Background: Paramyxovirus infection is initiated by coordinated action of the receptor binding (HN) and fusion (F) proteins.
Results: HN-F interaction is tracked in real time before and after receptor engagement. Association occurs before receptor engagement. Receptor-engaged HN drives formation of fusion clusters.
Conclusion: HN site II regulates HN-HN and HN-F interaction.
Significance: Strength of HN-F interaction modulates infection in the natural host.
Keywords: Confocal Microscopy, Fluorescence, Infectious Diseases, Protein-Protein Interactions, RNA Viruses, Paramyxovirus
Abstract
Parainfluenza viruses enter host cells by fusing the viral and target cell membranes via concerted action of their two envelope glycoproteins: the hemagglutinin-neuraminidase (HN) and the fusion protein (F). Receptor-bound HN triggers F to undergo conformational changes that render it fusion-competent. To address the role of receptor engagement and to elucidate how HN and F interact during the fusion process, we used bimolecular fluorescence complementation to follow the dynamics of human parainfluenza virus type 3 (HPIV3) HN/F pairs in living cells. We show that HN and F associate before receptor engagement. HN drives the formation of HN-F clusters at the site of fusion, and alterations in HN-F interaction determine the fusogenicity of the glycoprotein pair. An interactive site, at the HN dimer interface modulates HN fusion activation property, which is critical for infection of the natural host. This first evidence for the sequence of initial events that lead to viral entry may indicate a new paradigm for understanding Paramyxovirus infection.
Introduction
Paramyxoviruses, including parainfluenza viruses, enter their target cells by binding to a receptor molecule and fusing their viral envelope with the cell membrane to reach the cytoplasm. Binding to the target cell for human parainfluenza virus type 3 (HPIV3)2 is mediated via interaction of the viral receptor binding molecule (hemagglutinin-neuraminidase (HN)) with sialic acid-containing receptor molecules on the cell surface. HN then activates the viral fusion protein (F) to mediate direct fusion of the viral envelope with the plasma membrane of the cell (1–4). F is present on the surface of the virus as a trimer. Its cleavage by host cell proteases during transit to the cell surface yields the fusion-competent processed F, which consists of two disulfide-linked subunits (F1+F2) (5). The F1 C terminus is membrane-anchored and contains the cytoplasmic tail preceded by the transmembrane domain. The N terminus contains the hydrophobic residues constituting the hydrophobic “fusion peptide” (see Ref. 6). After HN binds to a sialic acid-containing receptor to initiate the fusion process, it activates the F protein, which rearranges to expose the previously buried fusion peptide for insertion into the target membrane (1). Importantly, the nature and timing of the HN role in F-activation and the role of receptor engagement remain unknown. We explored these questions with the pediatric pathogen HPIV3 as a model.
For HPIV3 and other paramyxoviruses, HN is also responsible for receptor binding and neuraminidase (receptor-cleaving) activities. These three activities are tightly regulated within a tetrameric type II membrane protein consisting of a cytoplasmic domain, a membrane-spanning region, a stalk region, and a globular head. The stalk region confers specificity for the homologous F in the fusion activation process. The globular domain contains two active sites; the primary sialic acid binding/neuraminidase active site and, as we recently discovered in HPIV3 and others have shown in other paramyxoviruses (8–10), a second active site that contributes to receptor binding and to activation of F (7). Using a variant HPIV3 HN containing a single mutation at site II, we examined the role of the site in HN activation of F.
Current models of HN-F interaction during the fusion process posit that either (a) HN-F interaction occurs in the absence of a receptor, or (b) HN-F interaction occurs only upon receptor binding (3, 4, 8–12). We have proposed that HN and F interact during fusion activation and that the role of HN extends beyond the initial F-triggering event (13). Standard experimental approaches for studying HN-F interactions in paramyxoviruses (2, 9, 14), including co-precipitation and co-purification, require the removal of the proteins from their normal environment. Instead, we choose an alternative, direct approach, visualization of protein complexes in living cells and at physiologically relevant temperatures, which allows us to study these interactions in their normal environment in real time (15). Specifically, we used a modification of bimolecular fluorescence complementation (BiFC) to explore how the HN and F proteins cooperate in a highly specific way to mediate fusion upon receptor binding and how this interaction is regulated during the viral life cycle.
BiFC is based on the reconstitution of a fluorophore when fragments of fluorescent proteins fused to putative interacting proteins associate (16). A recent study using BiFC (17) was affected by the issue of tag interaction; in the specific experimental conditions used in that study the glycoproteins bearing the fluorescent tags necessary for this method were attracted to interact more strongly by the tags, making biologically relevant data difficult to obtain. We, therefore, designed experiments that would minimize the natural tendency of tagged proteins to interact. By using an improved version of yellow fluorescent protein (YFP; “Venus”) that has been shown to produce reversible fluorescence and to reconstitute the fluorophore at higher temperatures, we could perform our analysis in live cells at 37 °C (18). This is in contrast to the previous study on PIV5 (17) in which the fluorescent reconstitution was analyzed at 4 °C in fixed cells. Our experimental setting also includes synchronization of protein synthesis in the glycoprotein-expressing cells as well as negative interacting partners (e.g. influenza HA-HPIV3 HN or influenza HA-HPIV3 F) that show minimal background interaction.
A schematic diagram of the BiFC strategy that we used for assessing the interaction between HN and F is shown in panel A of Fig. 1. To understand the role of HN active site II and determine the relevance of these findings to pathogenesis, we used an HN singly mutated at site II (HN Gln-552). HN site II region is shown in Fig. 1E along the dimer interface (7) highlighted by the red and blue colors in the picture of the crystal structure of HPIV3 HN. Site I is occupied by the small molecule zanamivir (the monomer on the right projected toward the reader) (19). The virus bearing this HN is highly fusogenic in tissue culture monolayers; the HN has higher avidity than wt HN for receptor, is highly efficient at activating F, and when co-expressed with F is highly efficient at promoting fusion (7, 20). As we will describe below, we visualized in real time and quantified the mechanisms of interaction at two critical stages during viral entry. To support the BiFC findings, we confirmed our quantitative fluorescent data using classical biochemical approaches.
FIGURE 1.
Schematic diagram of the use of BiFC to study HN-F interaction; the constructs used in our strategy and their expression. A, putatively interacting proteins X (orange) and Y (purple) were fused to complementary fragments (N-Venus and C-Venus) of the YFP. Upon interaction of X and Y, the complementary YFP-Venus fragments associate to produce a fluorescent bimolecular complex of YFP-Venus (excitation 488 nm; emission 529 nm). The same excitation and emission is used for N-Venus and C-Venus; for N-Venus and C-CFP the excitation is 488 nm and emission is 513 nm. B, shown is a schematic diagram of the constructs: I, HPIV3 HN pCAGGS; II, HPIV3 HN N-Venus; III, HPIV3 HN C-Venus; IV, HPIV3 HN pE GFP; V, HPIV3 F pCAGGS; VI, HPIV3 F C-CFP; VII, HPIV3 F N-Venus; VIII, influenza HA N-Venus; IX, influenza HA C-Venus. HN (type II glycoprotein) fusion construct tags are N-terminal; F and HA (type I glycoproteins) fusion construct tags are C-terminal. C and D, flow cytometric analysis of HN and F expression at the cell surface using anti-HN or anti-F monoclonal antibodies, respectively, is shown. The number of HN or F proteins on the cell surfaces was determined per cell. These data are representative of results from three experiments ± S.E. E, shown is a schematic structure of the HPIV3 HN dimer. The ribbon diagram shows site I complexed with zanamivir. In this structure site I projects toward the viewer for the monomer on the right, and zanamivir is shown in stick and ball conformation. Site II is located along the dimer interface with the amino acids between residues 551 and 559, highlighted in red (monomer on the right) and blue (monomer on the left). The image was produced using PDB file 1V3E (19) and analyzed with Chimera software (53)).
EXPERIMENTAL PROCEDURES
Expression Constructs
For eukaryotic expression of HN in BiFC constructs, GFP was removed from the pEGFP-C3 vector (Clontech) using restriction enzymes NheI and SacI (New England Biolabs). The N terminus (amino acids 1–157) or C terminus (amino acids 158–238) of Venus was cloned into the C3 vector using NheI and SacI. HN was subcloned C-terminally to the respective fluorescent tag via a 23-amino acid linker using EcoRI and BamHI. For eukaryotic expression of HA or F constructs, N-terminal Venus, C-terminal Venus, or C-terminal CFP (amino acids 158–238) were cloned into pCAGGS using NheI and BglII. HA or F were subcloned in frame N-terminally to the fluorescent tag via an 18-amino acid linker using XhoI and NheI.
