Abstract
The DR region of particular primate species may display allelic polymorphism and gene copy number variation (region configuration polymorphism). The sum of these distinct types of polymorphism is defined as complexity. To date, however, the DR region of cynomolgus macaques (Macaca fascicularis) has been poorly defined. Transcriptome analysis of a pedigreed colony, comprising animals from Indonesia and Indochina, revealed a total of 15 Mafa-DRA and 57 DRB alleles, specifying 28 different region configurations. The DRA alleles can be divided into two distinct lineages. One lineage is polymorphic, but the majority of the amino acid replacements map to the leader peptide. The second lineage is at best oligomorphic, and segregates with one specific Mafa-DRB allele. The number of Mafa-DRB genes ranges from two to five per haplotype. Due to the presence of pseudogenes, however, each haplotype encodes only one to three bona fide DRB transcripts. Depending on the region configuration in which the Mafa-DRB gene is embedded, identical alleles may display differential transcription levels. Region configurations appear to have been generated by recombination-like events. When genes or gene segments are relocated, it seems plausible that they may be placed in the context of distinct transcription control elements. As such, DRB region-related transcription level differences may add an extra layer of polymorphism to this section of the adaptive immune system.
Keywords: MHC, Transcriptome, Nonhuman primates, Comparative immunology
Introduction
Traditionally, Indian rhesus monkeys have been a prime species of choice for biomedical research. However, the import embargo on these animals has led to the search for alternatives; as a result, cynomolgus macaques (Macaca fascicularis) have become increasingly important as model species in recent years. To date, cynomolgus macaques have been used in studies for a variety of infectious diseases such as HIV/SHIV, tuberculosis, and dengue, as well as in transplantation and autoimmunity research (Aoyama et al. 2009; Benferhat et al. 2009; Capuano et al. 2003; Greene et al. 2010; Guirakhoo et al. 2004; Ma et al. 2009; Mee et al. 2009b; Wiseman et al. 2009). Since gene products of the major histocompatibility complex (MHC) play a crucial role in a variety of immune responses, detailed knowledge of the genetic background of these macaques has received increasing attention. The MHC class II region of Mauritian monkeys has been studied extensively, but due to a founder effect their DRB region shows limited levels of polymorphism and diversity (Blancher et al. 2006, 2008; Bonhomme et al. 2008; Mee et al. 2009a; O’Connor et al. 2007; Wojcechowskyj et al. 2007). In animals of other geographic origins, the diversity of the DR region is extensive (Aarnink et al. 2010; Doxiadis et al. 2006, 2010; Leuchte et al. 2004; Wei et al. 2007). For instance, in Philippine and Vietnamese cynomolgus macaque populations, up to 14 DRA alleles have been detected (Aarnink et al. 2010). Additionally, animals originating from Indochina and the Indonesian islands, showed a very high degree of DRB region polymorphism with 49 different Mafa-DRB regions described (Doxiadis et al. 2010).
In the latter study, a highly polymorphic microsatellite, D6S2878, had been used that maps to intron 2 of all DRB genes and pseudogenes with an intact exon 2–intron 2 segment. Most contemporary and more detailed information on Mafa-DRB genes is based on isolated exon 2 data. In the present report, we were keen to determine which DRB genes represent bona fide class II transcripts. In addition, we wanted to examine the level of polymorphism of DRA transcripts and their linkage to DRB haplotypes, in order to extend our knowledge of the DR region composition of monkeys of Indonesian and Indochinese origin.
Materials and methods
Animals and cell lines
The Biomedical Primate Research Centre (BPRC) houses a self-sustaining colony of cynomolgus macaques that have been pedigreed mainly by ethological observations and partially based on the segregation of defined MHC haplotypes (de Groot et al. 2008; Doxiadis et al. 2006). The animals originated from mainland Indochina and the Indonesian islands as proven by mtDNA analysis (de Groot et al. 2008). Two animals with Indonesian mtDNA profile were imported from Mauritius. The cynomolgus macaques analyzed for full-length DRA and DRB belong to an outbred breeding colony (52 out of 58), and are members of 12 pedigreed families with variable member sizes and generations, ranging from eight to 30 animals and from two to six generations. B-lymphoblastoid cell lines (BLCL) of the other six animals were received in collaboration with other European institutions, according to regulations approved by local ethics committees.
