specifically resulted from class II Arf depletion, the effect of individual siRNAs targeting Arf4 and/or Arf5 genes (Arf4i and Arf5i), respectively, was investigated and compared with that of the pooled siRNAs (Fig. 1C, lanes 6–8 and 2–4, respectively). Similar expression for GAPDH was observed for all conditions tested (Fig. 1C, lower panel). Combinations of either individual or pooled siRNAs targeting Arf4+5 led to a severe reduction of secreted RSPs (Fig. 1C, lanes 4 and 8, upper panel). Similar results were obtained with another set of individual siRNAs (data not shown). Altogether, our data demonstrate that blocking of dengue RSP secretion did not result from an off-targeting effect of siRNAs and, therefore, was specifically due to class II Arf depletion.
To verify the efficacy and specificity of siRNA knockdown, Arf4 and Arf5 expression was monitored by Western blotting using either anti-Arf4 or anti-Arf5 antibodies. A band with an apparent molecular mass of 21 kDa, corresponding to the predicted electrophoresis mobility of Arf4 or Arf5 protein, was readily observed in cells treated with NT siRNA but not in cells transfected with their specific siRNAs (Fig. 1C, 3rd and 4th panels). To further measure siRNA knockdown efficiency, we used a recombinant Arf protein fused at its C terminus to a GFP tag, so that siRNA targeting to Arf would also reduce expression of GFP. HeLa-prME-DENV1 cells were transfected with Arf-GFP in combination with siRNAs as indicated (Fig. 1D, upper panel). The percentage of GFP-positive cells was determined by flow cytometry analysis 48 h post-transfection. Treatment with Arf4 siRNAs significantly decreased the percentage of Arf4-GFP-positive cells (Fig. 1D, upper panel), from 31% (NT siRNA) to 3.4%, indicating that almost 90% of cells lost the expression of Arf4-GFP. As Arf4 and Arf5 proteins share 96% homology, Arf4-GFP cells were also treated with Arf5 siRNA. No decrease in the percentage of Arf4-GFP cells was observed after Arf5 siRNA treatment (Fig. 1D, upper panel, 31.2%), confirming the specificity of Arf4 siRNAs treatment. Similar results were obtained with Arf5-GFP cells and Arf5 siRNA. Arf5 siRNA treatment could reduce the percentage of Arf5-GFP-positive cells by over 90% (Fig. 1D, lower panel) but not that of Arf4-GFP. These results demonstrate that treatment with Arf4 and Arf5 siRNAs could efficiently knockdown expression of corresponding proteins.
Expression of siRNA-resistant Arf5 Rescues Secretion of Dengue Recombinant Subviral Particles
To further confirm the specific role of class II Arfs in secretion of DENV RSPs, we performed a rescue experiment. We engineered cells that express an siRNA-resistant form of Arf5 (Arf5SR), in which six nucleotides in the targeting site of Arf5i siRNA were substituted by introducing silent mutations that did not modify the G/C content (Fig. 2A). Under these conditions, treatment with siRNA on these cells would only target endogenous Arf5 without affecting the exogenous resistant form. Arf5SR or Arf5 encoding plasmids or an empty vector (pcDNA) were transfected into HeLa-prME-DENV1 cells, and stable cell lines expressing the specified transgenes were established. To test whether the Arf5SR could specifically rescue the depletion of endogenous Arf5 by Arf5 siRNA, these stable cell lines were transfected with Arf4 and Arf5 siRNAs, either separately or in combination. RSP secretion was assessed by monitoring the presence of E in cell supernatants. Expressions of E and Arf5 proteins in cell lysates were also analyzed. GAPDH was used as control, and similar levels were observed for each condition tested (Fig. 2B, lower panel). As expected, after Arf5 siRNA treatment, Arf5 was only detected in the Arf5SR cell line (Fig. 2B, lanes 10 and 11, 3rd panel) but not in empty vector cell lines (Fig. 2B, lanes 2 and 3, 3rd panel) and Arf5 cell lines (Fig. 2B, lanes 6 and 7, 3rd panel). Whereas class II Arfs double knockdown inhibited RSP secretion in empty vector and Arf5 cell lines (Fig. 2B, lanes 3 and 7, upper panel), RSP secretion was rescued in Arf5SR cells (Fig. 2B, lane 11, upper panel). Quantification of relative levels of RSP secretion indicated that class II Arf depletion reduced by over 90% RSP secretion in cell lines transfected with empty vector or overexpressing Arf5, but only by 33% in the Arf5SR cell line (Fig. 2C). These results further demonstrate that decrease of RSP secretion was not resulting from an off-targeting effect of siRNAs but was the direct consequence of the down-regulation of Arf4 and Arf5 proteins. Moreover, these experiments confirm that one member of class II Arf is sufficient to support the secretion of RSPs.
FIGURE 2.
Inhibitory effect of class II Arfs siRNAs can be rescued by expression of exogenous Arf5. A, siRNA-resistant Arf5 (Arf5SR) was created by mutating six nucleotides (highlighted and underlined) in the Arf5i siRNA targeting site without changing amino acid sequence and G/C content. B, HeLa-prME-DENV1 cells expressing empty vector (ev), Arf5, or Arf5SR were transfected with siRNA targeting class II Arfs. E protein expression was detected in cell lysate (CL) and supernatant (SN) by Western blot using the monoclonal antibody 4E11. GAPDH and Arf5 expression were assessed in parallel. C, mean density of each RSP band was measured using Photoshop, and the relative amount of RSP was then calculated by normalizing it to that of NT siRNA. Results are shown as mean ± S.D. of triplicate measurements from one experiment representative of three others.