Cell Culture
293T (human kidney epithelial) cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum and antibiotics in 5% CO2. Transient transfections were performed using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions.
Cell Surface Expression Assay
Monolayers of 293T cells were transfected for transient expression of HN or F constructs. Cells were washed twice in phosphate-buffered saline (PBS) then incubated with an anti-HPIV3 HN monoclonal antibody (77/5) supplied by Judy Beeler or anti HPIV3 F monoclonal antibody (CD9–4-3; Chemicon) in 3% BSA, 0.1% sodium azide in PBS for 1 h. Samples were then washed twice in PBS and incubated with 1:100 of anti-mouse IgG (H+L) R-Phycoerythrin conjugate (Caltag Laboratories). To quantify cell surface proteins in each sample, indirect immunofluorescence was performed with fluorescence-activated cell sorting (FACS) (FACSCalibur; BD Biosciences).
BiFC Assay
293T cells were transiently transfected on biocoated Delta TPG dishes (Fisher) with the indicated cDNA combinations and 10% (red fluorescence protein (RFP)) using Lipofectamine 2000 according to the manufacturer's instructions. For non-competition experiments the transfection ratio of the N-Venus:C-Venus/CFP constructs was 2:1, whereas in competition experiments the transfection ratio was 1:1:1 for N-Venus:cold competitor in pCAGGS:C-Venus/CFP. After 4 h of incubation at 37 °C, the transfection mixture was replaced with complete medium (DMEM; 10% FBS, 1% penicillin-streptomycin) supplemented with 10 mm zanamivir. Fifteen hours later the medium was replaced with Opti-MEM supplemented with 10 mm zanamivir and 100 ng/ml cycloheximide (Sigma) for 1 h at 37 °C. Fluorescent images and the mean fluorometric ratio (calculated as the fluorescence intensity produced by BiFC divided by the RFP (15, 21)) were acquired using a confocal laser scanning microscope (Nikon TE-2000U Digital Eclipse C1si equipped with a spectral detector) using EZ-C1 acquisition and analysis software and a 60× NA 1.4 oil objective. Venus, hybrid fluorescent complex, and RFP were excited at 488 nm (emission 529 nm), 488 nm (emission 513 nm), and 561 nm (emission 610 nm), respectively. Confocal images were collected in 0.1-μm intervals from the top to the base of the cell at a resolution of 512×512. Confocal images were collected for the time-lapse movie with a resolution of 265×265 at room temperature or at 37° every 60 s for 90 min. The same laser power and gain settings were used for all samples and for all replicate experiments.
Western Blot Analysis
After BiFC analysis, cells were lysed in n-dodecyl-β-maltoside (DDM) lysis buffer (20 mm HEPES, 300 mm NaCl, 0.8% DDM) containing protease inhibitors. The samples were then resolved by 4–20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a polyvinyl difluoride membrane by electroblotting. The membranes were immunoblotted with polyclonal peroxidase-conjugated anti-GFP antibodies (Abcam) in 3% bovine serum albumin in PBS (1:1000 dilution). Tetramethylbenzidine (Promega) was added as described previously (22), and the images from the stained membranes were acquired using a Kodak 2000 multimodal image station and analyzed using Kodak molecular imaging software, Version 4.0.
β-Galactosidase Complementation-based Fusion Assay
This assay was performed as previously described (23) using the various BiFC and untagged constructs.
Radiolabeling
293T cells were transiently transfected using Lipofectamine 2000 according to the manufacturer's instructions with the untagged HN cDNAs as indicated. The cells were incubated overnight in 30 milliunits of exogenous neuraminidase (Sigma). The cells were starved for 2 h using DMEM without l-methionine or l-cystine (Invitrogen) with 30 milliunits of neuraminidase. The cells were washed then incubated in medium containing 55 μCi of 35S-labeled methionine/cystine (PerkinElmer Life Sciences), 30 milliunits of neuraminidase for 2 h. Cycloheximide (100 ng/ml) was added to each well for 1 h. Cells were then lysed and immunoprecipitated as described below.
Immunoprecipitation (IP) and Capture of Native HN Complexes
For HN oligomerization experiments, cell lysis, and IP were done using the Pierce Cross-link IP kit (ThermoScientific) and a custom-made cDNA polyclonal HN antibody as described in the manufacturer's instructions.
Non-reducing Gel Analysis
Immunoprecipitated samples were resuspended and boiled in 2× Novex Tris-glycine SDS sample buffer (Invitrogen), resolved by 4–20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred onto a polyvinyl difluoride membrane by electroblotting. Membranes were dried then exposed to a storage phosphor screen for 5 h. The screen was scanned with a Typhoon imager (GE Healthcare).
Native Gel Analysis
Immunoprecipitated samples were resuspended in 4× Native PAGE Sample Buffer (Invitrogen), resolved by 4–16% Native PAGE Novex Bis-Tris gels (Invitrogen), and transferred onto a polyvinyl difluoride membrane by electroblotting. Membranes were dried and exposed to a storage phosphor screen for 5 h. The screen was scanned with a Typhoon imager (GE Healthcare).
RESULTS
Bimolecular Fluorescence Complementation for Analysis of Paramyxovirus Surface Glycoprotein Interactions
To investigate the interactions between HN and F, we used BiFC. Previous reports using this method for herpesvirus were successful (24). However for PIV5, results using a similar system in fixed cells were discouraging (17) due to two main problems. First, the presence of the tag altered the properties of the viral glycoproteins, increasing fusion. Second, fluorescence was observed even in the presence of nonspecific viral glycoproteins that should not interact with PIV5 HN and/or F. Our experimental approach using live cells at 37 °C overcame some of these issues and enabled us to successfully determine the interactions between HN and F.
The schematic diagram of the BiFC strategy that we used for assessing the interaction between HN and F is shown in panel A of Fig. 1. Putatively interacting proteins X (orange) and Y (purple) are fused to complementary fragments (N-Venus and C-Venus) of the yellow fluorescent protein (YFP Venus variant). Upon interaction of X and Y, the complementary Venus fragments associate to reconstitute fluorescence. The constructs used in the experiments are shown in panel B. In panel C the results of flow cytometric analysis of surface expression levels of the HN glycoprotein are shown, revealing that the presence of the tag alters surface expression, however within the range of 30–140% of untagged wt HN, and those for each tagged HN, wt, and H552Q variant glycoproteins are expressed at the same level. Panel D shows that adding N-Venus to F reduces surface expression to ∼60% of untagged F, whereas adding C-terminal C-CFP to F does not significantly alter the F surface expression. Because expression of the HN N-Venus and the F C-CFP was closest to the untagged protein surface expression levels, we chose this pair for our HN-F BiFC studies, and we use the HN C-Venus and the F N-Venus only for HN-HN and F-F interaction experiments.
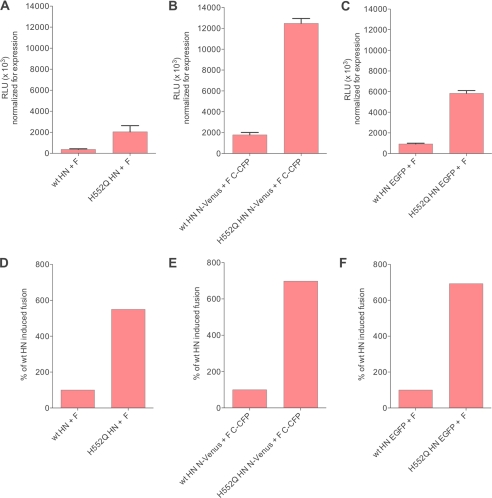
Fig. 2 (fusion in relative luminescence units, normalized for HN surface expression) shows that the tagged HN/F pairs produce four times more fusion than the untagged versions (Fig. 2, panel A compared with panels B and C). We observed an increase in the fusion mediated by tagged compared with untagged proteins; however, the relative fusion properties of our specific mutant glycoproteins were maintained in the tagged versions of the proteins (Fig. 2, panels D, E, and F). Moreover, the increase in fusion occurred also for HN molecules tagged with the entire enhanced green fluorescent protein (EGFP; Fig. 2, panels C and F), indicating that the increase in fusion was only in part due to interaction of complementary half-tags. It has been recently shown for Hendra F that the addition of an HA tag to the F increased fusion stimulation (25). Enhanced fusion by tagged receptor-binding proteins was previously observed for measles virus (26); the increase in fusion we observe is likely due to both a nonspecific effect of the tag and a specific increase in interaction. Because relative fusion properties of the mutant glycoproteins were maintained in the tagged proteins, we based our strategy on comparisons between expressed tagged proteins.
FIGURE 2.