Cloning, sequencing, and genotyping
RNA was isolated from BLCLs (Rneasy kit, Qiagen) and subjected to One-step reverse transcriptase polymerase chain reaction (RT-PCR), as recommended by the supplier (Promega). Full length Mafa-DRA sequences were amplified by PCR from DNA using primers specific for human DRA 5′ and 3′ untranslated sequences (Lekutis and Letvin 1995): 5′DRA-SalI, 5′-TCC CGT CGA CCG CCC AAG AAG AAA ATG GCC-3′ and 3′DRA-BamHI, 5′-CAT TGG ATC CGA AGT TTC TTC AGT GAT CTT-3′.
Likewise, Mafa-DRB sequences were amplified by PCR using primers specific for human 5′- and 3′ untranslated sequences (Lekutis and Letvin 1995): 5′DRB-SalI, 5′-GCC CGT CGA CCT GTC CTG TTC TCC AGC ATG-3′ and 3′DRB-BamHI, 5′-GGC GGG ATC CCT TTT CAT CCT GCA AAG CTG-3′. Primers were synthesised by Invitrogen (Paisley, UK). PCR was performed as earlier described by de Groot and co-workers (2004). The RT-PCR products were cloned using the Genejet cloning kit (Fermentas). After transformation, a minimum of 32 colonies were selected for plasmid isolations. Sequencing reactions were performed using the BigDye terminator cycle sequencing kit, and samples were run on an automated capillary sequencing system (ABI Genetic Analyzer 3130XL). The sequences were analysed using SeqMan Pro (DNASTAR, Lasergene 8.1.2.), and alleles are based on at least three clones with identical sequences from different monkeys or independent PCRs from one monkey. To define loci and lineages, alignments of the sequences were made using the MacVector™ version 11.1.2 (Oxford Molecular Group). DRB-STR typing was performed according to published methods (de Groot et al. 2008; Doxiadis et al. 2010). In total, 11 Mafa-DRA and 56 Mafa-DRB sequences have been deposited in the EMBL database (Accession numbers FR717360–FR717426, Table 1) and have been officially designated by the IPD-MHC database (Robinson et al. 2003).
Table 1.
Mafa full-length DRA and DRB alleles detected in 58 animals
| Allele | Animal | Accession number |
|---|---|---|
| Mafa-DRA | ||
| DRA*01:01:01 | Bilboa | EF208826 |
| DRA*01:01:09 | Kraa | FR717418 |
| DRA*01:02:01:01 | Yukka | EF208827cx |
| DRA*01:02:05 a | Yabaa | FR717422 |
| DRA*01:02:20 | Trespa | FR717419 |
| DRA*01:02:21 | Kraa | FR717420 |
| DRA*01:03:01 | Vivaa | AM943638 |
| DRA*01:03:02 a | Riva | FR717425 |
| DRA*01:03:03 a | Bilboa | FR717421 |
| DRA*01:03:07 | Cyn83 | FR717417 |
| DRA*01:03:08 | Hoeba | FR717416 |
| DRA*01:09 a | Blo | FR717423 |
| DRA*01:10:01 | Kippa | FR717424 |
| DRA*01:10:02 | Joshua | FR717426 |
| DRA*02:01:01:01 | Clint | EF208828 |
| Mafa-DRB | ||
| DRB1*03:06:01 a | Zazaa | FR717409 |
| DRB1*03:08:01 | Kippa | FR717406 |
| DRB1*03:08:02 | Alfa | FR717373 |
| DRB1*03:09 a | Zola | FR717393 |
| DRB1*03:12:01 a | Bufo | FR717381 |
| DRB1*03:14 | Cyn83 | FR717369 |
| DRB1*03:15 | Cyn83 | FR717368 |
| DRB1*03:16 a | Friko | FR717400 |
| DRB1*03:17 a | Roza | FR717396 |
| DRB1*03:21 a | Yukka | FR717399 |
| DRB1*04:03 a | Yabaa | FR717380 |
| DRB1*04:11 | Vivaa | FR717374 |
| DRB1*10:02 a | Indy | FR717386 |
| DRB1*10:04 | Kippa | AF492283 |
| DRB1*10:10 a | Yukka | FR717398 |
| DRB3*04:01 a | Kraa | FR717372 |
| DRB4*01:01 | Clint | FR717382 |
| DRB4*01:02 a | Cyn81 | FR717363 |
| DRB4*01:03 a | Joshua | FR717414 |
| DRB5*03:01:01 a | Gayo | FR717383 |
