Background: The antagonistic complex of SnRK2.6 and ABI1 regulates abscisic acid (ABA) signaling in plants.
Results: Presented here are the structure of SnRK2.6 kinase domain, and biochemical and computational characterizations of the ABI1-SnRK2.6 complex.
Conclusion: Our studies revealed the molecular basis for ABI1-mediated inhibition of SnRK2.6.
Significance: The studies advanced our understanding of the downstream signal transduction of ABA through SnRK2s and protein phosphatase type 2Cs.
Keywords: Abscisic Acid, Kinetics, Phosphatase, Protein Complexes, Protein Phosphorylation, Structural Biology, X-ray Crystallography
Abstract
Subclass III SnRK2s (SnRK2.6/2.3/2.2) are the key positive regulators of abscisic acid (ABA) signal transduction in Arabidopsis thaliana. The kinases, activated by ABA or osmotic stress, phosphorylate stress-related transcription factors and ion channels, which ultimately leads to the protection of plants from dehydration or high salinity. In the absence of stressors, SnRK2s are subject to negative regulation by group A protein phosphatase type 2Cs (PP2C), whereas the underlying molecular mechanism remains to be elucidated. Here we report the crystal structure of the kinase domain of SnRK2.6 at 2.6-Å resolution. Structure-guided biochemical analyses identified two distinct interfaces between SnRK2.6 and ABI1, a member of group A PP2Cs. Structural modeling suggested that the two interfaces lock SnRK2.6 and ABI1 in an orientation such that the activation loop of SnRK2.6 is posited to the catalytic site of ABI1 for dephosphorylation. These studies revealed the molecular basis for PP2Cs-mediated inhibition of SnRK2s and provided important insights into the downstream signal transduction of ABA.
Introduction
The phytohormone abscisic acid (ABA)4 protects plants in inclement environments, particularly during drought or high salinity. ABA also regulates seed dormancy, seedling development, stomatal aperture etc. (1–3). The molecular mechanism of ABA perception and signal transduction in Arabidopsis has been intensively studied, and a complex signaling network is being revealed (4, 5). Sitting in the central hub of the ABA signaling network are the antagonistic counterparts of subclass III SnRK2s (SNF1-related protein kinase 2) and group A PP2Cs (protein phosphatase type 2C), in addition to the ABA receptor PYL/PYR1/RACR family of proteins (PYLs for short) (4). SnRK2s are positive regulators of ABA signaling, whereas PP2Cs negatively affect the pathway by directly inhibiting SnRK2s.
Ten SnRK2s, SnRK2.1 through SnRK2.10, were identified in Arabidopsis and grouped to three subclasses (4, 6). Subclass I SnRK2s can be activated rapidly by osmotic stress, whereas II and III are activated by both osmotic stress and ABA, with subclass III proteins playing a more prominent role in ABA response (7–11). Subclass III consists of SnRK2.6 (SnRK2E), SnRK2.3 (SnRK2I), and SnRK2.2 (SnRK2D), which play a redundant role as positive regulators of ABA signaling. SnRK2-dei, the triple knock-out of subclass III SnRK2s gave rise to an ABA-insensitive phenotype (10–12). The substrates for SnRK2.6/3/2 include transcription factors responsible for the expression of the ABA-responsive genes (13–15), as well as ion channels that control the osmotic homeostasis, such as the anion channel SLAC1 (16) and potassium channel KAT1 (17). Activation of SnRK2s requires autophosphorylation (18), a process that is inhibited by group A PP2Cs, whose members include ABI1, ABI2, HAB1, HAB2, etc. (19). Genetic and biochemical characterizations revealed that group A PP2Cs are physically associated with and dephosphorylate SnRK2.6/3/2, hence impeding the kinase activities (20, 21).
The molecular mechanism of ABA-induced removal of the negative regulation by PP2Cs has been revealed (22–27). Upon ABA binding, PYLs bind to the active site of PP2Cs and inhibit the phosphatase activity of PP2Cs by blocking substrate entrance (25–27). A subfamily of PYLs was recently found to bind and inhibit PP2Cs even in the absence of ABA, although the physiological relevance remains elusive (28). Despite advancement in the understanding of ABA perception and ABA-regulated inhibition of PP2Cs, the molecular basis by which PP2Cs inhibit SnRK2s remains poorly understood. To address this important question, we focused on SnRK2.6 for structural and biochemical characterizations. Here we report the crystal structure of a carboxyl terminus-truncated SnRK2.6. On the basis of structural analysis, we performed extensive mutagenesis analyses, biochemical characterizations, and kinetic studies, which collectively revealed the molecular basis for the recognition and inhibition of SnRK2.6 by ABI1. Our studies provide important insight into the understanding of the downstream signal transduction of ABA.
EXPERIMENTAL PROCEDURES
Protein Preparation and Crystallization
SnRK2.6 (AT4G33950), ABI1 (AT4G26080), and ABF2 (AT1G45249) were subcloned from the Arabidopsis thaliana cDNA library using standard PCR-based protocol. All mutants of SnRK2.6 and ABI1 were generated with two-step PCR and verified by plasmid sequencing. The proteins were expressed and purified following the protocols reported in the supplementary data. Prior to crystallization, the His6 tag was removed by thrombin digestion. Crystals of SnRK2.6 (residues 1–317, C131A/C137A/C159A/S7A/S29A/S43A/S166A/T176A) were grown at 18 °C using the hanging drop vapor diffusion method. Typical crystals appeared after 1 to 2 days in the buffer containing 0.1 m Tris, pH 8.0, and 19% PEG4000.