Class II Arfs Inhibit Secretion of Recombinant Subviral Particles of Four Dengue Serotypes
To address whether class II Arfs are important for the secretion of RSPs of all four DENV serotypes, experiments with Arf4/Arf5 siRNAs were performed on stable cell lines producing DENV2, DENV3, and DENV4 RSPs (12). As observed for DENV1, a significant decrease in secretion could also be detected by Western blotting for all RSPs after down-regulation of class II Arfs (Fig. 3A, lanes 2, 4, 6, and 8, upper panel), albeit to a variable extent (Fig. 3B). This result demonstrates that class II Arfs play a critical and general role in dengue RSP secretion. The effect of class II Arf depletion was also tested on the production of RSPs of the SARS coronavirus, recently developed in our laboratory (30). As reported earlier, SARS coronavirus RSPs assemble in the ER-Golgi intermediary compartment and are exported through the secretion pathway to be released at the cell surface (30). Interestingly, no significant decrease in their production was observed (data not shown), pointing to a specific role for Arf4 and Arf5 during DENV secretion, rather than a general role in cell protein secretion.
FIGURE 3.
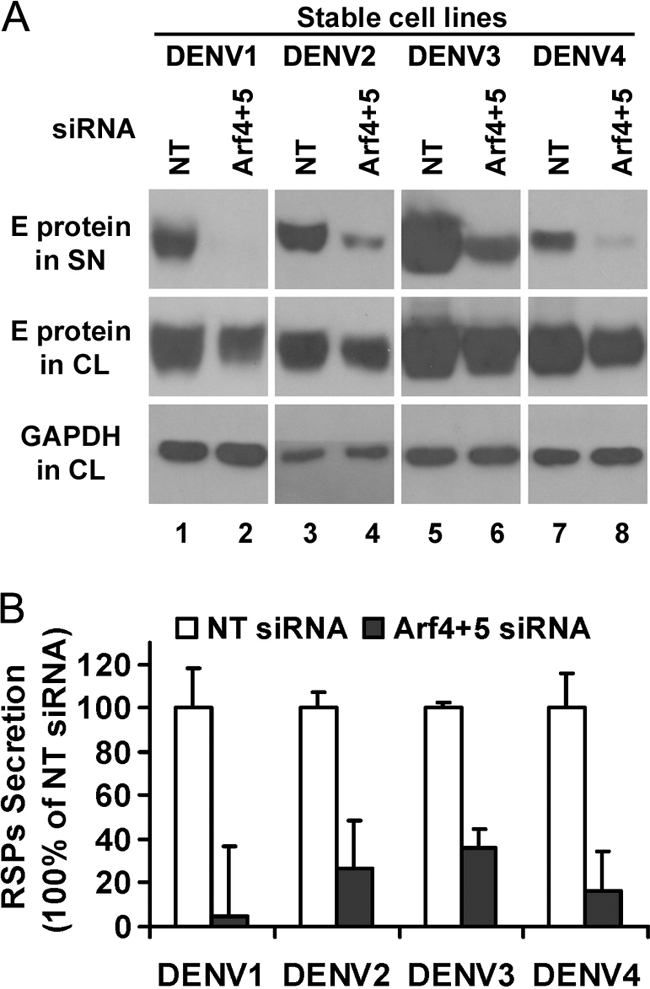
Combined knockdown of Arf4 and Arf5 decreases the release of the four dengue serotype RSPs. A, cell lysates (CL) and supernatants (SN) were prepared from HeLa-prME DENV1, DENV2, DENV3, and DENV4 cells transfected with either NT or siRNA targeting Arf4 + 5. Samples were analyzed by Western blot using the 4G2 monoclonal antibody that recognizes all four DENV serotypes (28). The results shown are representative of two independent experiments. B, mean density of each RSP band was measured using Photoshop, and the relative amount of RSPs was then calculated by normalizing it to that of NT siRNA. Results are shown as mean ± S.D. of triplicate measurements from one experiment representative of three others.
Silencing of Arf4 and Arf5 Does Not Alter Secretion of Proteins through the Constitutive Secretory Pathway
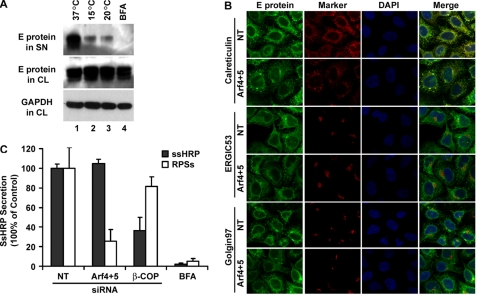
We have shown previously that dengue RSPs bud into the ER and transit through the Golgi apparatus for maturation before being released (12). The E protein is mainly localized to ER and partially to Golgi apparatus in HeLa-prME-DENV1 cells (12). To further confirm the use of this secretory route, we incubated HeLa-prME-DENV1 cells at 15 or 20 °C, which block cargo transport from ER to cis-Golgi and between cis- and trans-Golgi cisternae (31) or exit from the trans-Golgi, respectively (32). Brefeldin A (BFA), which inhibits the intracellular transport of secreted proteins (33), was used as control. Our results demonstrate that lower temperature treatment as well as BFA treatment significantly reduced RSP secretion into the supernatant (Fig. 4A, lanes 2–4, upper panel), indicating that RSPs followed the constitutive secretory pathway. We then investigated whether any accumulation of viral proteins could be observed along the secretory pathway when the secretion of RSPs was blocked by down-regulation of Arf4+5. We performed confocal microscopy analysis on HeLa-prME-DENV1 cells stained with the monoclonal anti-E 4E11 antibody in combination with antibodies targeting markers of the secretory pathway (calreticulin for ER, ERGIC53 for ER-Golgi intermediary compartment, and Golgin97 for Golgi apparatus). After treatment with siRNAs for class II Arfs, E protein was mainly localized within the ER where it co-localized with the calreticulin resident protein (Fig. 4B, upper panel). No co-localization of E with ERGIC53 or Golgin97 was observed (Fig. 4B, middle and lower panels), suggesting that RSPs do not accumulate in post-ER compartments. Thus, we hypothesized that the RSPs could remain mainly trapped inside the ER after knockdown of class II Arfs.