| DRB5*03:01:02 a | Cyn80 | FR717384 |
| DRB5*03:04 a | Joshua | FR717415 |
| DRB5*03:05 a | Vip | FR717385 |
| DRB5*03:06 a | Just-So | FR717371 |
| DRB5*03:09 a | Zazaa | FR717408 |
| DRB5*03:15 | Cyn81 | FR717387 |
| DRB5*03:16 | Tabasco | FR717413 |
| DRB*W1:07 | Cyn82 | FR717365 |
| DRB*W1:08 | Juanita | FR717403 |
| DRB*W3:03:01 | Hippo | FR717370 |
| DRB*W3:04:01 a | Cyn80 | FR717361 |
| DRB*W3:05 | Cyn80 | FR717362 |
| DRB*W3:06 | Cyn82 | FR717366 |
| DRB*W4:05 a | Dojo | FR717391 |
| DRB*W5:01 | Indy | FR717376 |
| DRB*W6:06 | Cyn80 | FR717360 |
| DRB*W6:07 | Kippa | FR717405 |
| DRB*W7:02 | Geisha | FR717404 |
| DRB*W7:07 | Cyn82 | FR717367 |
| DRB*W21:01 a | Zola | FR717394 |
| DRB*W21:01 | Clint | FR717375 |
| DRB*W20:02 a | Canada | FR717410 |
| DRB*W20:02 a | Bufo | FR717364 |
| DRB*W25:04 a | Dojo | FR717390 |
| DRB*W25:05 | Nanaea | FR717389 |
| DRB*W25:06 | Canada | FR717411 |
| DRB*W26:02:01 | Cyn82 | FR717388 |
| DRB*W36:01 a | Dojo | FR717395 |
| DRB*W36:04 | Alfa | FR717378 |
| DRB*W37:01 a | Yabaa | FR717379 |
| DRB*W40:01 | Friko | FR717401 |
| DRB*W49:01 | Hippo | FR717377 |
| DRB*W49:01:02 | Jura | FR717412 |
| DRB*W53:01 a | Jena | FR717407 |
| DRB*W66:01 | Rassoa | FR717402 |
| DRB*W67:01 a | Roza | FR717397 |
| DRB*W68:01 a | Nanaea | FR717392 |
aExtension of existing alleles. Previously unreported alleles are depicted in bold
Results and discussion
Definition of DR haplotypes
The 52 pedigreed cynomolgus macaques included in this study are members of 12 families comprising two to six generations, which belong to a self-sustaining breeding colony; an example of a pedigreed family has been provided (Fig. 1). Therefore, segregation analyses of the respective alleles within the macaque families allowed the definition of DR haplotypes. In some cases, DR haplotypes detected in family members have been confirmed by the presence of identical alleles defined by analyses of unrelated animals.
Fig. 1.
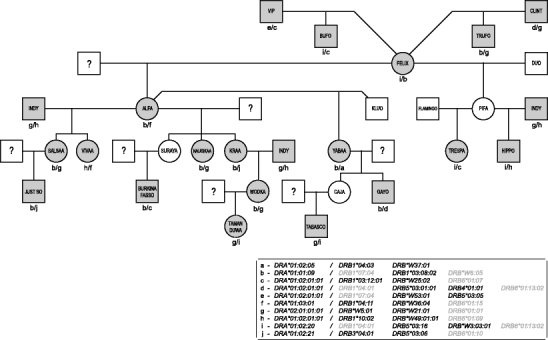
Pedigree of one cynomolgus family with the segregation of DRA/DRB haplotypes indicated. The analyzed animals are marked by shading; a question mark indicates that the sire could not be identified. Transcribed genes are depicted in bold
DRA polymorphism
Within the panel of Indonesian and Indochinese cynomolgus macaques, 15 different DRA alleles could be defined, seven of which had not been previously described (Table 1, bold). All but one of these alleles belong to one lineage: Mafa-DRA*01. In contrast to the HLA-DRA gene, of which only three alleles are documented, the Mafa-DRA gene is polymorphic, and the degree of its polymorphism appears also to be higher than in a thoroughly studied Indian rhesus macaque population (de Groot et al. 2004). However, the variations between these alleles are mainly due to synonymous substitutions, thus indicating a strong purifying selection operating on the gene and the exons specifying the antigen biding site (Hughes and Nei 1989). The DRA*01 alleles detected in our cohort give rise to only five different amino acids replacements, which are either situated in the leader sequence or within the transmembrane part of the molecule. These results are comparable to analyses of cynomolgus monkeys of other origins (Aarnink et al. 2010; O’Connor et al. 2007).