Data Collection, Structure Determination, and Refinement
The diffraction data were collected at the Shanghai Synchrotron Research Facility (SSRF), integrated, and scaled with the HKL2000 package (29). Structure of the N-terminal fragment SnRK2.6 (residues 1–317, denoted SnRK2.6ΔC hereafter) was determined using molecular replacement with PHASER (30). Structural refinement was performed with COOT (31) and PHENIX (32). Data collection and refinement statistics are summarized in supplemental Table S1.
GST-mediated Pull-down Assay
120 μg of GST fusion SnRK2.6 protein was added to 50 μl of glutathione S-Sepharose 4B (GS4B, GE Healthcare) resin. 120 μg of wild-type or mutant ABI1 proteins were incubated with the SnRK2.6-bound resin for 30 min at room temperature. The resin was then extensively rinsed with 25 mm Tris, pH 8.0, to remove unbound proteins. The resin was resuspended in 150 μl of buffer containing 25 mm Tris, pH 8.0, and 150 mm NaCl. 2.5 μl of suspension was applied to SDS-PAGE for analysis. The proteins were visualized with Coomassie Blue staining.
Size Exclusion Chromatography (SEC)
The SEC analyses were performed with a Superdex-200 HR10/300GL column (GE Healthcare) in a buffer containing 25 mm Tris, pH 8.0, 150 mm NaCl, and 4 mm MgCl2. To test the interaction of SnRK2.6 and ABI1, 0.5 mg of SnRK2.6 and 0.6 mg of ABI1 proteins were mixed in 200 μl and incubated at 18 °C for one-half hour before loading to Superdex-200 HR10/300GL.
Mass Spectrometry Analysis
For identification of phosphorylation sites of targeted protein, samples were separated with SDS-PAGE. The bands corresponding to the targeted proteins were excised from the gel, reduced with 10 mm DTT, and alkylated with 55 mm iodoacetamide. Then in gel digestion was carried out with sequence grade, modified trypsin (Promega, Fitchburg, WI) in 50 mm ammonium bicarbonate at 37 °C overnight. The peptides were extracted twice with 1% trifluoroacetic acid in 50% acetonitrile aqueous solution for 30 min. The extractions were then centrifuged in a SpeedVac to reduce the volume. For LC-MS/MS analysis, the digestion product was separated by a 65-min gradient elution at a flow rate of 0.250 μl/min with the EASY-nLCIITM integrated nano-HPLC system (Proxeon, Denmark), which is directly interfaced with the Thermo LTQ-Orbitrap mass spectrometer. The analytical column was a homemade fused silica capillary column (75 μm inner diameter, 150 mm length; Upchurch, Oak Harbor, WA) packed with C-18 resin (300 A, 5 μm, Varian, Lexington, MA). Mobile phase A consisted of 0.1% formic acid, and mobile phase B consisted of 100% acetonitrile and 0.1% formic acid. The LTQ-Orbitrap mass spectrometer was operated in the data-dependent acquisition mode using Xcalibur 2.0.7 software and there is a single full-scan mass spectrum in the Orbitrap (400–1800 m/z, 30,000 resolution) followed by 20 data-dependent MS/MS scans in the ion trap at 35% normalized collision energy. The MS/MS spectra from each LC-MS/MS run were searched against the selected data base using an in-house Mascot or Proteome Discovery searching algorithm. The ratios of phosphorylated peptides to unmodified peptides were determined based on ion intensities of targeted peptides at different time points.
Kinase Activity Assay
The kinase activity of SnRK2.6 was measured spectrophotometrically using ABF2 as substrates. This assay couples the production of ADP with the oxidation of NADH by pyruvate kinase (PK) and lactate dehydrogenase (LDH) (33). The standard assay was carried out at 25 °C in 1.8 ml of reaction mixture containing 50 mm MOPS, pH 7.0, 100 mm NaCl, 0.1 mm EDTA, 10 mm MgCl2, 0.2 mm NADH, 1.0 mm P-enolpyruvate, 20 units/ml of LDH, and 15 units/ml of PK, 1 mm ATP, 1.8 μm SnRK2.6 and varying amounts of ABF2. Progress of the reaction was monitored continuously by following the generation of NAD+ at 340 nm, on a PerkinElmer λ45 spectrophotometer equipped with a magnetic stirrer in the cuvette holder. The concentrations of ADP formed in the SnRK2.6-catalyzed reaction were determined using an extinction coefficient for NADH of 6220 cm−1 m−1 at 340 nm.
Phosphatase Activity Assay
Kinetic parameters for the dephosphorylation of SnRK2.6 by ABI1 were determined using a continuous spectrophotometric assay (34, 35). This assay incorporates a coupled enzyme system, which uses purine nucleoside phosphorylase (PNPase) and its chromogenic substrate 7-methyl-6-thioguanosine for the quantification of inorganic phosphate produced in the phosphatase reaction (36). All experiments were carried out at 25 °C in a 1.8-ml reaction mixture containing 50 mm MOPS, pH 7.0, 100 mm NaCl, 0.1 mm EDTA, 10 mm MgCl2, 100 μm 7-methyl-6-thioguanosine, 0.1 mg/ml of PNPase. The reactions were initiated by the addition of phosphatase unless indicated otherwise. The time courses of absorbance change at 360 nm were recorded on a PerkinElmer λ45 spectrophotometer equipped with a magnetic stirrer in the cuvette holder. Initial rates were determined from the linear slope of progress curves obtained and the experimental data were analyzed using a nonlinear regression analysis program. Quantitation of phosphate release was determined using the extinction coefficient of 11,200 m−1 cm−1 for the phosphate-dependent reaction at 360 nm at pH 7.0 (37). The concentration of 7-methyl-6-thioguanosine was determined at 331 nm, using a molar extinction coefficient of 32,000 m−1 cm−1. With p-nitrophenyl phosphate (pNPP) as a substrate, the reaction was initiated by the addition of the enzyme in a reaction mixture. The initial rate for hydrolysis of pNPP by phosphatase was measured at 25 °C in 1.8 ml of reaction mixture containing 50 mm MOPS, pH 7.0, 100 mm NaCl, 0.1 mm EDTA, and 10 mm MnCl2. Nonenzymatic hydrolysis of the substrate was corrected by measuring the control reaction in the absence of enzyme. The amount of product p-nitrophenol was determined with the absorbance at 405 nm using a molar extinction coefficient of 18,000 m−1 cm−1 (38).