Fusion is increased in the presence of the BiFC tags, but the relative fusion properties of wt and variant HNs are maintained. A–C, 293T cells were co-transfected with the indicated HN and F, and fusion was analyzed as described under “Experimental Procedures.” D–F, relative fusion between wt HN and H552Q HN is maintained irrespective of the tag. RLU, relative luminescence units; EGFP, enhanced green fluorescent protein.
In the series of experiments that follows using this set of expressed glycoproteins, cells were transfected with the HN and F molecules of interest and RFP. RFP was used for two reasons; (i) as reference for BiFC mean fluorometric ratio calculation (15) and (ii) for monitoring fusion. The cells were treated with zanamivir to block HN-receptor interaction when desired and treated with cycloheximide to block de novo protein synthesis. In experiments where we wanted to allow F-activation but block progression of fusion, we removed zanamivir and added specific fusion inhibitory peptides that permit F-triggering but inhibit the fusion that would otherwise follow triggering (27). Finally, in cases where we wished to allow F-activation and progression of fusion, we removed zanamivir and added medium without peptide. This series of manipulations allowed us to obtain a picture of the series of events that require HN-F interaction.
HPIV3 Viral Glycoprotein Homo-oligomerization Occurs before Receptor Engagement; Interactions of BiFC-labeled Proteins Are Specific
The crystal structure of PIV5 HN does not reveal significant changes in the HN structure when viewed alone or when complexed with receptor moiety (28). Although the dimer interface of NDV has been proposed to change in structure during the promotion of fusion (29), no significant conformational change at the dimer interface was identified for either PIV5 or HPIV3 HN in the presence of receptor moieties (19, 28). This finding has led to the hypothesis (4, 28) that changes in the oligomeric state of HN may mediate its ability to promote fusion. Although the existence of HPIV3 HN as a tetramer has been established, the role of oligomerization in HN fusion promotion activity is not understood (4). Newly published structural data for the NDV HN globular head with the stalk domain (30) suggests that in NDV HN an unstructured flexible linker is present between the structured helix stalk domain and the globular head and that the HN globular domain head is bent sideways. Based on these findings it was proposed that upon receptor engagement the HN elongates and that this transposition may be responsible for F-activation.
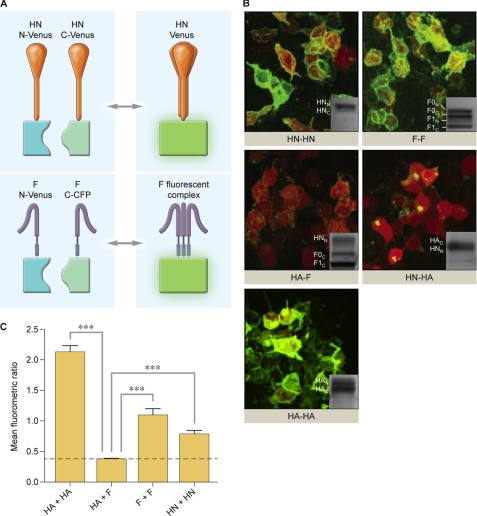
HPIV3 HN fusion proteins containing either the N-terminal half or the C-terminal half of Venus or CFP were co-expressed in cells to assess HN homo-oligomerization, and HPIV3 F fusion proteins containing F N-Venus and F C-CFP were co-expressed in cells to assess F homo-oligomerization. Panel A of Fig. 3 shows the strategy; if the two HNs or two Fs interact, the full fluorescent protein is reconstituted, and the molecule fluoresces. In these experiments zanamivir was used to completely block HN-receptor interaction (31) so that we can visualize interactions that occur before receptor engagement. The critical controls for this experiment were the following: assessments of interactions between influenza HA, expected to oligomerize upon co-expression of HA N-Venus and HA C-Venus; interactions between HPIV3 F and influenza HA, expected to fail to interact upon co-transfection of HA N-Venus and F C-CFP; interactions between HPIV3 HN and influenza HA, also expected to fail to interact upon co-transfection of HN N-Venus and HA C-Venus.
FIGURE 3.
Oligomerization of HPIV3 HN and F and control viral glycoproteins visualized by BiFC. A, shown is a schematic diagram of HN-HN and F-F oligomerization observed by BiFC. Upon interaction and oligomerization of monomers of HN or F, the halves of YFP-Venus complement to produce fluorescence. HN is tetrameric, and F is trimeric. B, 293T cells co-transfected with constructs encoding either HN N-Venus (HNN) and HN C-Venus (HNC), F N-Venus (FN) and F C-CFP (FC), HA N-Venus (HAN) and HA C-Venus (HAC), HA N-Venus and F C-CFP, or HN N-Venus and HA C-Venus. Receptor engagement is prevented by the addition of 10 mm zanamivir. These experiments were subjected to immunoblot analysis, shown as insets in each image, to confirm equal expression across all samples. Proteins are detected by goat anti-GFP HRP antibody. C, shown is the mean fluorometric ratio resulting from protein-protein interaction observed in panel B, measured in an average of at least three fields per experiment. The data are results from seven experiments, each with at least triplicate readings. Means with S.E. are reported. ***, p < 0.001 (one-way analysis of variance).
Fig. 3B, shows that the proteins anticipated to oligomerize, HN-HN, F-F, and HA-HA proteins, show intense fluorescence upon BiFC in the absence of receptor interaction, whereas proteins not expected to interact, HA-F and HN-HA, although bearing the same BiFC tags, show no fluorescent signal on the cell surface. Note that HN-HA shows clumps of fluorescence inside the cells but none evident on the cell surfaces, even more evident in supplemental Movie S3). Fig. 3C quantitates the experiment shown in Fig. 3B, providing the mean fluorometric ratio for each set of interacting proteins. Increases in fluorometric ratio indicate an increased protein-protein interaction. For the HN-HN, F-F, and HA-HA pairs shown in Fig. 3B, the fluorometric ratio is increased relative to the HA-F and HA-HN pairs. To ensure that the experiments were done on a basis of equal expression of all relevant proteins, the experiments shown in Fig. 3B were subjected to immunoblot analysis, shown as insets in each image. The corresponding movies of HA-HA, HA-F, and HA-HN (supplemental Movies S1, S2, and S3) show the cells in rotation so that the presence or absence of fluorescence is clearly visible.
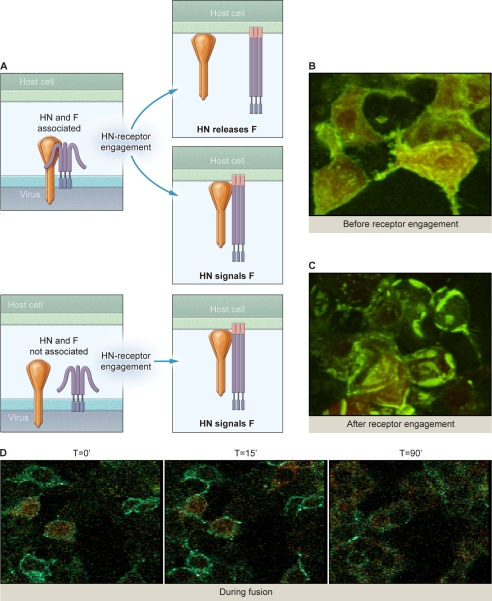
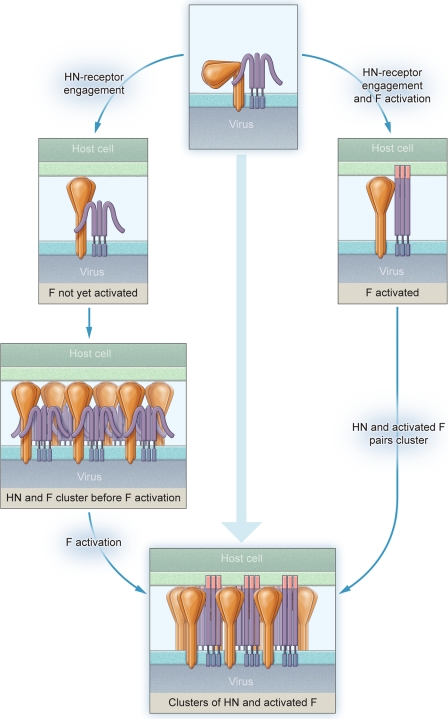
HN and F Interact on Membrane before Engaging Target Receptor and Aggregate into Clusters of HN-F Complexes upon Receptor Engagement
Current models for HN-F interaction during the process of fusion during viral entry are schematically diagrammed in Fig. 4A. If HN and F are associated before HN receptor engagement, then receptor binding may lead to separation of HN and F (4), freeing F to activate, or may induce HN to convey the activation signal to F during ongoing association. If HN and F are not associated before HN receptor engagement, then HN-F interaction would occur upon receptor binding (8), and HN may activate F via this interaction. To distinguish between these scenarios, we first assessed the interaction between HN N-Venus and F C-CFP in the absence of receptor interaction. Note that in contrast to the experiment shown in Fig. 3, where fluorescence indicates homo-oligomerization (HN-HN or F-F), in the experiment shown in Fig. 4B fluorescence indicates HN-F interaction. Only when HN and F associate is the fluorescent probe reconstituted. HN-receptor interaction is prevented by the use of zanamivir throughout the experiment; the cells co-expressing HN N-Venus and F C-CFP were treated overnight with zanamivir. One hour before observation the cells were treated with cycloheximide to block de novo protein synthesis (and thereby limit our observation to pre-made viral glycoproteins) and re-supplemented with zanamivir. Diffuse, evenly distributed fluorescence was visualized across the cell surfaces, indicating that HN and F are associated before receptor binding. The corresponding movie (supplemental Movie S4) shows the cells in rotation, where the diffuse fluorescence is clearly visible on the cell surfaces.