In contrast to the polymorphic DRA*01 lineage, only one allele of the second lineage, DRA*02, has been detected in our cohort. In contrast to the Mafa-DRA*01 allotypes, the DRA*02 lineage is typified by five amino acid replacements, two in the leader section and three that map to the alpha 1 domain that defines the scaffolding of the antigen-binding site (Table 2). An identical or similar allele has been detected in Chinese rhesus macaques (Doxiadis et al. 2008) and in the pigtailed macaque (Aarnink et al. 2010; O’Connor et al. 2007). Thus, DRA*02 seems to be an evolutionarily old entity. Within our cohort, the Mafa-DRA*02:01:01:01 allele is always present in cis configuration with a certain DRB haplotype (Table 3, #28) encoding a DRB*W21:01 and DRB*W5:01 allotype. This haplotype is present in monkeys from Indochina as well as in animals from the Indonesian islands (Doxiadis et al. 2010) and Mauritius (Aarnink et al. 2010). Furthermore, the Mafa-DRA*02 allele has been observed in animals originating from the Philippines together with another DRB configuration that also harbours a DRB*W5:01 allele (Aarnink et al. 2010). Therefore, a steric preference of the Mafa-DRA*02-encoded α chain for the DRB*W5:01-encoded β chain to form a stable molecule seems to be plausible. Although the amino acid changes within the alpha-1 domain of the DRA*02 chain are conservative, an additional possibility that the resulting DR molecule is able to present a set of peptides, which are advantageous in controlling certain pathogens, cannot be excluded.
Table 2.
Polymorphic amino acid sites of Mafa-DRA
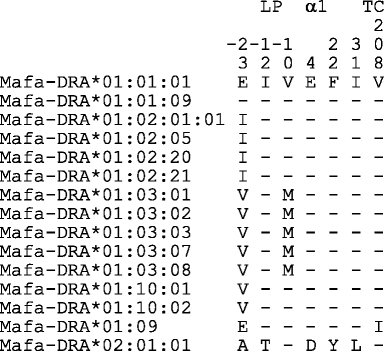
Table 3.
Mafa-DR haplotypes defined by exon 2 and full-length sequencing
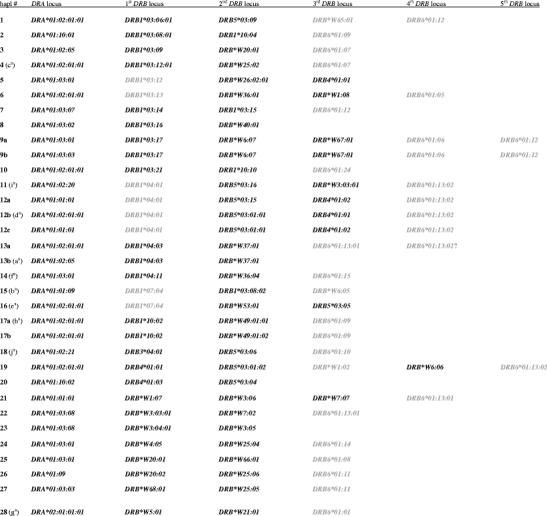
aDesignation of Fig. 1. Transcribed genes are depicted in bold
Mafa-DR haplotypes
As has been shown recently by means of microsatellite and exon 2 typing, cynomolgus macaques show abundant levels of DRB region configuration polymorphisms: that is, haplotypes that vary in the number and content of DRB genes (de Groot et al. 2008; Doxiadis et al. 2010). Most of the haplotypes encode three DRB genes or pseudogenes belonging to different loci/lineages, but haplotypes with two, four, and five loci are also observed. However, until now it had been unclear which of these alleles are transcribed and, as such, encode potential bona fide gene products. The subsequent full-length DRB sequencing of RT-PCR products of our cynomolgus macaque panel revealed a total of 57 DRB alleles. As can be expected, most of the alleles that were discovered are extensions of DRB alleles defined by exon 2 typing (Table 1). Additionally, 11 previously unreported alleles have been detected during the course of this study (Table 1, bold). Some alleles differ in exons other than exon 2 — e.g., DRB*W7:07 and DRB*W7:02 — demonstrating that exon 2 typing may not always be sufficient for an unambiguous allele definition. At this stage, it is not understood to what extent polymorphisms in exon 3 may affect actual peptide binding.