FIGURE 4.
Silencing of class II Arfs does not alter other secretion through the constitutive pathway. A, HeLa-prME-DENV1 cells were incubated at the indicated temperatures or in the presence of BFA for 8 h. RSP secretion was assessed by Western blot, visualizing dengue E protein expression in supernatant (SN) and cell lysate (CL) with the 4E11 monoclonal antibody. B, HeLa-prME-DENV1 cells treated with either NT or class II Arfs siRNAs were fixed, permeabilized, and stained for E protein and the indicated cellular markers. Calreticulin, ERGIC53, and Golgin97 were used to label ER, ER-Golgi intermediate compartment (ERGIC), and Golgi apparatus, respectively. Cell nuclei were stained with DAPI. C, HeLa cells were co-transfected with a plasmid coding for secretive horseradish peroxidase (ss-HRP) and with the specified siRNAs. HeLa-prME-DENV1 cells were transfected with specified siRNAs. BFA was used as the positive control. Secretion of ss-HRP was assessed by measuring horseradish peroxidase activity in the supernatant. Data are presented as the percentage of ss-HRP secretion relative to NT. Results are shown as mean ± S.D. of triplicate measurements from one experiment representative of three others.
To test whether the effect of class II Arfs on DENV1 RSP secretion was part of a general mechanism that would interfere with the secretion of protein through the constitutive pathway, we analyzed the secretion of the horseradish peroxidase fused to a signal sequence (ss-HRP). On synthesis, ss-HRP is translocated into the ER where after cleavage of the signal sequence, the soluble HRP is transported along the secretory pathway. The release of HRP into the medium is quantified by its enzymatic activity as described previously (34). HeLa cells were transiently transfected with ss-HRP in combination with various treatments, including siRNA targeting the β-COP subunit of the COP I coatomer, which interfere with the constitutive secretory pathway (34–36). HeLa-prME-DENV1 cells were also treated with the same siRNAs. BFA treatment was used as control. Two days post-transfection, peroxidase activity or RSPs in cell supernatants were measured by chemiluminescence or Western blot, respectively, and then normalized to those of control cells transfected with NT siRNA. No difference was observed in ss-HRP secretion after Arf4+5 down-regulation when compared with NT controls, whereas Arf4+5 siRNA reduced RSP secretion by 80% (Fig. 4C). BFA treatment induced a clear reduction of both ss-HRP and RSP secretion. Treatment with β-COP siRNA led to a drastic 60% reduction of secreted ss-HRP into the supernatant, whereas it only slightly affected RSP secretion by 20% (Fig. 4C). These results demonstrate that class II Arf proteins are not required for the generic constitutive secretory pathway and that they play a specific role in the secretion of dengue RSPs.
Dengue prM Interacts with Class II Arfs in HeLa-prME Cells
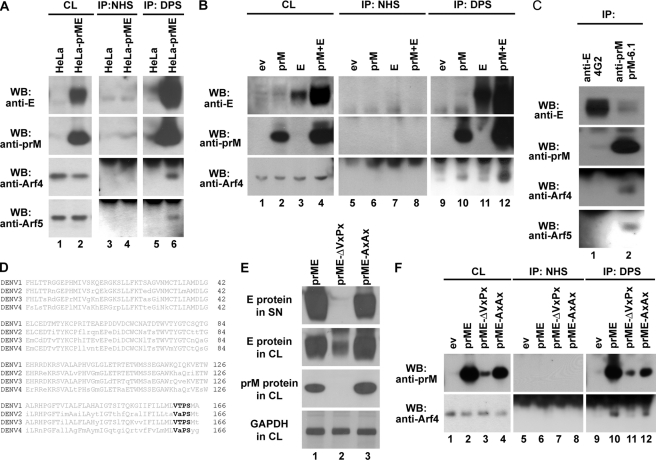
To establish whether DENV glycoproteins could interact with class II Arfs in mammalian cells, co-immunoprecipitation experiments were performed. Lysates from HeLa and HeLa-prME-DENV1 cells were incubated with either normal human serum or dengue patient serum (DPS), obtained from a patient infected by DENV1, and subjected to immunoprecipitation. The presence of DENV E, prM, Arf4, and Arf5 in immunoprecipitates was analyzed by Western blot. Anti-DENV1 serum specifically pulled down Arf4 and Arf5 proteins (Fig. 5A, lower two panels, lane 6) from HeLa-prME-DENV1 but not parental HeLa cells. Our results indicate an interaction between class II Arfs and DENV glycoproteins.
FIGURE 5.