Computational Modeling of the Complex Structure of SnRK2.6 and ABI1
The lack of a structure of SnRK2.6 domain II impeded the structural modeling of the complex between SnRK2.6 and ABI1. So we built the three-dimensional (3-D) structure model of full-length SnRK2.6 based on the protein sequence and structure of the SnRK2.6 kinase domain. The comparative modeling tool, Modeler 9.9 (39), was implemented on the in-house linux server. The results were compared with the online predictions by the I-TASSER server (40). Taken together, we picked up a model with a small helix for the residues 339–347 within the C-terminal long loop. The structure model of the complex between SnRK2.6 and ABI1 was predicted using the docking method HoDock (41), which incorporates an initial rigid docking and a refined semiflexible docking. In this work, the experimentally verified interactions were applied as restraints for conformational searching and model selecting. The one with the highest score was picked from over 11,000 results.
RESULTS
In Vitro Reconstitution of ABI1-mediated Dephosphorylation of SnRK2.6
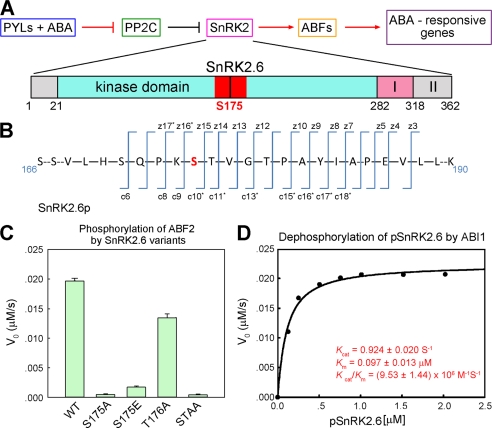
Subclass III SnRK2s play a redundant role in ABA signaling. To understand the inhibitory mechanism of SnRK2s by PP2Cs, we selected SnRK2.6 for in vitro structural and biochemical characterizations. SnRK2.6 (also known as OST1, open stomata 1) is functionally divided to three domains, the catalytic kinase domain (residues 21–282), the activation motif (residues 283–318, domain I), and the C-terminal ABI1 binding domain (residues 319–357, domain II) (Fig. 1A) (21). We expressed recombinant SnRK2.6 and ABI1 in Escherichia coli and sought to purify the proteins to homogeneity. During the purification of SnRK2.6, however, smeary bands with molecular masses close to 40 kDa were observed on SDS-PAGE (supplemental Fig. S1A, left lane). Incubation with the catalytic domain of ABI1 (residues 125–429) reduced the multiple bands to a single one corresponding to an apparent molecular masses of ∼41 kDa (supplemental Fig. S1A, right lane). These observations suggested that SnRK2.6 may be phosphorylated during recombinant expression and purification. Mass spectrometry (MS) analysis confirmed that multiple Ser/Thr residues were phosphorylated in the recombinantly expressed SnRK2.6, an observation similar to the protein obtained from plant cells (18, 20). Interestingly, when the dephosphorylated SnRK2.6 was further purified and then incubated with ATP and Mg2+, the dominant phosphorylation site was Ser175 (Fig. 1B, supplemental Figs. S1B and S2), although the reaction was not 100% complete even after incubation overnight.
FIGURE 1.
Reconstitution of ABI1-mediated dephosphorylation of pSnRK2.6 with purified recombinant proteins. A, a simplified schematic diagram of the ABA-signaling pathway regulated by PP2Cs and SnRK2s. PP2C refers to group A proteins and SnRK2 refers to SnRK2.6/2.3/2.2. ABFs stand for ABRE (ABA-responsive elements)-binding factors. The domain structure of SnRK2.6 is shown with the activation loop (residues 160–186) highlighted in red. B, MS analyses confirmed that Ser175 is autophosphorylated in the purified recombinant SnRK2.6. The experimental details and original data are presented in supplemental Figs. S1B and S2. C, Ser175 is essential for the kinase activity of SnRK2.6. The N-terminal fragment of ABF2 was used as substrate for the phosphorylation assay. The initial phosphorylation rates by SnRK2.6 variants were measured with ABF2 at 6 μm and SnRK2.6 at 1.8 μm. STAA: S175A and T176A. D, kinetic study of the dephosphorylation of phosphorylated SnRK2.6 (pSnRK2.6) by ABI1.
To examine the kinase activity of the recombinant pSnRK2.6, we reconstituted a coupled spectrophotometric assay using purified ABF2 (residues 1–199) as substrate (supplemental Fig. S3) (33). The kcat and Km values for the ABF2 phosphorylation by pSnRK2.6 were determined to be 0.04 ± 0.003 s−1 and 19.3 ± 2.52 μm, respectively. Substitution of Ser175 with Ala completely abolished the kinase activity of SnRK2.6 (Fig. 1C); by contrast, T176A only exhibited a moderate effect on the kinase activity. This observation corroborates the in vivo characterization that Ser175 plays a key role in the activity of SnRK2.6 (18, 42). Glutamate is generally used to mimic the phosphorylated serine. In SnRK2.6, however, the S175E mutant displayed significantly impaired activity (Fig. 1C).