FIGURE 4.
HPIV3 HN associates with F on the cell surface before receptor interaction. A, current models of HN and F interaction are shown. B, 293T cells co-transfected with constructs encoding HN N-Venus and F C-CFP were treated overnight with 10 mm zanamivir, washed, then 1 h before analysis replaced with 10 mm zanamivir and cycloheximide. Evenly distributed fluorescence was visualized across the cell surface. C, 293T cells co-transfected with constructs encoding HN N-Venus and F C-CFP were treated overnight with 10 mm zanamivir, washed, then 1 h before analysis the zanamivir was removed to allow HN to engage receptor and replaced with cycloheximide and 10 μm HRC peptides to inhibit progression of fusion beyond the stage of fusion peptide insertion. BiFC clustering of HN/F complexes was evident. D, 293T cells co-transfected with constructs encoding HN N-Venus and F C-CFP were treated overnight with 10 mm zanamivir, washed, then 1 h before analysis treated with 10 mm zanamivir and cycloheximide. Fusion was recorded over 90 min.
We next examined the effect of receptor engagement on HN-F interaction. Fig. 4C shows strong interaction between HN and F in the presence of receptor interaction just before fusion (in cells expressing HN-N-Venus and F-C-CFP). In this experiment, after zanamivir was removed to allow HN-receptor interaction, HRC F-inhibitory peptide was present to prevent progress of fusion beyond F-triggering and fusion peptide insertion. Note that the bright clusters of HN-F complexes (BiFC) that form upon receptor interaction are “frozen in place” in the presence of HRC F-inhibitory peptide. This peptide only inhibits fusion after F has been triggered; F has been activated in these HN-F complexes. These clusters are in marked distinction to the evenly distributed fluorescence observed as a consequence of HN-F interacting pairs in the absence of receptor interaction shown in Fig. 4B. The HN-F interaction is no longer evenly distributed but tightly clustered at sites of cell-cell contact. The corresponding movie shows the cells in rotation so that the patches are clearly visible in regions where the membrane abuts an adjacent cell (supplemental Movie S5). The complexes do not dissociate unless fusion is allowed to proceed beyond this point.
We asked what drives the formation of the bright receptor-induced HN-F clusters. We found that although F expressed alone does not form clusters, HN expressed alone forms clusters upon receptor interaction (supplemental Fig. S1). Thus, upon HN-receptor engagement, HN-F clusters form specifically in the areas of HN-F interaction, driven by HN, bringing F to the site of fusion.
Membrane Fusion Is Initiated Once HN-F Complexes Have Clustered into Patches
From the experiments in Fig. 4, B and C, it was evident that HN and F are associated together in clusters after HN engages receptor. We asked whether the HN-F interaction dissociates as fusion progresses. Fig. 4D (representing three still photos from a movie, shown in supplemental Movie S6) shows HN-F interaction detected by BiFC in cells expressing HN-N-Venus and F-C-CFP and RFP to monitor fusion and shows the sequence of events over 90 min from prereceptor interaction through initial receptor interaction and proceeding to cell-cell fusion. Immediately upon removal of the zanamivir block to receptor interaction (t = 0 min) we detected diffuse green fluorescence, indicating diffuse HN-F interaction. In the second photograph (t = 15 min), after receptor interaction and the onset of fusion, we noted intense, clustered specks of fluorescence at the onset of fusion; these are more evident in the movie. In the third photograph (t = 90 min), after progression of fusion to completion, we noted the decrease in fluorescent intensity, suggesting dissociated HN-F interaction as fusion progresses. As seen in the corresponding movie, we tracked HN-F interaction during fusion, starting with the onset of receptor interaction (as in the first photograph, at the moment of zanamivir removal). HN-F interaction initially intensified; bright specks of fluorescence moved out along the leading edge of fusion then dissipated as fusion progressed. As a control, we performed the same experiment but tracked HN-GFP coexpressed with F (data not shown). In this case the GFP fluorescence redistributed during fusion, but the intensity remained constant, confirming that the decrease in BiFC fluorescence in Fig. 4 and in the corresponding movie were most likely due to HN-F dissociation. All intensities were quantitated by measuring the mean fluorometric ratio of BiFC/RFP (see supplemental Table S1). We thus had quantitative validation of our visual assessment of this process.
Hyper-triggering HN Mutant with Mutation in Site II Shows Enhanced Interaction with F
Having shown that HN site II is important for the HN property of F-activation, we tested the hypothesis that HN site II modulates HN-F interaction. For these experiments we used the HN of a virus that is highly efficient at activating F and at promoting fusion in cell monolayers (32). The HN mutation (HN H552Q) is in site II at the HN interface.
We compared the site II-mutated HN with wt HN for their abilities to mediate HN-F interaction under identical conditions in the absence of receptor engagement. We first assessed the interaction between HN N-Venus and F C-CFP, as shown in Fig. 5A, for both HNs (wt and H552Q) coexpressed with wt F in the absence of receptor interaction. In this experiment fluorescence would indicate HN-F interaction, as only when HN and F associate, is the fluorescent probe reconstituted. HN-receptor interaction was prevented by the use of zanamivir throughout the experiment, as shown in Fig. 4, and cycloheximide was used to limit our observation to matured viral glycoproteins. Diffuse, evenly distributed fluorescence is visualized across the cell surfaces showing wt HN-F interaction and HN H552Q-F interaction before receptor binding, with more intense fluorescence for the HN H552Q-F interaction. The immunoblot in the same panel confirms that the expression levels were similar for wt and mutant glycoproteins and for F in both samples. The mean fluorometric ratios (Fig. 5B) reveal the significantly greater HN-F interaction mediated by the site II mutated HN (H552Q) compared with wt HN.
FIGURE 5.
Hyper-triggering HN mutant with mutation in site II at HN dimer interface (H552Q) shows enhanced interaction with F. A, 293T cells co-transfected with constructs encoding HN wt N-Venus and F C-CFP or HN H552Q N-Venus and F C-CFP were treated overnight with 10 mm zanamivir, and 1 h before analysis fresh 10 mm zanamivir and cycloheximide were added. Evenly distributed fluorescence is visualized across the cell surfaces in both representative fields. The immunoblot shows the expression level of HN and F for HN wt N-Venus, F C-CFP and HN H552Q N-Venus, F C-CFP. Anti-tubulin (55 kDa) loading controls are below. B, shown is the mean fluorometric ratio resulting from HN-F interaction observed in panel A, measured in an average of at least three fields per experiment, from seven separate experiments. ****, p < 0.0001 (one-tailed t test).
We next decided to provide more quantitative validation of our findings, determining whether and to what degree adding an untagged version of HN would diminish fluorescence levels. Reduced fluorescence would indicate that binding did occur, as the competing untagged HN would bind at the expense of binding by BiFC-tagged HN (24, 33) (Fig. 6A). We cotransfected cells with tagged wt HN and F along with untagged site II mutant (H552Q) HN; if the interaction of H552Q HN with F is more avid, then untagged H552Q HN will interact with F, and fluorescence should decrease as a function of competitor. This is exactly what we observed. Fig. 6B shows cells cotransfected with tagged F and tagged wt or H552Q HN. The presence of untagged H552Q HN decreases the BiFC seen with wt HN and F, as seen by comparing the top two images, revealing that H552Q HN effectively competes with wt HN for interaction with F. We next cotransfected cells with tagged H552Q HN and F along with untagged wt HN; if the interaction of H552Q HN with F is more avid, then untagged wt HN should not displace H552Q HN from its interaction with F, and fluorescence should not decrease as a function of competitor. This is exactly what we observed: the third image shows cells cotransfected with tagged H552Q HN and F, and in the fourth image untagged wt HN has been cotransfected and does not decrease the BiFC seen with H552Q HN and F, indicating that the interaction of the site II mutated HN with F is not out-competed by wt HN (the photographs in supplemental Fig. S2A show higher magnification of the fields shown in Fig. 6B). To further evaluate whether HN-F interaction is site II-specific, we used an untagged HN with a mutation in site I that enhances receptor avidity (HN T193A). This site I mutant HN does not decrease BiFC in the presence of tagged H552Q HN and F, and it competed similarly to wt HN in the presence of tagged wt HN and F.