In family studies, the segregation of alleles on one chromosome has been determined, and 28 DR region configurations have been defined (Table 3; letters in brackets refer to the respective haplotype of Fig. 1). As observed in rhesus macaques, only a few region configurations show limited allelic variation for their DRA and/or DRB genes (Table 3, #9a/9b; #12a/12b/12c; #13a/13b; #17a/17b). In humans, HLA class II-mediated immune responses may differ between individuals due to allelic polymorphism. In macaque populations, the strategy is fundamentally different, as allelic variation within a region configuration is virtually absent. The actual outcome is more or less the same, as macaques display abundant region configuration polymorphism at the population level.
With one exception, all Mafa-DRB haplotypes are linked to alleles of the DRA*01 lineage, and only one DRB region configuration is associated with the DRA*02 lineage (Table 3, #28). Per haplotype, one to three DRB genes are transcribed, resembling the situation observed in rhesus macaques (de Groot et al. 2004). The DRB transcription products of a certain haplotype belong mostly to alleles of different loci/lineages. There are, however, region configurations (e.g., Table 3, #7), which encode two allelic transcripts of the same DRB lineage and are therefore probably the result of a recombination process.
As in other primate species such as rhesus macaques, humans, and chimpanzees, DRB6 always remains untranscribed, and thus is confirmed to be a pseudogene. However, alleles from various other loci/lineages are also not detected at the transcription level (Table 3, grey). As has been shown in previous studies of the rhesus macaque, some alleles that group in the same lineage as, for example, DRB1*03 (Table 3, #1–10), may be transcribed, whereas others are not detected at the transcription level. Notably, however, is the observation that Mafa-DRB alleles, which are identical for exon 2, may be either transcribed (Table 3, #6, DRB*W1:08) or untranscribed (Table 3, #19, DRB*W1:02). Additionally, a certain allele may be observed as a transcript in the context of one region configuration (Table 3, #4, DRB1*03:12:01), whereas it remains untranscribed as a member of another configuration (Table 3, #5, DRB1*03:12). A further example is provided by allele DRB1*04:01, which is detected in two different region configurations (Table 3, #11 and 12) in our cohort and appears to be untranscribed. In another study (Blancher et al. 2006), the same allele is defined on a cDNA level; here too, the region configuration, in which the authors detected the DRB1*04:01 allele, is different from configurations #11 and 12 (a, b, c) (Table 3) of our cohort. In configuration #12, we cannot exclude the possibility that the discussed alleles may have mutations outside exon 2. However, the fact that the same or closely related alleles are either pseudogenes or encode bona fide transcripts appears to be dependent on the region configuration in which they are situated: for instance, their surroundings on the genome. In cynomolgus macaques, an undocumented high level of DRB region configuration-associated diversity has been described (Doxiadis et al. 2010). Since these region configurations appear to be generated by recombination-like events, it seems plausible that genes may be placed next to or far away from a promotor/enhancer region so that transcription may be switched on or off. Future studies relating to the whole genome sequencing of several macaque MHC haplotypes will help to answer these questions.