Addition of ABI1 led to dephosphorylation of pSnRK2.6 (supplemental Fig. S1). We determined the kinetic parameters for the ABI1-catalyzed dephosphorylation of SnRK2.6 using the continuous spectrophotometric assay for protein phosphatases (34, 35). ABI1 exhibited a highly specific and efficient phosphatase activity for pSnRK2.6 with Km 0.097 ± 0.013 μm and kcat 0.924 ± 0.020 s−1 (Fig. 1D). MS analysis revealed that phospho-Ser175 was the dominant substrate for ABI1 within the first 5 min of the reaction (supplemental Fig. S1C).
Phosphorylation of Ser175, which is located on the activation loop (also known as T-loop) of SnRK2.6, was reported to be essential to the kinase activity of SnRK2.6 (18, 20). Experiments with purified recombinant proteins here demonstrated that activation of SnRK2.6 can be achieved by autophosphorylation, whereas ABI1 inhibits SnRK2.6 by the effective dephosphorylation of SnRK2.6-Ser(P)175.
Crystal Structure of the C terminus-truncated SnRK2.6
To better understand the recognition and inhibition of SnRK2.6 by ABI1, we sought to obtain the crystal structures of SnRK2.6 or the complex of SnRK2.6 and ABI1. After many trials, the structure of the C terminus-truncated SnRK2.6 (SnRK2.6ΔC) was determined and refined to 2.6-Å resolution (Fig. 2 and supplemental Table S1). To obtain better diffracting crystals, three cysteine residues were replaced by Ala to avoid unwanted oxidation. The Ser and Thr residues that may be heterogeneously phosphorylated during expression and purification, including Ser7, Ser29, Ser43, Ser166, and Thr176, were also mutated to Ala.
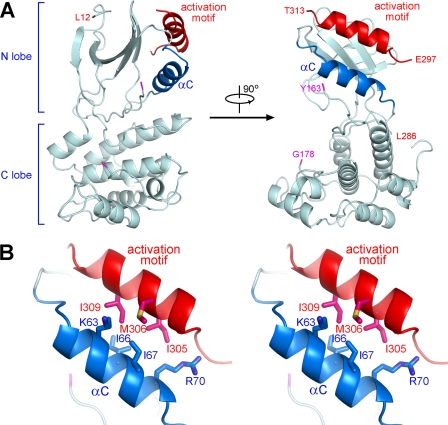
FIGURE 2.
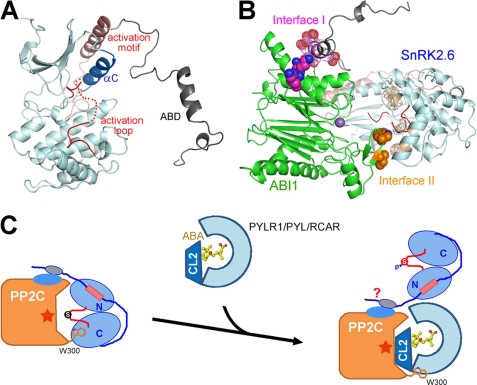
Crystal structure of SnRK2.6ΔC. A, two perpendicular views of the overall structure of SnRK2.6 (1–317). The fragments containing residues 164–177 and 287–296 are invisible in the structure. B, a stereo view of the interaction between helix αC and the activation motif of SnRK2.6. The residues that are involved in the extensive van der Waals contacts are shown in sticks. All structure figures were prepared with PyMol (45).
The catalytic core of SnRK2.6 exhibits a canonical kinase fold with distinct N- and C-lobes. We included ATP or AMPPNP in the crystallization trials; nevertheless, the structure is in an apo-form. As in the case of a number of apo-kinase structures, the activation loop (residues 164–177) is invisible, probably due to the intrinsic flexibility in the absence of a co-factor (Fig. 2A). Residues 287–296 of the activation motif are invisible, whereas residues 301–313 form a helix that lies against the N-lobe. The activation helix is positioned approximately in parallel with helix αC through extensive van der Waals contacts (Fig. 2B). It is known that the mobility of helix αC is an important regulating factor to the kinase activity (43). Placement of the activation helix in contact with helix αC suggests that the activation motif may regulate the catalytic activity of SnRK2.6 through modulation of the spatial position of helix αC, a caveat requiring further investigations.
Biochemical Identification of the Interface between SnRK2.6 and ABI1
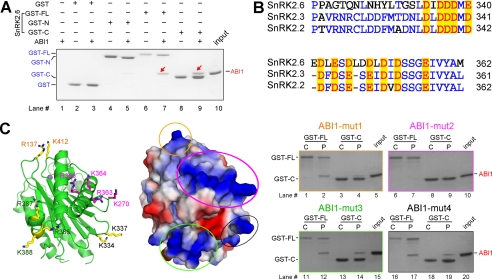
Cell-based co-immunoprecipitation, BiFC, and Y2H screenings showed that SnRK2.6 and ABI1 are physically associated, and domain II of SnRK2.6 is required for the interaction (20, 21). With the purified recombinant proteins, we attempted to identify the interface between the kinase and phosphatase. Glutathione S-transferase (GST)-fused SnRK2.6 variants, including the full-length protein (GST-FL), the N-terminal fragment (residues 1–317, GST-N), and domain II (residue 318–362, GST-C) were immobilized on GS4B resin and used as bait to pull down ABI1 (Fig. 3A). Consistent with the in vivo observations (21), GST-FL and GST-C of SnRK2.6 could successfully pull down the catalytic domain of ABI1, whereas GST-N of SnRK2.6 could only pull down a barely visible band of ABI1 (Fig. 3A). We then sought to identify the motif on ABI1 that recognizes SnRK2.6.
FIGURE 3.
