Abstract
DNA microarrays have become one of the most powerful tools in the field of genomics and medical diagnosis. Recently, there has been increased interest in combining microfluidics with microarrays since this approach offers advantages in terms of portability, reduced analysis time, low consumption of reagents, and increased system integration. Polymers are widely used for microfluidic systems, but fabrication of microarrays on such materials often requires complicated chemical surface modifications, which hinders the integration of microarrays into microfluidic systems. In this paper, we demonstrate that simple UV irradiation can be used to directly immobilize poly(T)poly(C)-tagged DNA oligonucleotide probes on many different types of plastics without any surface modification. On average, five- and fourfold improvement in immobilization and hybridization efficiency have been achieved compared to surface-modified slides with aminated DNA probes. Moreover, the TC tag only costs 30% of the commonly used amino group modifications. Using this microarray fabrication technique, a portable cyclic olefin copolymer biochip containing eight individually addressable microfluidic channels was developed and used for rapid and parallel identification of Avian Influenza Virus by DNA hybridization. The one-step, cost-effective DNA-linking method on non-modified polymers significantly simplifies microarray fabrication procedures and permits great flexibility to plastic material selection, thus making it convenient to integrate microarrays into plastic microfluidic systems.
Electronic supplementary material
The online version of this article (doi:10.1007/s00216-011-5459-4) contains supplementary material, which is available to authorized users.
Keywords: Microarray, Microfluidic, Non-modified plastic, UV, DNA immobilization
Introduction
DNA microarray, in which tens to thousands of different molecular reporter groups are attached at preset locations on a solid support, has become one of the most powerful tools in the field of genomics and medical diagnosis as it enables molecular analysis in a highly parallel fashion [1]. Microfluidics, as another emerging technology in the last decade, have also received significant attention lately due to their ability to analyze small volumes of samples and reduce reaction and processing times [2]. Recently, there has been increased interest in merging the two technologies since this approach offers advantages in terms of portability, analysis speed, system integration and automation [3]. To facilitate the integration of microarrays with microfluidic devices, materials must be carefully chosen so that they can be easily fabricated on the size-scale demanded by the microfluidic application and at the same time, allows for stable attachment of oligonucleotide probes. Glass is a well-established substrate material for microarrays [4, 5], however, microfabrication of glass devices is costly and time-consuming [6]. Developing polymeric materials as alternative solid supports for making DNA microarrays is then of particular interest since plastics are of low-cost and amenable to high volume manufacturing processes [7].
Several polymers have been investigated as potential supports for microarray production including cyclic olefin copolymer (COC) [8], poly (methyl methacrylate) (PMMA) [9–11], poly(dimethylsiloxane) (PDMS) [12], polycarbonate (PC) [13], polystyrene and poly(ethylene terephthalate) (PET) [14]. As most plastics do not possess a suitable interfacial chemical structure required for immobilizing DNA molecules on their surfaces, different attachment methodologies have to be implemented to immobilize probes to these supports. Typical methods involve the generation of active functional groups on the surface that react covalently with a chemical modification of the probe DNA. Plastic-based microarrays have been demonstrated on amine-modified or copolymerized substrates using thiol-, amino- or acrylamide-labeled capture probes [8, 15]. To avoid the extra steps involved in surface modification, alternative methods have also been developed for fabricating arrays with non-modified plastic surfaces. DNA oligonucleotide probes were shown to be immobilized on non-modified plastic substrates through SN2 reaction [16], commercial binding buffers [17] or more conveniently, direct attachment by UV exposure [9, 13, 14]. Li et al. [13] irradiated PC with UV/ozone to facilitate the attachment of amino-modified DNA probes. Kimura et al. [14] reported UV-induced attachment of DNA strands modified with poly(dT) and an undisclosed linker sequence, to PC, PMMA, and PET. These studies demonstrate that UV irradiation could successfully convert inert plastics into bio-reactive substrates for DNA immobilization/hybridization. However, amino modification of DNA oligonucleotides makes DNA probes approximately three times as expensive and the linker sequence by Kimura et al. [14] was not disclosed, making replication of the method impossible.
Our previous studies have shown that oligonucleotide probes containing a poly(T)10-poly(C)10 tag (TC tag) could be directly immobilized onto unmodified and agarose-coated glass [18–20] and unmodified PMMA surfaces [9] by UV linking and still function as capture probes for microarray genotyping analysis. A notable advantage of this immobilization strategy is that the TC tags only cost 30% of the commonly used amino group modifications. The mechanism for binding DNA to non-modified surfaces is not clear, but it is highly dependent on the combination of both T and C in the tag as no or very weak signals were observed when using oligonucleotide probes with either no tag, a T20 tag or a C20 tag [20]. It is known that UV irradiation can activate substrates with a high surface density of functional groups [13]. We assume that the glass or plastic surfaces were activated and the TC tags were immobilized to the substrates during UV exposure.