Acknowledgements
The authors wish to thank Donna Devine for editing the manuscript and Henk van Westbroek for preparing the figures. This study was supported in part by NIH/NIAID contract numbers HHSN266200400088C/NOI-AI-0088 and 5R24RR016038-05 (CFA:03.9389).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
G. Doxiadis and A. de Vos-Rouweler contributed equally to the manuscript.
References
- Aarnink A, Estrade L, Apoil PA, Kita YF, Saitou N, Shiina T, Blancher A. Study of cynomolgus monkey (Macaca fascicularis) DRA polymorphism in four populations. Immunogenetics. 2010;62:123–136. doi: 10.1007/s00251-009-0421-8. [DOI] [PubMed] [Google Scholar]
- Aoyama A, Ng CY, Millington TM, Boskovic S, Murakami T, Wain JC, Houser SL, Madsen JC, Kawai T, Allan JS. Comparison of lung and kidney allografts in induction of tolerance by a mixed-chimerism approach in cynomolgus monkeys. Transplant Proc. 2009;41:429–430. doi: 10.1016/j.transproceed.2008.08.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benferhat R, Martinon F, Krust B, Le Grand R, Hovanessian AG. The CBD1 peptide corresponding to the caveolin-1 binding domain of HIV-1 glycoprotein gp41 elicits neutralizing antibodies in cynomolgus macaques when administered with the tetanus T helper epitope. Mol Immunol. 2009;46:705–712. doi: 10.1016/j.molimm.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Blancher A, Tisseyre P, Dutaur M, Apoil PA, Maurer C, Quesniaux V, Raulf F, Bigaud M, Abbal M. Study of Cynomolgus monkey (Macaca fascicularis) MhcDRB (Mafa-DRB) polymorphism in two populations. Immunogenetics. 2006;58:269–282. doi: 10.1007/s00251-006-0102-9. [DOI] [PubMed] [Google Scholar]
- Blancher A, Bonhomme M, Crouau-Roy B, Terao K, Kitano T, Saitou N. Mitochondrial DNA sequence phylogeny of 4 populations of the widely distributed cynomolgus macaque (Macaca fascicularis fascicularis) J Hered. 2008;99:254–264. doi: 10.1093/jhered/esn003. [DOI] [PubMed] [Google Scholar]
- Bonhomme M, Doxiadis GG, Heijmans CM, Vervoort V, Otting N, Bontrop RE, Crouau-Roy B. Genomic plasticity of the immune-related Mhc class I B region in macaque species. BMC Genom. 2008;9:514. doi: 10.1186/1471-2164-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuano SV, 3rd, Croix DA, Pawar S, Zinovik A, Myers A, Lin PL, Bissel S, Fuhrman C, Klein E, Flynn JL. Experimental Mycobacterium tuberculosis infection of cynomolgus macaques closely resembles the various manifestations of human M. tuberculosis infection. Infect Immun. 2003;71:5831–5844. doi: 10.1128/IAI.71.10.5831-5844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot N, Doxiadis GG, De Groot NG, Otting N, Heijmans C, Rouweler AJ, Bontrop RE. Genetic makeup of the DR region in rhesus macaques: gene content, transcripts, and pseudogenes. J Immunol. 2004;172:6152–6157. doi: 10.4049/jimmunol.172.10.6152. [DOI] [PubMed] [Google Scholar]
- de Groot N, Doxiadis GG, de Vos-Rouweler AJ, de Groot NG, Verschoor EJ, Bontrop RE. Comparative genetics of a highly divergent DRB microsatellite in different macaque species. Immunogenetics. 2008;60:737–748. doi: 10.1007/s00251-008-0333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxiadis GG, Rouweler AJ, de Groot NG, Louwerse A, Otting N, Verschoor EJ, Bontrop RE. Extensive sharing of MHC class II alleles between rhesus and cynomolgus macaques. Immunogenetics. 2006;58:259–268. doi: 10.1007/s00251-006-0083-8. [DOI] [PubMed] [Google Scholar]