Identification of the polar interface between SnRK2.6 and ABI1. A, domain II of SnRK2.6 is necessary for binding to ABI1. GST-fused SnRK2.6 variants, including the full-length protein (GST-FL), the N-terminal fragment (residues 1–317, GST-N), and domain II (residue 318–362, GST-C) were immobilized on GS4B resin and used as bait to pull down ABI1. B, sequence alignment of domain II of subclass III SnRK2s. Conserved acidic residues were highlighted in red. C, identification of a surface patch of ABI1 enriched of positively charged residues that are involved in SnRK2.6 recognition. Left panel, four positively charged clusters are highlighted on the crystal structure of ABI1 (PDB accession code 3KDJ). The metal ion is shown as a purple sphere to indicate the active site of the phosphatase. Middle panel, electrostatic surface potential of ABI1, calculated by PyMol, is shown in the same view as on the left panel. Right panel, pull-down experiment identified one positively charged surface patch that is involved in the interaction of SnRK2.6 and ABI1. Each mutant contains two to four point mutations of the residues labeled in the corresponding colors on the structure.
Notably, domain II of all the subclass III SnRK2s is highly enriched of acidic residues (Fig. 3B and supplemental Fig. S4). We reasoned that the counterpart of ABI1 may involve a cluster of positively charged residues. Therefore, we carefully examined the crystal structure of ABI1 (PDB accession code 3KDJ) and identified four positively charged surface patches (Fig. 3C). To examine whether any of them are involved in SnRK2 recognition, we generated four ABI1 variants, each containing two to four point mutations and named mut1 through mut4 (mut1, R137D/K412D; mut2, K270D/R363A/K364A/R365A; mut3, R386A/R387A/K388A; and mut4, K334A/K337A). We then tested the binding of these variants to both SnRK2.6 FL and domain II. Although mut1, mut3, and mut4 of ABI1 retained interaction with FL or domain II of SnRK2.6, mut2 failed to do so. Therefore ABI1 residues Lys270, Arg363, Lys364, and Arg365 are involved in association to the Glu/Asp-rich domain II of SnRK2.6. The polar interface is likely to be conserved for subclass III SnRK2s and most of group A PP2Cs, as the positively charged residues are conserved among four PP2Cs, ABI1, ABI2, HAB1, and HAB2 (supplemental Fig. S5).
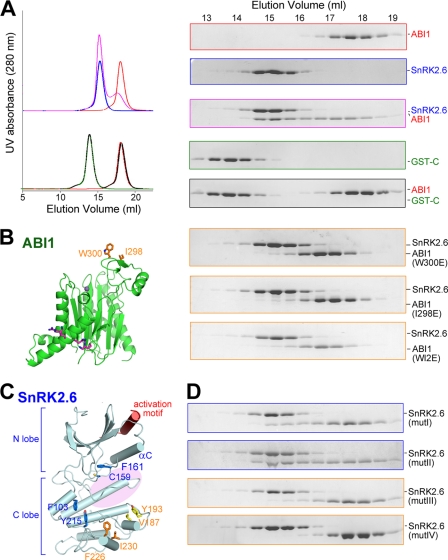
We also examined the effect of the interface mutations on the dephosphorylation of pSnRK2.6 by ABI1. The Km values were both increased by 2–3-fold for ABI1-mut2 or domain II-truncated SnRK2.6 (Table 1). The activity assay supported the identification of the interface. On the other hand, the moderate effect on Km by the interface mutations may suggest the existence of an additional interface other than the identified one. This notion was supported by an extra line of evidence. When SnRK2.6 and ABI1 were subjected to SEC for the binding test, they were co-eluted, indicating formation of a stable SnRK2-ABI1 complex (Fig. 4A, top three panels on the right). By contrast, the interaction between SnRK2.6 domain II and ABI1, which were detected by the GST-mediated pulldown assay, could not sustain SEC (Fig. 4A, bottom two panels on the right). These observations demonstrated that SnRK2.6 FL exhibits a higher binding affinity with ABI1 than domain II, and that the N-terminal fragment of SnRK2.6 (residues 1–317) may possess a second recognition site for ABI1.
TABLE 1.
Summary of the kinetic parameters of ABI1 variants
| Phosphatase | Substrate | kcat | Km | kcat/Km |
|---|---|---|---|---|
| s−1 | μm | m−1s−1 | ||
| ABI1 WT | SnRK2.6 | 0.924 ± 0.020 | 0.097 ± 0.013 | (9.53 ± 1.44) × 106 |
| SnRK2.6ΔC | 0.962 ± 0.009 | 0.260 ± 0.012 | (3.70 ± 0.21) × 106 | |
| ABI1 (RKR363A,K270D) | SnRK2.6 | 1.03 ± 0.03 | 0.358 ± 0.035 | (2.88 ± 0.37) × 106 |
| SnRK2.6ΔC | 1.11 ± 0.03 | 0.632 ± 0.048 | (1.76 ± 0.18) × 106 | |
| ABI1 (I298E,W300E) | SnRK2.6 | 0.939 ± 0.013 | 5.59 ± 0.23 | (1.68 ± 0.08) × 105 |
| SnRK2.6ΔC | NAa | >10 | NA | |
| ABI1 (W300E) | SnRK2.6 | 1.17 ± 0.02 | 0.751 ± 0.057 | (1.57 ± 0.15) × 106 |
| ABI1 (I298E) | SnRK2.6 | 1.03 ± 0.02 | 0.803 ± 0.071 | (1.28 ± 0.14) × 106 |
| ABI1 (I298E,W300E, RKR363A,K270D) | SnRK2.6 | NA | >10 | NA |
a NA, not applicable.
FIGURE 4.