FIGURE 6.
Hyper-triggering HN mutant with mutation in site II at HN dimer interface (H552Q) out-competes wt HN for interaction with F. A, shown is a schematic of cold competition BiFC experiments using untagged HN to compete with tagged HN. B, 293T cells co-transfected with constructs encoding: HN wt N-Venus, F C-CFP, and pCAGGS, HN H552Q N-Venus, F C-CFP, and pCAGGS, HN wt N-Venus, F C-CFP and HN H552Q pCAGGS, or HN H552Q N-Venus, F C-CFP, and HN wt pCAGGS were treated overnight with 10 mm zanamivir, and 1 h before analysis fresh 10 mm zanamivir and cycloheximide were added. Representative images are provided. C and D, shown is the mean fluorometric ratio resulting from HN-F interaction observed in panel B, measured in an average of at least three fields per experiment, from seven separate experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (one-way analysis of variance).
We carried out several specificity controls to further validate our results. Our specificity control for interaction, shown in supplemental Fig. S2B), showed that tagged influenza HA and HPIV3 F are not brought together non-specifically by their BiFC tags. We also carried out a specificity control for the competitive binding experiments (shown in supplemental Fig. S2B). We cotransfected cells with tagged influenza HA to obtain homo-oligomerization (as in Fig. 3), then cotransfected untagged HPIV3 H552Q HN; the BiFC produced by HA homo-oligomerization is not non-specifically decreased by competition with untagged HPIV3 H552Q HN.
Our competitive binding experiments (Figs. 6, C and D) confirmed that site II-mutated HN out-competes wt HN for interaction with F. The efficient F-activation property of this mutated HN correlates with an increased physical interaction with F even before HN engages the receptor, suggesting that the HN site II modulates HN-F interaction upon receptor engagement.
Co-immunoprecipitation Experiments Confirm That BiFC Fluorescence Is Not Due to Nonspecific Interaction Driven by Tags
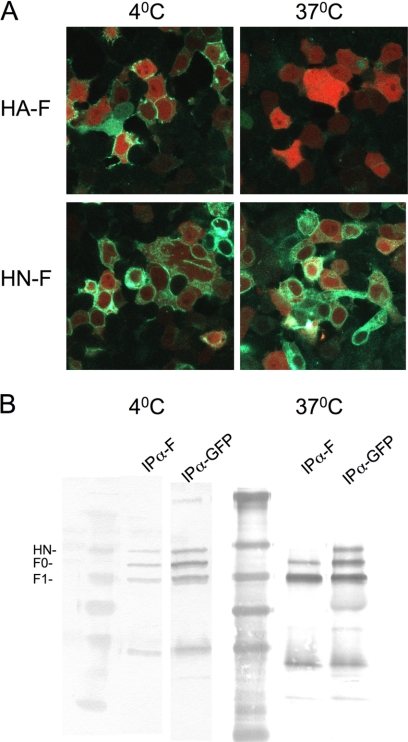
Paramyxovirus envelope glycoprotein interactions have been studied by coimmunoprecipitation of the fusion and the receptor-binding proteins. For Nipah, Hendra, measles, and NDV the coimmunoprecipitation technique has been successful (26, 34–39). In the case of PIV5, only the presence of complementary BiFC tags resulted in efficient co-immunoprecipitation of HN and F (17). The coimmunoprecipitation required stringent maintenance of a cold chain at 4 °C (17, 38, 40), not a physiologically relevant temperature for the function of the envelope glycoproteins. By using BiFC tags from superfolder fluorescent protein variants (41, 42), we investigated protein-protein interaction at 37 °C but with the trade-off of potential self-association. Self-association is amplified at lower temperatures (43). To assess nonspecific association due to the BiFC tags, we lysed cells co-expressing HN and F with the BiFC tags either at 4 °C (as previously described (17)) or at 37 °C and performed coimmunoprecipitation. To lyse the cells we used a previously described buffer (38). We postulated that at the low temperature the natural tendency of fluorophores to reconstitute would force co-immunoprecipitation of HN and F, whereas at 37 °C the tag effect should be minimal.
For the experiment shown in Fig. 7 cells were co-transfected with either HN-F or HA-F with the BiFC complementary tags as shown in Fig. 1. For Fig. 7A the cells were cycloheximide-treated for 1 h at either 23 or 37 °C and then incubated at either 4 or 37 °C for 1 h before image acquisition. For Fig. 7B the cells were cycloheximide-treated for 1 h at 37 °C, transferred to 4 °C or 37 °C, and lysed. The clarified cell lysates were immunoprecipitated overnight at 4 °C in the presence of anti-F antibodies or control anti-GFP antibodies. The captured complexes were washed and subjected to SDS-PAGE and Western blotted with anti-GFP antibodies. The images of cells expressing HA-F at 37 °C show minimal fluorescence. In striking contrast, the cells expressing HA-F incubated at 4 °C show extensive green fluorescence. This indicates that at the lower temperature, non-interacting partners can produce BiFC fluorescence as expected. The HN-F pairs, expected to interact, show fluorescence at both temperatures.
FIGURE 7.
Temperature dependence of BiFC and co-immunoprecipitation. 293T cells were co-transfected with constructs encoding either HN wt N-Venus and F C-CFP or HA N-Venus and F C-CFP and treated overnight with 10 mm zanamivir and before analysis fresh 10 mm zanamivir and cycloheximide were added. A, the cells were incubated for 1 h at either 23 °C or 37 °C to allow protein maturation and then transferred to either 4 or 37 °C. Green fluorescence indicates BiFC for HA-F at 4 °C, but not at 37 °C, and BiFC for HN-F at both temperatures. B, the cells were lysed at either 4 or 37 °C. The figure shows the Western blot after immunoprecipitation (IP) with either anti-F (to show coimmunoprecipitation of the BiFC partners) antibodies or anti-GFP antibodies (control for expression and immunoprecipitation).
The experiment shown in Fig. 7B reveals that using anti-F antibodies at 4 °C, F and HN co-immunoprecipitate. However, at 37 °C, no co-immunoprecipitation was detected. The lack of co-immunoprecipitation for the 37 °C samples shows that at the physiologically relevant temperature at which we performed our quantitative imaging, the BiFC tags did not form a stable structure. These data confirm that the fluorescence that we observed in the BiFC experiments results from HN-F interaction, which is likely too weak to withstand the lysing process at 37 °C but can be investigated with the BiFC method.
Hyper-triggering HN with Mutation in Site II at Dimer Interface Shows Enhanced Homo-oligomerization
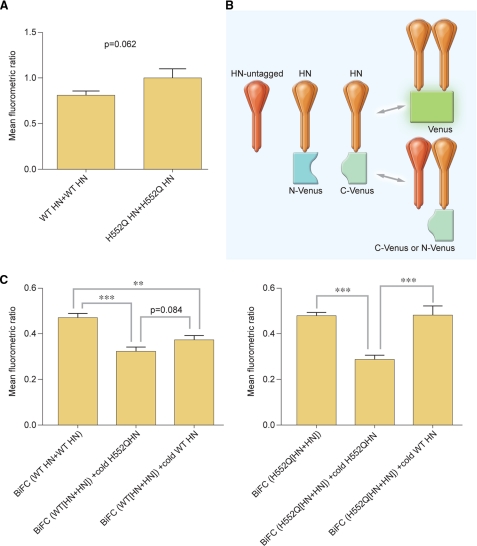
Because site II is at the dimer interface of HPIV3 HN, this site might regulate HN oligomerization, and oligomer alterations might modulate F-triggering. The variant HN molecule (H552Q) studied in Figs. 5 and 6, with a mutation at the dimer interface (F-triggering site), allowed us to test this hypothesis. We compared the HN homo-oligomerization of the H552Q HN and wt HN. Comparing homo-oligomerization by BiFC, we observed enhancement of homo-oligomerization for the mutated HN (shown in Fig. 8A), but the difference was not statistically significant. We then conducted additional competition experiments, diagrammed in Fig. 8B, examining evidence of homo-oligomerization in HN by comparing competitive interaction trials run on mutant HN versus wt HN. If the oligomerization of H552Q HN with wt HN was more avid, then fluorescence should decrease as a function of competitor. Cells were also cotransfected with tagged mutant H552Q HN pairs along with untagged wt HN; if the oligomerization of H552Q HN with wt HN was more avid, then fluorescence should not decrease as a function of wt HN competitor. These results are exactly what was observed. Controls included attempts to compete tagged mutant HN with untagged mutant HN and to compete tagged wt HN with untagged wt HN; all quantitative data are shown in Fig. 8C. Untagged H552Q HN successfully outcompeted wt HN for homo-oligomerization with tagged wt HN, decreasing BiFC fluorescence significantly more than untagged wt HN. Untagged wt HN did not outcompete H552Q HN for homo-oligomerization and failed to decrease the H552Q HN BiFC fluorescence compared with the decrease seen in the presence of untagged H552Q HN. Comparison of the HN homo-oligomerization of the H552Q HN and wt HN thus reveals that the site II mutant HN homo-oligomerizes more efficiently than wt HN. These results indicate that HN oligomer alterations modulate F-activation and fusion and that HN site II is critical for these properties.