Interaction of dengue prM with human class II Arf. A, lysates of HeLa and HeLa-prME-DENV1 cells were immunoprecipitated (IP) with normal human sera (NHS, lanes 3 and 4) or DPS (lanes 5 and 6). E, prM, Arf4, and Arf5 expression (lanes 1 and 2) was tested in all cell lysates (CL) as input control. Immune complexes were detected by Western blot (WB) using anti-prME, anti-Arf4, or anti-Arf5 antibody. The bands on the top of the Western blot results using anti-Arf4 or anti-Arf5 antibodies are light chains of IgG used for immunoprecipitation and can also be observed in B, C, and F. B, 293T cells were transfected with prM and E, either individually or in combination, and cell lysates were immunoprecipitated with normal human sera (lanes 5–8) or DPS (lanes 9–12). Empty vector (ev) served as the control. Arf4 expression was tested in all cell lysates as input control (lanes 1–4). Immune complexes were detected by Western blot using anti-prME or anti-Arf4 antibody. C, lysates of HeLa-prME-DENV1 cells were immunoprecipitated with anti-E monoclonal antibody 4G2 (lane 1) or anti-prM monoclonal antibody prM-6.1 (lane 2). Immune complexes were detected by Western blot using 4E11 conjugated with HRP, anti-prME, anti-Arf4, or anti-Arf5 antibody. D, alignment of prM of four DENV serotypes, with the consensus sequence below, revealed a VXPX motif in the C terminus, shown in boldface and highlighted. E, 293T cells were transfected with prME, prME-ΔVXPX, or prME-AXAX. Glycoprotein prM or E protein in both supernatant and cell lysates were analyzed by Western blot using mouse anti-prME IgG. GAPDH was used as control. F, 293T cells were transfected with prME, prME-ΔVXPX, prME-AXAX, or empty vector, and cell lysates were immunoprecipitated with normal human sera (lanes 5–8) or DPS (lanes 9–12). Arf4 expression was tested in all cell lysates as input control (lanes 1–4). Immune complexes were detected by Western blot analysis using anti-prME or anti-Arf4 antibody.
To test which glycoprotein, prM or E, could be involved in this interaction with class II Arfs, we first subcloned prM and E into two separate constructs and transfected them into 293T cells individually or in combination. Cell lysates were subjected to immunoprecipitation using the DPS anti-DENV1 serum, and co-immunoprecipitation of Arf4 was analyzed by Western blotting. A clear band corresponding to Arf4 was detected in cells transfected with prM and E in combination (Fig. 5B, lane 12, lower panel), and a weak band was also observed in cells expressing prM (Fig. 5B, lane 10, lower panel). We noted that E and prM were expressed at much lower levels when these proteins were expressed individually than in combination (Fig. 5B, lanes 2–4, upper and middle panels). To exclude the possibility that the failure to precipitate Arf4 in cells expressing E individually may have resulted from the low expression level of E, we used two monoclonal antibodies, prM-6.1 and 4G2, which recognize prM and E, respectively (26, 28), for immunoprecipitation from HeLa-prME-DENV1 cell lysate. These two monoclonal antibodies efficiently pulled down their specific glycoproteins and a trace amount of others (Fig. 5C, upper two panels). We found that both class II Arfs were immunoprecipitated with anti-prM antibody but not with 4G2 (Fig. 5C, lower two panels), indicating that prM but not E protein interacts with class II Arfs.
To gain insight into the molecular mechanism underlying this interaction, we inspected prM sequence. It has been reported that a VXPX motif in the C-terminal portion of rhodopsin was recognized by Arf4 (24, 25). We found a similar motif at the C-terminal end of prM, viz. V161XP163X, which was conserved in all four serotypes used in our experiments (Fig. 5D). To test whether this sequence was important for interaction of DENV prM with class II Arfs, we produced mutated forms of prME with either a deletion of the VXPX motif (prME-ΔVXPX) or a substitution of valine 161 and proline 163 with alanines (prME-AXAX) and transfected them into 293T cells. No RSPs could be detected in supernatants of cells transfected with prME-ΔVXPX, whereas RSPs were detected after transfection with prME-AXAX (Fig. 5E, upper panel). The absence of RSPs in the supernatant of prME-ΔVXPX-expressing cells could be explained by the significant reduction of expression levels of prM and E in corresponding cell lysates (Fig. 5E, lane 2, middle two panels). This result indicates that the VXPX motif has an important role for expression and/or stability of the prM protein, but the Val-161 and Pro-163 were not critical residues. Cell lysates of cells transfected with prME-ΔVXPX and prME-AXAX were then subjected to immunoprecipitation with anti-DENV1 sera. We found that Arf4 could be pulled down from cells transfected with either prME of prME-AXAX (Fig. 5F, lower panel, lanes 10 and 12) but not from those expressing the deletion mutant (Fig. 5F, lower panel, lane 11), which also resulted in a much reduced amount of prM protein in the cell lysate (Fig. 5F, lane 3, upper panel). These results show that although the VXPX in the C-terminal portion of prM is important for the expression of prM, it is not the motif recognized by Arf4.
Dengue Viruses Are Sensitive to Class II Arf Depletion
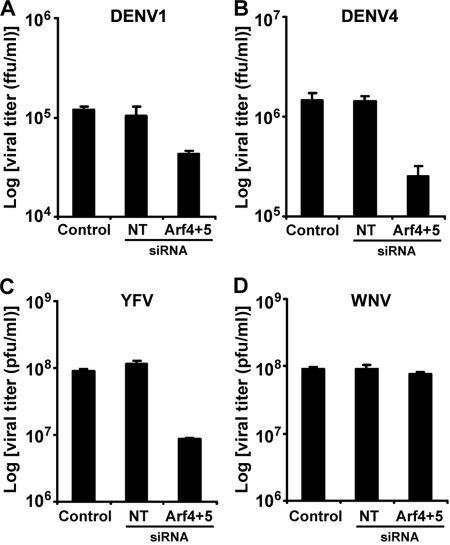
RSPs are not fully representative of the DENV viral life cycle as only the structural viral genes are expressed and not the viral replication machinery. To determine whether class II Arf proteins would affect the life cycle of fully replicative DENV, we infected human hepatic HepG2 cells with DENV1 (d1d FGA/NA strain) or DENV4 (63632/76 strain) viruses, which are the parent viral strains used to design DENV1 and DENV4 RSPs, respectively. HepG2 cells were used in these experiments because they are highly susceptible to infection with the 63632/76 DENV4 viral strain, as this virus was isolated from a patient who died from liver failure due to dengue infection. Cells were first transfected with the specified siRNAs, which did not cause any detectable cytotoxicity (data not shown). Two days post-transfection, HepG2 cells were infected with DENV1 or DENV4. An input of 20 pfu/cell, i.e. multiplicity of infection of 20 for DENV1 or of 1 for DENV4, was required to infect HepG2 cells at a significant level. Production of progeny viruses was evaluated by titration of culture supernatants collected on day 2 post-infection on AP61 cells. A significant albeit moderate reduction in virus progeny production was observed in infected HepG2 cells treated concomitantly with Arf4 and Arf5 siRNAs, when compared with control (≈2.5-fold for DENV1, Fig. 6A; ≈5-fold for DENV4, Fig. 6B). As expected, Arf5 was efficiently silenced in double knockdown cells, and expression levels of the cellular housekeeping protein GAPDH were similar for all conditions tested (data not shown). Taken together, our results extend the findings obtained with RSPs to the parental viruses and suggest that Arf4 and Arf5 are crucial host cell factors that play a role during dengue infection.