Identification of the second interface between SnRK2.6 and ABI1. A, SEC analyses of the interactions between ABI1 and the FL or domain II of SnRK2.6. The same elution fractions of each SEC injection were applied to SDS-PAGE followed by Coomassie Blue staining. FL SnRK2.6 is non-tagged, whereas domain II is GST-fused. B, SEC analyses showed that ABI1 variants containing point mutations of W300E, or I298E, or W300E/I298E (WI2E) were unable to form stable complex with SnRK2.6. C, selection of hydrophobic residue clusters on the surface of SnRK2.6 that may be involved in ABI1 interaction. Four clusters, whose residues are shown in sticks, were selected on the basis of the crystal structure of SnRK2.6. The likely position of the activation loop, which is invisible in the crystal structure, is indicated by the pink shadow. D, SEC analyses of the interactions between ABI1 and SnRK2.6 variants. Refer to the main text for the annotation of mutI–IV.
Ile298 and Trp300 of ABI1 Are Involved in SnRK2.6 Recognition
During an attempt to identify the second interface between SnRK2.6 and ABI1, when an ABI1 variant containing the Trp300 to Glu mutation (ABI1-W300E) was incubated with SnRK2.6 and subjected to SEC analysis, their interaction was completely abolished (Fig. 4B). In addition, mutation of an adjacent hydrophobic residue, Ile298, of ABI1 to Glu also led to complete loss of interaction between SnRK2.6 and ABI1 on SEC. Supporting this observation, in the phosphatase assay, ABI1 variants containing single point mutations of I298E or W300E both displayed increased Km values by about 8-fold with respect to the WT protein (Table 1). Moreover, the Km value of double mutations (I298E/W300E) was increased by ∼60-fold. These observations suggested that Ile298 and Trp300 are involved in the association of ABI1 with SnRK2.6.
Interestingly, these ABI1 mutants retained interaction with FL or domain II of SnRK2.6 in the GST-mediated pulldown assay (supplemental Fig. S6), indicating that their interaction counterpart is not Glu/Asp-rich domain II. Supporting this analysis, Ile298 and Trp300 are located at a loop region about 40 Å away from the positive patch of ABI1 recognized by the SnRK2.6 domain II (Fig. 4B, left panel). It is likely that the hydrophobic region comprised of Ile298 and Trp300 represents a separate binding site for SnRK2.6.
C-lobe of Kinase Domain of SnRK2.6 Is Recognized by ABI1
We examined the structure of SnRK2.6 to look for hydrophobic clusters that may be recognized by Ile298 and Trp300 of ABI1. Notably, Ile298 and Trp300 are located in the vicinity of the catalytic site of ABI1. We reasoned that their recognition site on SnRK2.6 was likely to be on the same side of the structure as the activation loop, because phospho-Ser175 was readily dephosphorylated by ABI1. Based on structural analysis, we generated four variants of SnRK2.6, each containing double point mutations surrounding the potential position for the activation loop (mutI, F103D/Y215D; mutII, C159A/F161A; mutIII, V187D/Y193D; and mutIV, F226D/I230D) (Fig. 4C). SEC was exploited to test the interactions between the SnRK2.6 variants and ABI1 (Fig. 4D). Although mutI and mutII retained a stable interaction with ABI1 as WT SnRK2.6 does (Fig. 4D, top two panels), mutIII showed a decreased binding affinity (Fig. 4D, panel 3). In particular, mutIV was completely dissociated from ABI1 (Fig. 4D, bottom panel). Val187/Tyr193 and Phe226/Ile230 are located at the adjacent structural motifs on the C-lobe of SnRK2.6. Therefore, this region may represent a second surface patch that is recognized by ABI1, most likely through van der Waals contacts with Ile298 and Trp300. Notably, Val187, Tyr193, Phe226, and Ile230 are highly conserved among the subclass III SnRK2s, and Ile298 and Trp300 are invariant among group A PP2Cs (supplemental Fig. S5).
Modeling Complex Structure of SnRK2.6 and ABI1
The biochemical evidence presented above led to the conclusion that there are at least two separate interfaces between SnRK2.6 and ABI1, one through polar interactions between the negatively charged domain II of SnRK2.6 and a positively charged surface patch of ABI1, the other potentially involving van der Waals contacts between a few hydrophobic residues on the C-lobe of SnRK2.6 and Ile298 and Trp300 of ABI1.
On the basis of the biochemical analyses as well as the available structures of SnRK2.6 and ABI1, we were able to generate a structural model of the SnRK2-ABI1 complex through computational approaches. First, we generated a structural model with the program Modeler 9.9 (39) for domain II of SnRK2.6 (Fig. 5A). Residues 339–347 were calculated as a helix, whereas the other residues were loops. Then we exploited the docking method Hodock (44) to predict the structure of the complex. The two biochemically identified interfaces were set as restraints for conformational searching and model selecting. In total, 11,000 models were generated, and the one with the highest score was picked up (Fig. 5B). We are fully aware of the speculative nature of this model, which mainly serves to examine the spatial compatibility of the two biochemically identified interfaces. Interestingly, this model showed that the two interfaces between ABI1 and SnRK2.6 anchor the proteins in an orientation that the activation loop of SnRK2.6 is positioned into the active site of ABI1 (Fig. 5B), and thus provided a plausible molecular basis for the efficient dephosphorylation of phospho-Ser175 of SnRK2.6 by ABI1.
FIGURE 5.
Molecular basis of group A PP2Cs-mediated inhibition of class III SnRK2s. A, a computational model of the full-length SnRK2.6. Domain II is predicted to be in an extended conformation with a short α-helix for residues 339–347. B, a docking model of the complex between SnRK2.6 and ABI1. The experimentally identified surfaces are highlighted. The polar interface is labeled Interface I and the hydrophobic one is labeled Interface II. C, a working model to illustrate the molecular basis of PYLs-mediated activation of SnRK2.6. It remains to be seen whether SnRK2.6 can bind to the PYL-PP2C complex through its elongated domain II.