FIGURE 8.
Hyper-triggering HN mutant with mutation in site II at dimer interface (H552Q) shows enhanced homo-oligomerization. A, shown are mean fluorometric ratios of BiFC resulting from oligomerization of HN WT-HN WT and HN H552Q-HN H552Q. Data are the means of three-four readings from triplicate experiments with S.E. B, shown is a schematic of untagged competition BiFC experiments using untagged HN to displace tagged HN in HN-HN oligomerization. C, 293T cells were co-transfected with HN WT N-Venus, HN WT C-Venus, and pCAGGS (BiFC (WT HN+WT HN)), HN WT N-Venus, HN WT C-Venus, and HN H552Q pCAGGS (BiFC (WT [HN+HN]) + cold HN H552Q), or HN WT N-Venus, HN WT C-Venus, and HN WT pCAGGS (BiFC (WT [HN+HN] + cold HN WT) or HN H552Q N-Venus, HN H552Q C-Venus, and pCAGGS (BiFC (H552Q[HN+HN])), HN WT N-Venus, HN WT C-Venus, and HN H552Q pCAGGS (BiFC (H552Q[HN+HN]) + cold HN H552Q), or HN H552Q N-Venus, HN H552Q C-Venus and HN WT pCAGGS (BiFC (H552Q[HN+HN]) + cold HN WT). Zanamivir (10 mm) was added to prevent HN-receptor interaction. One hour before analysis, fresh 10 mm zanamivir and cycloheximide were added. **p < 0.01, ***,p < 0.001 (one-way analysis of variance). Data are the means of three-four readings from triplicate experiments with S.E.
The BiFC fluorescence results of the HN-oligomerization could be due to the interaction of different ratios of the half-tagged monomer in each tetramer. Tetramers with 1:3, 2:2, or 3:1 ratios would all fluoresce. Although our competition experiments showed a decrease in fluorescence in the presence of untagged HN H552Q, it was not possible to distinguish whether the alteration was due to a decrease in either dimer and/or tetramer formation. Although transfection stoichiometry was used to solve similar issues for measles (44), we chose a direct approach, described in the next section, to determine whether the increased HN H552Q oligomerization was due to a change in the ratio of dimer or monomer states.
The Enhanced Oligomerization of the H552Q HN Is Due to Increased Dimer Formation
The H552Q mutation is at the dimer interface in the HN HPIV3 crystal structure (45), and we propose that it enhances the stability of the HN-HN dimer. For PIV5 and measles, the oligomeric state of the receptor-binding protein was revealed by blue native PAGE (44, 46). We applied this method to investigate the oligomeric state of the H552Q HN compared with the wt HN. We also took advantage of a mutation (S554C) in HPIV3 HN that leads to irreversible HN-HN interaction and provides a positive control for the dimer state (47). Cells were transfected with wt HN, S554C HN, H552Q HN, or the double mutant H552Q/S554C HN, radiolabeled for 3 h, and synchronized by adding cycloheximide. The cells were also treated with neuraminidase to observe HN oligomerization in the absence of receptor interaction. The cells were lysed and purified as described under “Experimental Procedures.” Fig. 9 shows the native (Fig. 9A) and non-reducing (Fig. 9B) migration pattern of the four different HN molecules. As expected, HN carrying the S554C mutation formed a stable dimer. In the native gel HN-H552Q is mainly in dimeric form compared with wt HN, indicating that, as expected, the HN-H552Q forms a more stable dimer. It is interesting to note that even for the mutant HN S554C, the addition of H552Q (double mutant) results in a higher dimer to monomer ratio than the single mutant. We speculate that the H552Q mutation stabilizes the dimer and permits more efficient disulfide linkage at 554C. H552 and S554 are located along the HN dimer interface (highlighted in red and blue in Fig. 1E).
FIGURE 9.
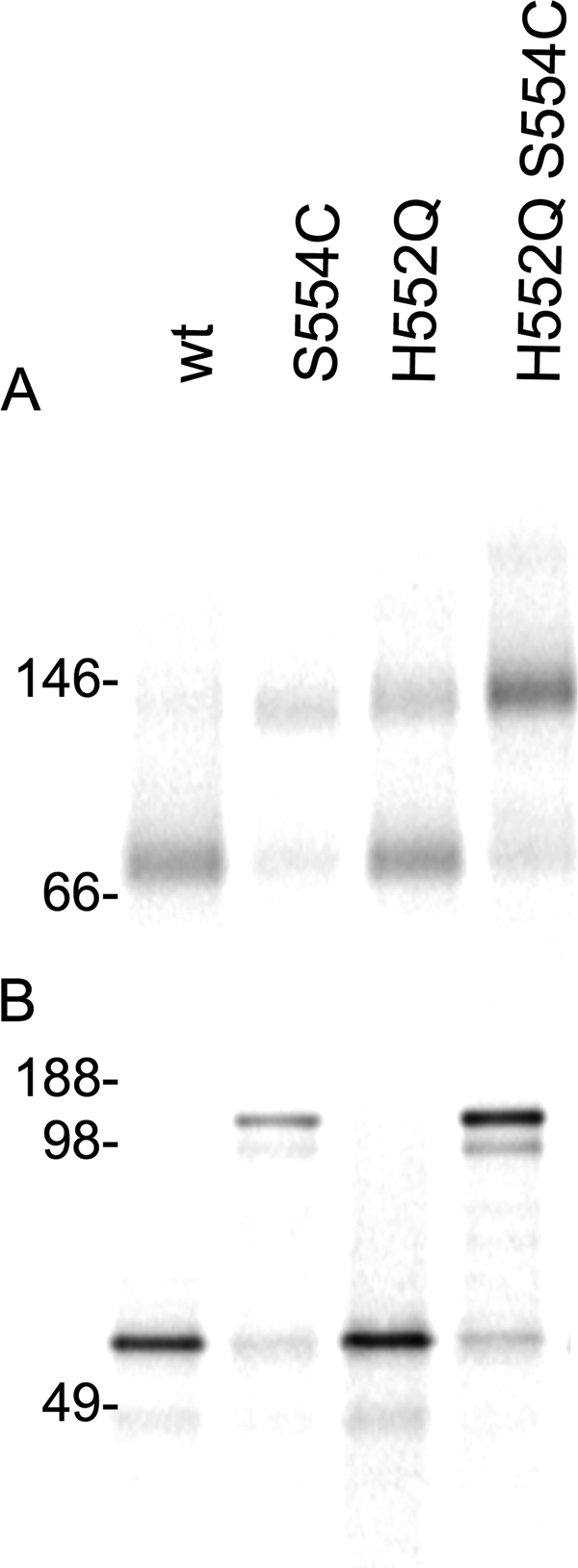
Hyper-triggering HN mutant with mutation in site II at dimer interface H552Q shows enhanced dimerization. A, shown is blue native gel autoradiography of the indicated untagged HNs. B, non-reducing SDS-PAGE autoradiography of the indicated HNs is shown.