FIGURE 6.
Differential implication of class II Arf proteins in flavivirus secretion. The effect of Arf4+Arf5 down-regulation was evaluated on replication-competent viruses by measuring viral titers in the supernatants of infected cells 2 days post-infection using either focus immunodetection assay for DENV1 (A), DENV4 (B), and WNV (D) or plaque assay for yellow fever virus (C). Results are shown as mean ± S.D. of triplicate measurements from one experiment. Similar results were obtained in two other infections.
As HepG2 cells are permissive to hepatotropic YFV and neurotropic WNV, these cells were exposed to YFV strain Asibi and WNV strain IS-98-ST1 at an multiplicity of infection of 1. To address the role of class II Arfs in virus progeny production, HepG2 cells were treated during 48 h with specific siRNAs prior to YFV and WNV infection. Interestingly, siRNA treatment reduced YFV release by ≈10-fold (Fig. 6C), whereas inhibition of class II Arf proteins was ineffective in reducing WNV progeny production (Fig. 6D). These results suggest that Arf4 and Arf5 may play a role in YFV but not in WNV release from HepG2 cells. Altogether our data indicate that class II Arf proteins exhibit a differential involvement in the secretion of flaviviruses from infected cells.
DISCUSSION
Virus-host interactions during flavivirus secretion are poorly understood, and very few cellular factors involved in this process have been described so far (13, 14). We have used an original approach, based on the development of dengue RSPs, to identify two novel host cellular factors, Arf4 and Arf5, involved in DENV secretion. Our results demonstrate that simultaneous depletion of Arf4 and Arf5 blocks RSP secretion for all four dengue serotypes. Experiments with parental viruses used to construct the RSPs show that the life cycle of DENV is significantly affected by targeting class II Arfs. Interestingly, we also tried Arf4 and Arf5 depletion on cells infected with yellow fever flavivirus and found it partially blocks virus production as with DENV1 and -4 viruses, suggesting a role for Arf4 and Arf5 in the life cycle of hepatotropic flaviviruses. Moreover, our results show that class II Arfs are functionally redundant and can complement each other, as previously reported for some other members of the family (29). By using antibodies specific for prM and E, or by expressing prM and E individually in mammalian cells, we have found that it is the prM protein that interacts with class II Arfs. Dengue prM protein has previously been reported to help the correct folding of E protein in ER (37) and then protect it from fusing within the host cell before progeny viruses are released (38, 39). Here, we demonstrate another important role of prM during the secretion process, i.e. recruitment of class II Arfs, which are crucial factors for intracellular trafficking. Taken together, our data uncover a crucial role for a novel class of cellular factors during the late stages of the flavivirus life cycle.
DENV is thought to be released from infected cells through the constitutive secretory pathway (6, 12, 40). This is in agreement with our results with low temperature and BFA treatments. Interestingly, by using horseradish peroxidase protein fused to a signal peptide, a soluble protein described as a model for constitutive secretion (34), we find that this secretory pathway is not disrupted by down-regulation of class II Arfs. Therefore, the inhibition in RSP secretion through class II Arfs does not result from a general perturbation of the constitutive pathway but appears to inhibit DENV secretion in a specific manner. This result suggests that the secretion of DENV is a process more complex than the constitutive secretion.
Our study demonstrates that there is no overlapping role between class I and II Arfs during dengue RSP secretion. We find that treatment with Arf1 siRNA partially decreases the secretion of RSPs. Arf1 plays a key role in traffic through the Golgi apparatus where it is involved in vesicle formation (41), thus explaining the inhibitory effect of Arf1 siRNA on dengue RSP secretion. Targeting of Arf1 in combination with any member of class II Arfs, however, does not induce stronger inhibition when compared with Arf1 siRNA alone, indicating that class II Arfs and Arf1 participate in dengue RSP secretion at different steps. Previous studies have shown that both class I and II Arfs co-localized with the Golgi and ER-Golgi intermediate compartment, but only class II Arfs showed partial co-localization with ER (42, 43). The differential role of class I and II Arf proteins during DENV secretion is therefore in accordance with their differential cellular localization. Our results with immunostaining show that the RSPs do not accumulate in post-ER compartments after down-regulation of Arf4 and Arf5. Taken together, our results suggest that class II Arfs are required for the secretion of RSPs at the early pre-Golgi steps.