Molecular Basis of PYLs-mediated Activation of SnRK2.6
SnRK2.6 can be activated by both ABA and osmotic stress in planta. Despite the poorly understood ABA-independent activation mechanism, the biochemical and structural investigations presented here, together with previous reports, provide the molecular basis for PYL-mediated activation of SnRK2.6 (Fig. 5C). PYL, upon ABA binding, or in some cases independent of ABA (28), bind to group A PP2Cs and directly inhibit their phosphatase activity by blocking the substrate entrance (25–27). Notably, the conserved residue ABI1-Trp300 plays an important role in the recognition between ABI1 and PYLs (25–27). Here we further identified that ABI1-Trp300 represented an essential binding site for SnRK2.6 (Fig. 3B). Therefore, in addition to directly blocking the catalytic site of PP2Cs, PYLs also compete with SnRK2.6 for binding to Trp300 of ABI1 (Fig. 5C), thus disrupting the hydrophobic interface between ABI1 and SnRK2.6.
It is noteworthy that disruption of the hydrophobic interface between ABI1 and SnRK2.6 resulted in a nearly 60-fold increase of Km value for the dephosphorylation of SnRK2.6 by ABI1. By contrast, loss of the polar interface only led to a slight augment of the Km value (Table 1). The hydrophobic interface is closer to the active site of ABI1 and phospho-Ser175 of SnRK2.6, and may directly facilitate a stable association between the enzyme and the substrate. On the other hand, domain II of SnRK2.6 may be rather flexible as suggested by structural prediction (Fig. 5A). Domain II may serve as a tethering factor for the initial recognition by PP2Cs, which is indispensable for complex formation (Fig. 2A). Once the proteins were pulled close enough, the hydrophobic interface may help anchor the enzyme and the substrate with the appropriate orientation so that phospho-Ser175 of SnRK2.6 is subject to dephosphorylation by ABI1. When ABA-bound or ABA-independent PYLs arrive, the hydrophobic interface is disrupted and the catalytic site of ABI1 is blocked. Our preliminary analyses showed that PYLs, ABI1, and SnRK2.6 may be able to form a ternary complex (supplemental Fig. S7), as the polar interface between SnRK2.6 and ABI1 is not significantly affected by PYLs. Nonetheless, due to the long and flexible linker between the catalytic domain and domain II, the kinase domain of SnRK2.6 is relieved and activated in response to ABA (Fig. 5C).
DISCUSSION
In summary, the biochemical and structural characterizations reported here revealed the molecular basis for the recognition and inhibition of subclass III SnRK2s by group A PP2Cs, and served as an important platform to the understanding of the downstream signal transduction of ABA. Nevertheless, an important question awaits further investigations. Subclass III SnRK2s can be activated in both ABA-dependent and -independent manners. Our current studies revealed the molecular mechanism for ABA-dependent activation of SnRK2s. It remains elusive how osmotic stress activates SnRK2s. As reported previously, a subclass of PYLs inhibit PP2Cs independent of ABA (28). Further studies are required to elucidate whether osmotic stress directly regulates SnRK2s, or through inhibition of PP2Cs by ABA-independent PYLs. It is also plausible that unidentified factors or mechanisms are involved in the activation of SnRK2s by osmotic stress.
Supplementary Material
This work was supported by Projects 91017011 and 31070644 of the National Natural Science Foundation of China, Tsinghua University 985 Phase II funds, and the Yuyuan Foundation.

This article contains supplemental Figs. S1–S7 and Table S1.
- ABA
- abscisic acid
- PP2C
- protein phosphatase type 2C
- SEC
- size exclusion chromatography
- AMPPNP
- 5′-adenylyl-β,γ-imidodiphosphate
- PYL
- PYL/PYR1/RACR family of proteins
- FL
- full-length.
REFERENCES
- 1. Fedoroff N. V. (2002) Sci. STKE 2002, re10. [DOI] [PubMed] [Google Scholar]
- 2. Finkelstein R., Reeves W., Ariizumi T., Steber C. (2008) Annu. Rev. Plant Biol. 59, 387–415 [DOI] [PubMed] [Google Scholar]
- 3. Schroeder J. I., Nambara E. (2006) Cell 126, 1023–1025 [DOI] [PubMed] [Google Scholar]
- 4. Umezawa T., Nakashima K., Miyakawa T., Kuromori T., Tanokura M., Shinozaki K., Yamaguchi-Shinozaki K. (2010) Plant Cell Physiol. 51, 1821–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Klingler J. P., Batelli G., Zhu J. K. (2010) J. Exp. Bot. 61, 3199–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fujii H., Verslues P. E., Zhu J. K. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mustilli A. C., Merlot S., Vavasseur A., Fenzi F., Giraudat J. (2002) Plant Cell 14, 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fujii H., Verslues P. E., Zhu J. K. (2007) Plant Cell 19, 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoshida R., Hobo T., Ichimura K., Mizoguchi T., Takahashi F., Aronso J., Ecker J. R., Shinozaki K. (2002) Plant Cell Physiol. 43, 1473–1483 [DOI] [PubMed] [Google Scholar]