- Doxiadis GG, de Groot N, de Groot NG, Doxiadis II, Bontrop RE. Reshuffling of ancient peptide binding motifs between HLA-DRB multigene family members: old wine served in new skins. Mol Immunol. 2008;45:2743–2751. doi: 10.1016/j.molimm.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Doxiadis GG, de Groot N, de Groot NG, Rotmans G, de Vos-Rouweler AJ, Bontrop RE. Extensive DRB region diversity in cynomolgus macaques: recombination as a driving force. Immunogenetics. 2010;62:137–147. doi: 10.1007/s00251-010-0422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene JM, Lhost JJ, Burwitz BJ, Budde ML, Macnair CE, Weiker MK, Gostick E, Friedrich TC, Broman KW, Price DA, O’Connor SL, O’Connor DH. Extralymphoid CD8+ T cells resident in tissue from simian immunodeficiency virus SIVmac239{Delta}nef-vaccinated macaques suppress SIVmac239 replication ex vivo. J Virol. 2010;84:3362–3372. doi: 10.1128/JVI.02028-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirakhoo F, Pugachev K, Zhang Z, Myers G, Levenbook I, Draper K, Lang J, Ocran S, Mitchell F, Parsons M, Brown N, Brandler S, Fournier C, Barrere B, Rizvi F, Travassos A, Nichols R, Trent D, Monath T. Safety and efficacy of chimeric yellow Fever–dengue virus tetravalent vaccine formulations in nonhuman primates. J Virol. 2004;78:4761–4775. doi: 10.1128/JVI.78.9.4761-4775.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc Natl Acad Sci USA. 1989;86:958–962. doi: 10.1073/pnas.86.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekutis C, Letvin NL. Biochemical and molecular characterization of rhesus monkey major histocompatibility complex class II DR. Hum Immunol. 1995;43:72–80. doi: 10.1016/0198-8859(94)00155-J. [DOI] [PubMed] [Google Scholar]
- Leuchte N, Berry N, Kohler B, Almond N, LeGrand R, Thorstensson R, Titti F, Sauermann U. MhcDRB-sequences from cynomolgus macaques (Macaca fascicularis) of different origin. Tissue Antigens. 2004;63:529–537. doi: 10.1111/j.0001-2815.2004.0222.x. [DOI] [PubMed] [Google Scholar]
- Ma A, Xiong Z, Hu Y, Qi S, Song L, Dun H, Zhang L, Lou D, Yang P, Zhao Z, Wang X, Zhang D, Daloze P, Chen H. Dysfunction of IL-10-producing type 1 regulatory T cells and CD4(+)CD25(+) regulatory T cells in a mimic model of human multiple sclerosis in Cynomolgus monkeys. Int Immunopharmacol. 2009;9:599–608. doi: 10.1016/j.intimp.2009.01.034. [DOI] [PubMed] [Google Scholar]
- Mee ET, Badhan A, Karl JA, Wiseman RW, Cutler K, Knapp LA, Almond N, O’Connor DH, Rose NJ. MHC haplotype frequencies in a UK breeding colony of Mauritian cynomolgus macaques mirror those found in a distinct population from the same geographic origin. J Med Primatol. 2009;38:1–14. doi: 10.1111/j.1600-0684.2008.00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mee ET, Berry N, Ham C, Sauermann U, Maggiorella MT, Martinon F, Verschoor EJ, Heeney JL, Le Grand R, Titti F, Almond N, Rose NJ. Mhc haplotype H6 is associated with sustained control of SIVmac251 infection in Mauritian cynomolgus macaques. Immunogenetics. 2009;61:327–339. doi: 10.1007/s00251-009-0369-8. [DOI] [PubMed] [Google Scholar]
- O’Connor SL, Blasky AJ, Pendley CJ, Becker EA, Wiseman RW, Karl JA, Hughes AL, O’Connor DH. Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics. 2007;59:449–462. doi: 10.1007/s00251-007-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SG. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–314. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Wang H, Hou S, Hu S, Fan K, Fan X, Zhu T, Guo Y. DRB genotyping in cynomolgus monkeys from China using polymerase chain reaction-sequence-specific primers. Hum Immunol. 2007;68:135–144. doi: 10.1016/j.humimm.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Wiseman RW, Karl JA, Bimber BN, O’Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, Szekeres E, Jr, Wright C, Harkins T, O’Connor DH. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med. 2009;15:1322–1326. doi: 10.1038/nm.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcechowskyj JA, Yant LJ, Wiseman RW, O’Connor SL, O’Connor DH. Control of simian immunodeficiency virus SIVmac239 is not predicted by inheritance of Mamu-B*17-containing haplotypes. J Virol. 2007;81:406–410. doi: 10.1128/JVI.01636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