During the formation of trafficking vesicles, Arf proteins are recruited by cargo protein to cellular membranes where they trigger membrane curvature necessary for budding of trafficking vesicles, in which the cargo protein is incorporated (41, 44–46). For example, frog (Rana berlandieri) Arf4 has been shown to be recruited to membrane compartments by binding to the C-terminal VXPX motif of the photoreceptor rhodopsin and thus regulate its incorporation into specialized post-Golgi trafficking vesicles that target rhodopsin to the sensory cilia (24, 25). In mammalian cells, class II Arfs down-regulation has been reported to block retrograde transport from the Golgi apparatus to ER (29), suggesting their participation in the trafficking between these compartments. Vesicles containing dengue RSPs (12) or infectious virus particles (40) have been observed at the site of the Golgi apparatus, thus, one possible role for class II Arf in dengue release is that they are critical for the formation of RSPs trafficking vesicle from ER to Golgi. Besides Arf4, some other factors such as Rab11 are also required to form transport vesicles for rhodopsin (25). Intriguingly, our previous study demonstrated that the down-regulation of Rab11 could reduce the secretion of dengue RSPs (12). With the identification of class II Arfs, further studies are needed to establish whether they interact with Rab11 to promote the formation of transport vesicle containing DENV. In addition, recent studies have shown that, during DENV secretion, curved membranes have been observed at the assembly and budding sites of viral particles (40) and that class II but not class I Arfs were partially co-localized with the ER protein calreticulin (43). Because our results with co-immunoprecipitation demonstrate that prM proteins, which were mainly localized in the ER, can interact with Arf5, it is also tempting to speculate that class II Arfs are recruited to the ER and facilitate the membrane curvature necessary for inward bud formation, which is structurally similar to that occurring with cellular trafficking vesicles. Further studies are needed to precisely characterize the mechanism by which class II Arfs are involved.
A VXPX motif that mediates the interaction between rhodopsin and Arf4 was found conserved in the C terminus of the prM protein of all four DENV serotypes. Our results indicate that the presence of VXPX is very important for the efficient expression of prM and E. However, our results further show that the substitution of Val-161 and Pro-163 does not disrupt prM interaction with Arf4. Therefore, the VXPX motif in prM protein is not the sequence recognized by class II Arfs. The result is consistent with the fact that although VXPX is present in the C terminus of WNV prM (data not shown), WNV replication is resistant to class II Arf down-regulation. Indeed, the VXPX sequence in prM has a different localization from that of rhodopsin, which is exposed to the cytosol (24, 25), being adjacent to the signalase cleavage site and thus on the lumen side of ER (38). It is therefore unlikely that this portion of prM is accessible to Arf proteins that are recruited from the cytosol to the ER membranes (41). Further experiments are needed to reveal the mechanism for the interaction between prM and class II Arfs.
The results obtained with dengue RSPs have been confirmed with replication-competent flaviviruses, albeit to a lesser extent. Intriguingly, unlike DENV and YFV, WNV release was not altered by siRNA depletion of Arf4 and Arf5, suggesting a differential role for class II Arfs among flaviviruses. An RNA interference screen performed on WNV and DENV (48) revealed that these two flaviviruses have developed both overlapping and specific interaction strategies with intracellular host factors. Even if this study was designed only to identify cellular host factors associated with early stages of infection, it is likely that a number of virus-host interactions are also essential for successful virus secretion. Our results give an example of the unique molecular steps developed by WNV and DENV/YFV during late stages. Of note, although the phylogenetic distance between WNV and DENV is slightly shorter than that between YFV and DENV (49), the latter two are more similar in the severe human symptoms they cause. Thus, YFV and DENV are mainly responsible for hemorrhagic fevers, whereas WNV causes encephalitis (4). The characterization and comparison of host genes such as class II Arfs involved during flavivirus infection remain important questions to be further addressed and could help to elucidate the diversity observed in pathogenesis.
We note that the effects of siRNAs targeting both Arf4 and Arf5 are not as efficient in the virus infection experiments as opposed to the RSPs. One possible reason may be the lower expression level of prME in DENV-infected cells than that in HeLa-prME cells, in which a codon-optimized prME gene is continually expressed. Codon optimization enhances the expression of the prME gene in mammalian cells without changing the amino acid of the prME protein (12). As more particles form in HeLa-prME cells, their secretion may need more Arf4/5 proteins, which makes HeLa-prME cells a sensitive tool to identify cellular factors required for DENV secretion. In our experiment, although the siRNA reduced almost 90% Arf4/5 protein, there were detectable levels of Arf4/5 proteins in host cells. The remaining Arf4/5 proteins were not sufficient to support the secretion of RSPs but might still be able to partially support the secretion of replicative dengue virus because their prME level is lower and does not require high levels of Arf4/5 proteins. In the future, we will design some approaches other than siRNA to test this hypothesis.
The function of class II Arfs has so far remained elusive. Our results reveal a novel function for these proteins, the less studied and known members of the Arf family. As there are no antiviral therapies approved for use against flaviviruses, a possible strategy is to consider the new characterized host cell components as potential therapeutic targets for drug development (47).
Acknowledgments
We thank Dr. Vivek Malhorta (Center for Genomic Regulation, Barcelona, Spain) for the gift of soluble horseradish peroxidase construct, Dr. Philippe Buchy (Institut Pasteur du Cambodge, Kingdom of Cambodia) for kindly providing us with sera from patients infected by dengue and mouse anti-prME sera, and Dr. Sittisombut Nopporn (Chiang Mai University, Chiang Mai, Thailand) for kindly providing us anti-prM monoclonal antibody prM-6.1.
This work was supported by Research Fund for Control of Infectious Diseases of Hong Kong Grants RFCID 08070952 and RFCID 10091312 and by a donation from BNP Paribas Corporate and Investment Banking.
- DENV
- dengue virus
- YFV
- yellow fever virus
- RSP
- recombinant subviral particle
- NT
- nontargeting
- WNV
- West Nile virus
- DPS
- dengue patient serum
- ER
- endoplasmic reticulum
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- E
- envelope
- TRITC
- tetramethylrhodamine isothiocyanate
- BFA
- brefeldin A.