- 10. Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., Maruyama K., Yoshida T., Ishiyama K., Kobayashi M., Shinozaki K., Yamaguchi-Shinozaki K. (2009) Plant Cell Physiol. 50, 1345–1363 [DOI] [PubMed] [Google Scholar]
- 11. Fujita Y., Nakashima K., Yoshida T., Katagiri T., Kidokoro S., Kanamori N., Umezawa T., Fujita M., Maruyama K., Ishiyama K., Kobayashi M., Nakasone S., Yamada K., Ito T., Shinozaki K., Yamaguchi-Shinozaki K. (2009) Plant Cell Physiol. 50, 2123–2132 [DOI] [PubMed] [Google Scholar]
- 12. Fujii H., Zhu J. K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson R. R., Wagner R. L., Verhey S. D., Walker-Simmons M. K. (2002) Plant Physiol. 130, 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Uno Y., Furihata T., Abe H., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geiger D., Scherzer S., Mumm P., Stange A., Marten I., Bauer H., Ache P., Matschi S., Liese A., Al-Rasheid K. A., Romeis T., Hedrich R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sato A., Sato Y., Fukao Y., Fujiwara M., Umezawa T., Shinozaki K., Hibi T., Taniguchi M., Miyake H., Goto D. B., Uozumi N. (2009) Biochem. J. 424, 439–448 [DOI] [PubMed] [Google Scholar]
- 18. Vlad F., Droillard M. J., Valot B., Khafif M., Rodrigues A., Brault M., Zivy M., Rodriguez P. L., Merlot S., Laurière C. (2010) Plant J. 63, 778–790 [DOI] [PubMed] [Google Scholar]
- 19. Hirayama T., Shinozaki K. (2007) Trends Plant Sci. 12, 343–351 [DOI] [PubMed] [Google Scholar]
- 20. Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., Ishihama Y., Hirayama T., Shinozaki K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yoshida R., Umezawa T., Mizoguchi T., Takahashi S., Takahashi F., Shinozaki K. (2006) J. Biol. Chem. 281, 5310–5318 [DOI] [PubMed] [Google Scholar]
- 22. Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009) Science 324, 1064–1068 [DOI] [PubMed] [Google Scholar]
- 23. Park S. Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T. F., Alfred S. E., Bonetta D., Finkelstein R., Provart N. J., Desveaux D., Rodriguez P. L., McCourt P., Zhu J. K., Schroeder J. I., Volkman B. F., Cutler S. R. (2009) Science 324, 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Santiago J., Rodrigues A., Saez A., Rubio S., Antoni R., Dupeux F., Park S. Y., Marquez J. A., Cutler S. R., Rodriguez P. L. (2009) Plant J. 60, 575–588 [DOI] [PubMed] [Google Scholar]
- 25. Yin P., Fan H., Hao Q., Yuan X., Wu D., Pang Y., Yan C., Li W., Wang J., Yan N. (2009) Nat. Struct. Mol. Biol. 16, 1230–1236 [DOI] [PubMed] [Google Scholar]
- 26. Melcher K., Ng L. M., Zhou X. E., Soon F. F., Xu Y., Suino-Powell K. M., Park S. Y., Weiner J. J., Fujii H., Chinnusamy V., Kovach A., Li J., Wang Y., Li J., Peterson F. C., Jensen D. R., Yong E. L., Volkman B. F., Cutler S. R., Zhu J. K., Xu H. E. (2009) Nature 462, 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miyazono K., Miyakawa T., Sawano Y., Kubota K., Kang H. J., Asano A., Miyauchi Y., Takahashi M., Zhi Y., Fujita Y., Yoshida T., Kodaira K. S., Yamaguchi-Shinozaki K., Tanokura M. (2009) Nature 462, 609–614 [DOI] [PubMed] [Google Scholar]
- 28. Hao Q., Yin P., Li W., Wang L., Yan C., Lin Z., Wu J. Z., Wang J., Yan S. F., Yan N. (2011) Mol. Cell 42, 662–672 [DOI] [PubMed] [Google Scholar]
- 29. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 30. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emsley P., Cowtan K. (2004) Acta Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 33. Roskoski R., Jr. (1983) Methods Enzymol. 99, 3–6 [DOI] [PubMed] [Google Scholar]
- 34. Cheng Q., Wang Z. X., Killilea S. D. (1995) Anal. Biochem. 226, 68–73 [DOI] [PubMed] [Google Scholar]
- 35. Zhang Y. Y., Mei Z. Q., Wu J. W., Wang Z. X. (2008) J. Biol. Chem. 283, 26591–26601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Webb M. R. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4884–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sergienko E. A., Srivastava D. K. (1994) Anal. Biochem. 221, 348–355 [DOI] [PubMed] [Google Scholar]
- 38. Zhou B., Zhang Z. Y. (1999) J. Biol. Chem. 274, 35526–35534 [DOI] [PubMed] [Google Scholar]
- 39. Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., Sali A. (2007) Current Protocols in Protein Science 50, 2.9.1–2.9.31 [DOI] [PubMed] [Google Scholar]
- 40. Roy A., Kucukural A., Zhang Y. (2010) Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gong X., Wang P., Yang F., Chang S., Liu B., He H., Cao L., Xu X., Li C., Chen W., Wang C. (2010) Proteins Struct. Funct. Bioinformat. 78, 3150–3155 [DOI] [PubMed] [Google Scholar]
- 42. Belin C., de Franco P. O., Bourbousse C., Chaignepain S., Schmitter J. M., Vavasseur A., Giraudat J., Barbier-Brygoo H., Thomine S. (2006) Plant Physiol. 141, 1316–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen L., Jiao Z. H., Zheng L. S., Zhang Y. Y., Xie S. T., Wang Z. X., Wu J. W. (2009) Nature 459, 1146–1149 [DOI] [PubMed] [Google Scholar]
- 44. Gong X., Liu B., Chang S., Li C., Chen W., Wang C. (2010) SCIENCE CHINA Life Sciences 53, 1152–1161 [DOI] [PubMed] [Google Scholar]
- 45. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Schrödinger, LLC, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.