DISCUSSION
Having directly observed and analyzed the sequence of events leading up to HN-F mediated membrane fusion, we propose a model for the role of receptor interaction. The first step in Paramyxovirus infection is binding of the receptor-binding protein (HN for HPIV) to cell surface receptors (sialic acid-containing molecules). Receptor engagement activates the viral fusion proteins (F) to their fusion-ready conformation, and F then inserts and fuses the viral envelope with the target cell membrane, allowing viral entry and infection. Using the strategies presented here, this series of events can now be clearly observed and experimentally manipulated for HPIV3. Receptor interaction is manipulated with zanamivir (7) to block HN-receptor interaction, HN-F interaction is monitored by the use of specific BiFC strategies, and progress of fusion after F-activation is controlled with the use of specific inhibitory peptides corresponding to the HRC domain of F that only inhibit fusion after F has been activated (9). Using this set of strategies, we examined the sequence of events leading up to HN-F-mediated membrane fusion. Taken together, our data allow us to propose a new model for HN-F interaction during fusion and the role of receptor interaction, shown in Fig. 10. HN and F are associated before receptor binding. Based on the structure of the NDV HN stalk domain that was recently solved (30), it is reasonable to propose that the HN globular domain lies sideways and that upon receptor engagement HN elongates and drives the formation of HN-F clusters at the site of fusion. F-triggering is accomplished within these clusters, whereas HN and F remain associated. The oligomeric status of HN impacts the HN-F association and triggering. As fusion progresses further, the fluorescence due to HN-F interaction decreases; this may be due to dissociation of the HN-F interaction or due to dispersion of the clusters of HN-F complexes. In future work we will aim to address whether the physical relationship between the stalk and head suggested by the recent NDV crystal structure is also observable in the native form of the HN on the viral membrane. We hope to address this question by imaging the viral fusion machinery using electron cryotomography. Electron cryotomography is uniquely positioned at the crossroad between high resolution structures and live cell imaging and is suited for integrating information from both directions (48–50).
FIGURE 10.
Model for Paramyxovirus surface glycoprotein interaction during fusion and the role of receptor interaction.
We have previously identified a key role for HN site II in F triggering and fusion promotion. Here we demonstrate that alterations at HN site II modulate HN-F interaction. The comparison in these experiments between wt HN and the HN site II mutant, HN-H552Q, allowed us to provide answers about whether and to what degree the site II mutation that confers an F-triggering advantage also confers an advantage in terms of interacting with F. If site II, upon receptor engagement, mediates HN-F interaction and clustering as we hypothesize, then the site II mutant should respond to a receptor stimulus by inducing HN-F complexing more efficiently than wt HN (i.e. gain of BiFC). We detected this difference in the presence of F-inhibitory peptides that inhibit progression of fusion (not shown). We thus provide further validation of a role for site II in fusion based on finding that the site II variant has enhanced HN-F interaction, which implicates this site in HN-F association and in F triggering.
We hypothesized that site II, positioned in the dimer interface of HPIV3 HN, may exert its influence on F-triggering by regulating HN homo-oligomerization. It has been suggested that high order oligomers are important for fusion promotion mediated by the receptor-binding protein of measles virus (H) (51). The data we present for HPIV3 support our hypothesis, as alterations in site II that affect oligomerization also modulate fusion promotion.
In a previous study using BiFC to investigate HN-F interaction (17), it was noted that fluorescently tagged glycoproteins interacted with each other more strongly and promoted more fusion than untagged glycoproteins, implicating the tags themselves in enhancing HN-F function. Here we find that although BiFC-tagged HN and F interact more strongly than their untagged counterparts, the functional properties of each molecule and their respective mutants are preserved in the tagged molecules. Thus, comparison of wt and variant BiFC-tagged molecules under the same conditions and with the appropriate controls produces biologically relevant data.
The communication between the two molecules that comprise the Paramyxovirus fusion machine is biologically relevant, with a clear impact on infection in the natural host. Alterations in site II at the dimer interface of HPIV3 HN influence growth, implicating site II in pathogenesis (32). The virus bearing the mutated H552Q HN binds its receptor more avidly, activates F more efficiently, and fuses in monolayer culture more extensively (7, 20), and we have now shown that the efficient F-activation property of this mutated HN correlates with an increased physical interaction with F. However, this virus is unfit in vivo (cotton rat model) and in human airway epithelium, a system that mimics the natural host (32), suggesting that an increase in F-activation efficiency and the increased physical interaction with F we show here is a disadvantage in the natural host and that the properties under study are physiologically relevant (32). This virus produces inactive viral particles in the natural host in which the F protein may have been triggered before contacting target cells (32). Taken together with the present results, these findings suggest that avid HN-F interaction may lead to premature and, therefore, ineffective activation of the fusion process (52). In contrast, the virus bearing the site I mutated HN (T193A) binds its receptor more avidly and on that basis alone fuses in monolayer culture more extensively but does not activate F more efficiently; this mutated HN has an interaction with F similar to that of wt HN. The T193A HN-bearing virus grows well in the natural host (32). The comparison between these two viruses, in which both bind receptor avidly but only the H552Q HN exhibits increased HN-F interaction, indicates that enhanced HN-F interaction is detrimental to infectivity in natural host.
Supplementary Material
Acknowledgments
We are grateful to Ashton Kutcher and Jonathan Ledecky for support without which the microscopy critical to this work would not have been possible, to Dan and Nancy Paduano for their essential support of innovative research projects, and to the Friedman Family Foundation for our laboratories at Weill Cornell Medical College. We thank Tim McGraw for helpful scientific discussions and advice, Eric M. Jurgens for help with statistical analysis, and Christine C. Yokoyama for critical reading of the manuscript. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health Grant P41 RR001081). We acknowledge the Northeast Center of Excellence for Bio-defense and Emerging Infections Disease Research Proteomics Core for peptide synthesis and purification.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI31971 (to A. M.), 3R01AI031971-19S1 (Research Supplement to Promote Diversity in Health-Related Research Program; to A. M.), and U54AI057158 (Northeast Center of Excellence for Biodefense and Emerging Infectious Disease Research (NIAID; to M. P. and A. M. (Principal Investigator of Center of Excellence grant to W. I. Lipkin)).

This article contains supplemental Movies 1–6, Figs. 1 and 2, and Table S1.
- HPIV3
- human parainfluenza virus type 3
- HN
- hemagglutinin-neuraminidase
- F
- fusion protein
- BiFC
- bimolecular fluorescence complementation
- RFP
- red fluorescent protein
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- CFP
- cyan fluorescent protein
- NDV
- Newcastle disease virus.
REFERENCES
- 1. Moscona A. (2005) Entry of parainfluenza virus into cells as a target for interrupting childhood respiratory disease. J. Clin. Invest. 115, 1688–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith E. C., Popa A., Chang A., Masante C., Dutch R. E. (2009) Viral entry mechanisms. The increasing diversity of Paramyxovirus entry. FEBS J. 276, 7217–7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee B., Ataman Z. A. (2011) Modes of Paramyxovirus fusion. A Henipavirus perspective. Trends Microbiol. 19, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lamb R. A., Paterson R. G., Jardetzky T. S. (2006) Paramyxovirus membrane fusion. Lessons from the F and HN atomic structures. Virology 344, 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scheid A., Choppin P. (1974) Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity of proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology 57, 475–490 [DOI] [PubMed] [Google Scholar]
- 6. Hernandez L. D., Hoffman L. R., Wolfsberg T. G., White J. M. (1996) Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12, 627–661 [DOI] [PubMed] [Google Scholar]
- 7. Porotto M., Fornabaio M., Kellogg G. E., Moscona A. (2007) A second receptor binding site on human parainfluenza virus type 3 hemagglutinin-neuraminidase contributes to activation of the fusion mechanism. J. Virol. 81, 3216–3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li J., Quinlan E., Mirza A., Iorio R. M. (2004) Mutated form of the Newcastle disease virus hemagglutinin-neuraminidase interacts with the homologous fusion protein despite deficiencies in both receptor recognition and fusion promotion. J. Virol. 78, 5299–5310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iorio R. M., Melanson V. R., Mahon P. J. (2009) Glycoprotein interactions in Paramyxovirus fusion. Future virol. 4, 335–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dutch R. E. (2010) Entry and fusion of emerging paramyxoviruses. PLoS Pathog. 6, e1000881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saphire E. O., Oldstone M. B. (2011) Measles virus fusion shifts into gear. Nat. Struct. Mol. Biol. 18, 115–116 [DOI] [PubMed] [Google Scholar]