REFERENCES
- 1. Gubler D. J. (2002) Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 10, 100–103 [DOI] [PubMed] [Google Scholar]
- 2. Halstead S. B. (1988) Pathogenesis of dengue: challenges to molecular biology. Science 239, 476–481 [DOI] [PubMed] [Google Scholar]
- 3. Halstead S. B. (2007) Dengue. Lancet 370, 1644–1652 [DOI] [PubMed] [Google Scholar]
- 4. Lindenbach B. D., Rice C. M. (2001) in Fields Virology (Knipe D. M., Howley P. M., eds) 4th Ed., pp. 991–1041, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 5. Stadler K., Allison S. L., Schalich J., Heinz F. X. (1997) Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71, 8475–8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li L., Lok S. M., Yu I. M., Zhang Y., Kuhn R. J., Chen J., Rossmann M. G. (2008) The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science 319, 1830–1834 [DOI] [PubMed] [Google Scholar]
- 7. Yu I. M., Zhang W., Holdaway H. A., Li L., Kostyuchenko V. A., Chipman P. R., Kuhn R. J., Rossmann M. G., Chen J. (2008) Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319, 1834–1837 [DOI] [PubMed] [Google Scholar]
- 8. Mukhopadhyay S., Kuhn R. J., Rossmann M. G. (2005) A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3, 13–22 [DOI] [PubMed] [Google Scholar]
- 9. Allison S. L., Stadler K., Mandl C. W., Kunz C., Heinz F. X. (1995) Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J. Virol. 69, 5816–5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferlenghi I., Clarke M., Ruttan T., Allison S. L., Schalich J., Heinz F. X., Harrison S. C., Rey F. A., Fuller S. D. (2001) Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7, 593–602 [DOI] [PubMed] [Google Scholar]
- 11. Hunt A. R., Cropp C. B., Chang G. J. (2001) A recombinant particulate antigen of Japanese encephalitis virus produced in stably transformed cells is an effective noninfectious antigen and subunit immunogen. J. Virol. Methods 97, 133–149 [DOI] [PubMed] [Google Scholar]
- 12. Wang P. G., Kudelko M., Lo J., Siu L. Y., Kwok K. T., Sachse M., Nicholls J. M., Bruzzone R., Altmeyer R. M., Nal B. (2009) Efficient assembly and secretion of recombinant subviral particles of the four dengue serotypes using native prM and E proteins. PLoS One 4, e8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chu J. J., Yang P. L. (2007) c-Src protein kinase inhibitors block assembly and maturation of dengue virus. Proc. Natl. Acad. Sci. U.S.A. 104, 3520–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hirsch A. J., Medigeshi G. R., Meyers H. L., DeFilippis V., Früh K., Briese T., Lipkin W. I., Nelson J. A. (2005) The Src family kinase c-Yes is required for maturation of West Nile virus particles. J. Virol. 79, 11943–11951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gillingham A. K., Munro S. (2007) The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 23, 579–611 [DOI] [PubMed] [Google Scholar]
- 16. Donaldson J. G., Cassel D., Kahn R. A., Klausner R. D. (1992) ADP-ribosylation factor, a small GTP-binding protein, is required for binding of the coatomer protein β-COP to Golgi membranes. Proc. Natl. Acad. Sci. U.S.A. 89, 6408–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stamnes M. A., Rothman J. E. (1993) The binding of AP-1 clathrin adaptor particles to Golgi membranes requires ADP-ribosylation factor, a small GTP-binding protein. Cell 73, 999–1005 [DOI] [PubMed] [Google Scholar]
- 18. Beck R., Sun Z., Adolf F., Rutz C., Bassler J., Wild K., Sinning I., Hurt E., Brügger B., Béthune J., Wieland F. (2008) Membrane curvature induced by Arf1-GTP is essential for vesicle formation. Proc. Natl. Acad. Sci. U.S.A. 105, 11731–11736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brown H. A., Gutowski S., Moomaw C. R., Slaughter C., Sternweis P. C. (1993) ADP-ribosylation factor, a small GTP-dependent regulatory protein, stimulates phospholipase D activity. Cell 75, 1137–1144 [DOI] [PubMed] [Google Scholar]
- 20. Cockcroft S., Thomas G. M., Fensome A., Geny B., Cunningham E., Gout I., Hiles I., Totty N. F., Truong O., Hsuan J. J. (1994) Phospholipase D: a downstream effector of ARF in granulocytes. Science 263, 523–526 [DOI] [PubMed] [Google Scholar]
- 21. Kahn R. A., Cherfils J., Elias M., Lovering R. C., Munro S., Schurmann A. (2006) Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J. Cell Biol. 172, 645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bonifacino J. S., Glick B. S. (2004) The mechanisms of vesicle budding and fusion. Cell 116, 153–166 [DOI] [PubMed] [Google Scholar]
- 23. Donaldson J. G. (2003) Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J. Biol. Chem. 278, 41573–41576 [DOI] [PubMed] [Google Scholar]
- 24. Deretic D., Williams A. H., Ransom N., Morel V., Hargrave P. A., Arendt A. (2005) Rhodopsin C terminus, the site of mutations causing retinal disease, regulates trafficking by binding to ADP-ribosylation factor 4 (ARF4). Proc. Natl. Acad. Sci. U.S.A. 102, 3301–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mazelova J., Astuto-Gribble L., Inoue H., Tam B. M., Schonteich E., Prekeris R., Moritz O. L., Randazzo P. A., Deretic D. (2009) Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. EMBO J. 28, 183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Junjhon J., Lausumpao M., Supasa S., Noisakran S., Songjaeng A., Saraithong P., Chaichoun K., Utaipat U., Keelapang P., Kanjanahaluethai A., Puttikhunt C., Kasinrerk W., Malasit P., Sittisombut N. (2008) Differential modulation of prM cleavage, extracellular particle distribution, and virus infectivity by conserved residues at nonfurin consensus positions of the dengue virus pr-M junction. J. Virol. 82, 10776–10791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Desprès P., Frenkiel M. P., Deubel V. (1993) Differences between cell membrane fusion activities of two dengue type-1 isolates reflect modifications of viral structure. Virology 196, 209–219 [DOI] [PubMed] [Google Scholar]