In this paper, we further demonstrated that this simple and cost-effective DNA immobilization technique could be expanded as a general method to produce microarrays on various native plastic surfaces. Characterization on four popular plastics including COC, PC, PMMA and PDMS showed that the method offered higher immobilization as well as higher hybridization efficiency than conventional immobilization techniques. As combining microarrays and microfluidics can offer attractive advantages to bioassays, a portable COC microarray device containing eight individually addressable microfluidic channels were manufactured for fast and parallel identification of Avian Influenza Virus (AIV) by DNA hybridization. The one-step DNA-linking method significantly simplifies microarray fabrication procedures and permits great flexibility with respect to material selection, thus facilitating the integration of DNA microarrays with polymeric microfluidic devices and providing enormous potential for the wide applications in biological research and clinical diagnosis.
Experimental
Viral strains
Three inactivated AIV strains namely H1N1 A/DK/ALB 35/76, H5N1 A/CK/Scotland/59 06.04.67 and H7N5 A/Chick/Nether/2993–17/03 AV 506/03, and one strain of Newcastle diseases virus (NDV) were used to check the specificity of the DNA array. Hemagglutination (HA) titre values were measured to be 1:64. All strains were kindly supplied by the National Veterinary Institute, Technical University of Denmark.
Primer and probes
Table 1 shows a list of primers and probes used for AIV identification and subtyping. The Matrix (M) gene-specific primers and probes were designed for general screening of all type A influenza viruses [21]. H5 and H7 primers and probes target at the H5 and H7 regions on hemagglutinin (HA) gene and were designed to specify the subtypes of influenza virus with avian origin. Each forward primer was Cy3-labeled at the 5′ end for visualization. The probes were modified at the 5′ end with a poly (T)10-poly (C)10 tail to facilitate the attachment to the plastic substrates as previously described by Dufva et al. [20]. All the oligonucleotide primers and probes were synthesized at DNA Technology A/S, Denmark.
Table 1.
List of specific primers and probes for AIV identification and subtyping
| Type or subtype | Target gene | Primers and probes | Sequences (5′–3′) | Amplicon (bp) |
|---|---|---|---|---|
| A | Matrix | Forward DB-MF | Cy3-AGA TGA GTC TTC TAA CCG AGG TCG | 100 |
| Reverse DB-MR | TGC AAA AAC ATC TTC AAG TCT CTG | |||
| M gene probea | TTTTTTTTTTCCCCCCCCCC TCA GGC CCC CTC AAA GCC GA | |||
| H5 | HA | Forward DB-H5LH1 | Cy3-ACA TAT GAC TAC CCA CAR TAT TCA G | 151 |
| Reverse DB-H5RH1 | AGA CCA GCT AYC ATG ATT GC | |||
| H5 probe | TTTTTTTTTTCCCCCCCCCC TCW ACA GTG GCG AGT TCC CTA GCA | |||
| H7 | HA | Forward DB-LH6H7 | Cy3-GGC CAG TAT TAG AAA CAA CAC CTA TGA | 131 |
| Reverse DB-R4H7 | GCC CCG AAG CTA AAC CAA AGT AT | |||
| H7 probe | TTTTTTTTTTCCCCCCCCCC CCG CTG CTT AGT TTG ACT GGG TCA ATC T |
aProbe used in immobilization and hybridization quantification: Cy5-TTTTTTTTTTCCCCCCCCCC TCA GGC CCC CTC AAA GCC GA
RNA extraction and RT-PCR
Isolation of RNAs from virus strains was performed using the RNeasy Kit (Qiagen, Germany) according to the manufacturer’s instruction. The RNAs were eluded using RNase free water. The isolated RNAs were amplified by reverse transcription (RT) PCR. Multiplex RT-PCR was performed using a RT-PCR kit (Qiagen, Hilden, Germany). Briefly, a 25-μl reaction mixture was prepared with 5 μl of 5× RT-PCR buffer, 1 μl of 10 mM DNTP mix, 1 μl of enzyme mix, 5 μl of RNA sample, and three pairs of primers (Table 1), each at a final concentration of 0.2 μM. PCR was carried out in a thermal cycler (MJ Research Inc., MA, USA) and the cycling protocol consisted of 30 min at 50 °C for reverse transcription, 15 min at 95 °C for enzyme activation, followed by 40 cycles of 10 s at 95 °C, 30 s at 54 °C and 10 s at 72 °C, finally 5 min at 72 °C for extension.