- 12. Harrison S. C. (2008) Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Porotto M., Devito I., Palmer S. G., Jurgens E. M., Yee J. L., Yokoyama C. C., Pessi A., Moscona A. (2011) Spring-loaded model revisited: Paramyxovirus fusion requires engagement of a receptor binding protein beyond initial triggering of the fusion protein. J. Virol. 85, 12867–12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vigant F., Lee B. (2011) Hendra and nipah infection. Pathology, models, and potential therapies. Infect. Disord. Drug Targets 11, 315–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kerppola T. K. (2006) Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 7, 449–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu C. D., Kerppola T. K. (2003) Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21, 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Connolly S. A., Leser G. P., Jardetzky T. S., Lamb R. A. (2009) Bimolecular complementation of Paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion. Implications for the mechanism of fusion triggering. J. Virol. 83, 10857–10868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo Y., Rebecchi M., Scarlata S. (2005) Phospholipase Cβ2 binds to and inhibits phospholipase Cð1. J. Biol. Chem. 280, 1438–1447 [DOI] [PubMed] [Google Scholar]
- 19. Lawrence M. C., Borg N. A., Streltsov V. A., Pilling P. A., Epa V. C., Varghese J. N., McKimm-Breschkin J. L., Colman P. M. (2004) Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III. J. Mol. Biol. 335, 1343–1357 [DOI] [PubMed] [Google Scholar]
- 20. Moscona A., Peluso R. W. (1993) Relative affinity of the human parainfluenza virus type 3 hemagglutinin-neuraminidase for sialic acid correlates with virus-induced fusion activity. J. Virol. 67, 6463–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin H. P., Vincenz C., Eliceiri K. W., Kerppola T. K., Ogle B. M. (2010) Bimolecular fluorescence complementation analysis of eukaryotic fusion products. Biol. Cell 102, 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McKimm-Breschkin J. L. (1990) The use of tetramethylbenzidine for solid phase immunoassays. J. Immunol. Methods 135, 277–280 [DOI] [PubMed] [Google Scholar]
- 23. Palermo L. M., Porotto M., Greengard O., Moscona A. (2007) Fusion promotion by a Paramyxovirus hemagglutinin-neuraminidase protein. pH modulation of receptor avidity of binding sites I and II. J. Virol. 81, 9152–9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Atanasiu D., Whitbeck J. C., Cairns T. M., Reilly B., Cohen G. H., Eisenberg R. J. (2007) Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc. Natl. Acad. Sci. 104, 18718–18723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Popa A., Pager C. T., Dutch R. E. (2011) C-terminal tyrosine residues modulate the fusion activity of the Hendra virus fusion protein. Biochemistry 50, 945–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Plemper R. K., Hammond A. L., Gerlier D., Fielding A. K., Cattaneo R. (2002) Strength of envelope protein interaction modulates cytopathicity of measles virus. J. Virol. 76, 5051–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Porotto M., Rockx B., Yokoyama C. C., Talekar A., Devito I., Palermo L. M., Liu J., Cortese R., Lu M., Feldmann H., Pessi A., Moscona A. (2010) Inhibition of Nipah virus infection in vivo. Targeting an early stage of Paramyxovirus fusion activation during viral entry. PLoS Pathog. 6, e1001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yuan P., Thompson T. B., Wurzburg B. A., Paterson R. G., Lamb R. A., Jardetzky T. S. (2005) Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13, 803–815 [DOI] [PubMed] [Google Scholar]
- 29. Zaitsev V., von Itzstein M., Groves D., Kiefel M., Takimoto T., Portner A., Taylor G. (2004) Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase. Implications for fusion. J. Virol. 78, 3733–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yuan P., Swanson K. A., Leser G. P., Paterson R. G., Lamb R. A., Jardetzky T. S. (2011) Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc. Natl. Acad. Sci. 108, 14920–14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Greengard O., Poltoratskaia N., Leikina E., Zimmerberg J., Moscona A. (2000) The anti-influenza virus agent 4-GU-DANA (zanamivir) inhibits cell fusion mediated by human parainfluenza virus and influenza virus HA. J. Virol. 74, 11108–11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Palermo L. M., Porotto M., Yokoyama C. C., Palmer S. G., Mungall B. A., Greengard O., Niewiesk S., Moscona A. (2009) Human parainfluenza virus infection of the airway epithelium. Viral hemagglutinin-neuraminidase regulates fusion protein activation and modulates infectivity. J. Virol. 83, 6900–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Farnsworth A., Wisner T. W., Webb M., Roller R., Cohen G., Eisenberg R., Johnson D. C. (2007) Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. 104, 10187–10192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aguilar H. C., Matreyek K. A., Filone C. M., Hashimi S. T., Levroney E. L., Negrete O. A., Bertolotti-Ciarlet A., Choi D. Y., McHardy I., Fulcher J. A., Su S. V., Wolf M. C., Kohatsu L., Baum L. G., Lee B. (2006) N-Glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J. Virol. 80, 4878–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bishop K. A., Hickey A. C., Khetawat D., Patch J. R., Bossart K. N., Zhu Z., Wang L. F., Dimitrov D. S., Broder C. C. (2008) Residues in the Stalk Domain of the Hendra Virus G glycoprotein modulate conformational changes associated with receptor binding. J. Virol. 82, 11398–11409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bishop K. A., Stantchev T. S., Hickey A. C., Khetawat D., Bossart K. N., Krasnoperov V., Gill P., Feng Y. R., Wang L., Eaton B. T., Wang L. F., Broder C. C. (2007) Identification of Hendra virus G glycoprotein residues that are critical for receptor binding. J. Virol. 81, 5893–5901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Corey E. A., Iorio R. M. (2007) Mutations in the stalk of the measles virus hemagglutinin protein decrease fusion but do not interfere with virus-specific interaction with the homologous fusion protein. J. Virol. 81, 9900–9910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Melanson V. R., Iorio R. M. (2006) Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J. Virol. 80, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mahon P. J., Mirza A. M., Iorio R. M. (2011) Role of the two sialic acid binding sites on the newcastle disease virus HN protein in triggering the interaction with the F protein required for the promotion of fusion. J. Virol. 85, 12079–12082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Melanson V. R., Iorio R. M. (2004) Amino acid substitutions in the F-specific domain in the stalk of the newcastle disease virus HN protein modulate fusion and interfere with its interaction with the F protein. J. Virol. 78, 13053–13061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rekas A., Alattia J. R., Nagai T., Miyawaki A., Ikura M. (2002) Crystal structure of venus, a yellow fluorescent protein with improved maturation and reduced environmental sensitivity. J. Biol. Chem. 277, 50573–50578 [DOI] [PubMed] [Google Scholar]
- 42. Ottmann C., Weyand M., Wolf A., Kuhlmann J. (2009) Applicability of superfolder YFP bimolecular fluorescence complementation in vitro. Biol. Chem. 390, 81–90 [DOI] [PubMed] [Google Scholar]
- 43. Rose R. H., Briddon S. J., Holliday N. D. (2010) Bimolecular fluorescence complementation. Lighting up seven transmembrane domain receptor signaling networks. Br. J. Pharmacol. 159, 738–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brindley M. A., Plemper R. K. (2010) Blue native PAGE and biomolecular complementation reveal a tetrameric or higher order oligomer organization of the physiological measles virus attachment protein H. J. Virol. 84, 12174–12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Porotto M., Murrell M., Greengard O., Lawrence M. C, McKimm-Breschkin J. L., Moscona A. (2004) Inhibition of parainfluenza virus type 3 and Newcastle disease virus hemagglutinin-neuraminidase receptor binding. Effect of receptor avidity and steric hindrance at the inhibitor binding sites. J. Virol. 78, 13911–13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Robach J. G., Lamb R. A. (2010) Analysis of parainfluenza virus-5 hemagglutinin-neuraminidase protein mutants that are blocked in internalization and degradation. Virology 406, 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mahon P. J., Mirza A. M., Musich T. A., Iorio R. M. (2008) Engineered intermonomeric disulfide bonds in the globular domain of Newcastle disease virus hemagglutinin-neuraminidase protein. Implications for the mechanism of fusion promotion. J. Virol. 82, 10386–10396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Luci V., Förster F., Baumeister W. (2005) Structural studies by electron tomography. From cells to molecules. Annu. Rev. Biochem. 74, 833–865 [DOI] [PubMed] [Google Scholar]
- 49. Luci V., Yang T., Schweikert G., Förster F., Baumeister W. (2005) Morphological characterization of molecular complexes present in the synaptic cleft. Structure 13, 423–434 [DOI] [PubMed] [Google Scholar]
- 50. Maurer U. E., Sodeik B., Grünewald K. (2008) Native three-dimensional intermediates of membrane fusion in herpes simplex virus 1 entry. Proc. Natl. Acad. Sci. 105, 10559–10564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Plemper R. K., Brindley M. A., Iorio R. M. (2011) Structural and mechanistic studies of measles virus illuminate Paramyxovirus entry. PLoS Pathog. 7, e1002058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Farzan S. F., Palermo L. M., Yokoyama C. C., Orefice G., Fornabaio M., Sarkar A., Kellogg G. E., Greengard O., Porotto M., Moscona A. (2011) Premature activation of the Paramyxovirus fusion protein before target cell attachment with corruption of the viral fusion machinery. J. Biol. Chem. 286, 37945–37954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.