- 28. Henchal E. A., Gentry M. K., McCown J. M., Brandt W. E. (1982) Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am. J. Trop. Med. Hyg. 31, 830–836 [DOI] [PubMed] [Google Scholar]
- 29. Volpicelli-Daley L. A., Li Y., Zhang C. J., Kahn R. A. (2005) Isoform-selective effects of the depletion of ADP-ribosylation factors 1–5 on membrane traffic. Mol. Biol. Cell 16, 4495–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Siu Y. L., Teoh K. T., Lo J., Chan C. M., Kien F., Escriou N., Tsao S. W., Nicholls J. M., Altmeyer R., Peiris J. S., Bruzzone R., Nal B. (2008) The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 82, 11318–11330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saraste J., Palade G. E., Farquhar M. G. (1986) Temperature-sensitive steps in the transport of secretory proteins through the Golgi complex in exocrine pancreatic cells. Proc. Natl. Acad. Sci. U.S.A. 83, 6425–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saraste J., Kuismanen E. (1984) Pre- and post-Golgi vacuoles operate in the transport of Semliki Forest virus membrane glycoproteins to the cell surface. Cell 38, 535–549 [DOI] [PubMed] [Google Scholar]
- 33. Misumi Y., Misumi Y., Miki K., Takatsuki A., Tamura G., Ikehara Y. (1986) Novel blockade by brefeldin A of intracellular transport of secretory proteins in cultured rat hepatocytes. J. Biol. Chem. 261, 11398–11403 [PubMed] [Google Scholar]
- 34. Bard F., Casano L., Mallabiabarrena A., Wallace E., Saito K., Kitayama H., Guizzunti G., Hu Y., Wendler F., Dasgupta R., Perrimon N., Malhotra V. (2006) Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature 439, 604–607 [DOI] [PubMed] [Google Scholar]
- 35. Pepperkok R., Scheel J., Horstmann H., Hauri H. P., Griffiths G., Kreis T. E. (1993) β-COP is essential for biosynthetic membrane transport from the endoplasmic reticulum to the Golgi complex in vivo. Cell 74, 71–82 [DOI] [PubMed] [Google Scholar]
- 36. Peter F., Plutner H., Zhu H., Kreis T. E., Balch W. E. (1993) β-COP is essential for transport of protein from the endoplasmic reticulum to the Golgi in vitro. J. Cell Biol. 122, 1155–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Courageot M. P., Frenkiel M. P., Dos Santos C. D., Deubel V., Desprès P. (2000) α-Glucosidase inhibitors reduce dengue virus production by affecting the initial steps of virion morphogenesis in the endoplasmic reticulum. J. Virol. 74, 564–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Y., Corver J., Chipman P. R., Zhang W., Pletnev S. V., Sedlak D., Baker T. S., Strauss J. H., Kuhn R. J., Rossmann M. G. (2003) Structures of immature flavivirus particles. EMBO J. 22, 2604–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu I. M., Holdaway H. A., Chipman P. R., Kuhn R. J., Rossmann M. G., Chen J. (2009) Association of the pr peptides with dengue virus at acidic pH blocks membrane fusion. J. Virol. 83, 12101–12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C. K., Walther P., Fuller S. D., Antony C., Krijnse-Locker J., Bartenschlager R. (2009) Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5, 365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. D'Souza-Schorey C., Chavrier P. (2006) ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 42. Chun J., Shapovalova Z., Dejgaard S. Y., Presley J. F., Melançon P. (2008) Characterization of class I and II ADP-ribosylation factors (Arfs) in live cells: GDP-bound class II Arfs associate with the ER-Golgi intermediate compartment independently of GBF1. Mol. Biol. Cell 19, 3488–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Duijsings D., Lanke K. H., van Dooren S. H., van Dommelen M. M., Wetzels R., de Mattia F., Wessels E., van Kuppeveld F. J. (2009) Differential membrane association properties and regulation of class I and class II Arfs. Traffic 10, 316–323 [DOI] [PubMed] [Google Scholar]
- 44. Kahn R. A., Volpicelli-Daley L., Bowzard B., Shrivastava-Ranjan P., Li Y., Zhou C., Cunningham L. (2005) Arf family GTPases: roles in membrane traffic and microtubule dynamics. Biochem. Soc. Trans. 33, 1269–1272 [DOI] [PubMed] [Google Scholar]
- 45. Nie Z., Hirsch D. S., Randazzo P. A. (2003) Arf and its many interactors. Curr. Opin. Cell Biol. 15, 396–404 [DOI] [PubMed] [Google Scholar]
- 46. Donaldson J. G., Jackson C. L. (2000) Regulators and effectors of the ARF GTPases. Curr. Opin. Cell Biol. 12, 475–482 [DOI] [PubMed] [Google Scholar]
- 47. Pastorino B., Nougairède A., Wurtz N., Gould E., de Lamballerie X. (2010) Role of host cell factors in flavivirus infection: Implications for pathogenesis and development of antiviral drugs. Antiviral Res. 87, 281–294 [DOI] [PubMed] [Google Scholar]
- 48. Krishnan M. N., Ng A., Sukumaran B., Gilfoy F. D., Uchil P. D., Sultana H., Brass A. L., Adametz R., Tsui M., Qian F., Montgomery R. R., Lev S., Mason P. W., Koski R. A., Elledge S. J., Xavier R. J., Agaisse H., Fikrig E. (2008) RNA interference screen for human genes associated with West Nile virus infection. Nature 455, 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kuno G., Chang G. J., Tsuchiya K. R., Karabatsos N., Cropp C. B. (1998) Phylogeny of the genus Flavivirus. J. Virol. 72, 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]