Preparation of DNA microarrays on plastic substrates and in the microfluidic chip
One-millimeter-thick non-modified PC (SUSTANAT PC, Röchling Engineering Plastics, Dallas, NC, USA) and PMMA (PLEXIGLAS® XT, Evonik Industries AG, Germany) sheets were purchased from Nordiskplast, Denmark and cut into 76 × 26-mm-sized slides. COC slides were injection molded using Topas COC resin with glass transition temperature of 130° (5013, TOPAS Advanced Polymers GmbH, Germany). PDMS slides were fabricated by spin coating a 10:1 mixture of PDMS prepolymer and curing agent (Sylgard 184, Dow Corning, MI, USA) on a glass slide and then curing for 1 h at 65 °C.
DNA microarrays were produced on the four non-modified plastic slides by UV cross-linking. The Cy5-labeled M probe with a poly(T) 10-poly(C) 10 tail was diluted in 150 mM sodium phosphate buffer (pH 8.5) containing 0.004% Triton X to a final concentration from 2.5 to 25 μM. Spotting was performed using a non-contact array nano-plotter 2.1 (GeSim, Dresden, Germany) with a Picoliter pin that deposited 100 pl/spot. For each experiment condition tested on the microarrays, ten replica spots of each solution were spotted on the same substrate and the experiment was repeated five times. The spots were allowed to dry and then exposed to UV irradiation at 254 nm with power of 3 mW/cm2 for 10 min (Stratalinker 2400, Stratagene, CA, USA). Subsequently, the plastic slides were washed under agitation in 0.1× standard saline citrate (SSC) with 0.1% (w/v) sodium dodecyl sulfate (SDS; Promega, WI, USA) solution for 10 min, then rinsed in deionized water and dried by nitrogen.
To integrate DNA microarrays within a microfluidic device, an eight-chamber microfluidic biochip was fabricated on a COC slide by computer controlled milling (Folken Ind, Glendale, California, USA). Each chamber had dimensions of 5 mm (length) × 4 mm (width) × 0.5 mm (height), corresponding to a volume of 10 μL. Microfluidic channels with widths of 500 μm and depths of 200 μm led from the chambers and connected these to 0.8 mm diameter inlet and outlet through holes. Using the same protocol as described above, DNA microarrays containing probes specific to the M gene of AIV type A as well as H5 and H7 subtypes were immobilized within the microchambers of the COC chip. The three probes were diluted in the same spotting buffer to a final concentration of 20 μM. The array was spotted at the center of the microchamber and each probe was repeated four times for easy identification of the reaction products. After immobilizing the DNA microarrays in each chamber, the COC slide was lidded with a 200-μm-thin COC film using a bonding press (P/O/Weber, Remshalden, Germany) at 120 °C and 2 bars for 10 min.
Hybridization
In passive hybridization, Cy3-labeled complementary strand of the M probe was used as target DNA and was diluted to a final concentration ranging from 1 pM to 1 μM in the Perfect Hybridization Buffer (Sigma-Aldrich, MO, USA). 20 μl samples were loaded directly on the oligonucleotide microarrays and cover slips were mounted to seal the reaction droplets. The plastic slides were then transferred into an oven where the hybridization was performed for 1 h at 37 °C. After hybridization, the slides were washed with agitation in 0.1× SSC/0.1% SDS for 10 min, followed by a short rinse in deionized water.
For microfluidic flow-through hybridization, the COC microchip was interfaced to MainSTREAM microfluidic platform components [22] including micropump [23], reservoir chip, and outlet interconnection component solutions [24]. RT-PCR products of different viral strains were run in parallel through the microchambers at a flow rate of 1 μL/min using the eight-channel micropump. In each microchamber, PCR product of the individual viral strain was hybridized with the DNA microarray at 37 °C for 10 min. Fluorescent images of the microchambers after hybridization were taken with a microscope (Olympus Corp, Japan).
DNA immobilization and hybridization quantification
After each step of spotting, immobilization and hybridization, the microarrays on non-modified plastic slides were scanned by ScanArray Lite (Packard Bioscience, MA, USA), with the same laser power and PMT settings. ScanArray software (Packard Bioscience, MA, USA) was used to quantify the spots by calculating the average pixel intensity inside the defined spots. Standard curves for probe immobilization were prepared by diluting fluorescent Cy5-labeled DNA probes in MilliQ water to a final concentration ranging from 1 to 25 μM. Each dilution was spotted in ten replicates on non-modified plastics and the fluorescence signals were recorded after UV-treatment but before washing away unbound probes. The same procedure was adopted for the hybridization standard curves while the range of dilutions for the Cy3-labeled DNA target was from 0.025 to 5 μM and the fluorescence signals were obtained directly after spotting (See Electronic Supplementary Material Fig. S1). To quantify the immobilization and hybridization, the amount of immobilized and hybridized DNA was obtained from the respective calibration curves.
Results and discussion
Immobilization of TC-tagged probes on non-modified plastic slides
Immobilization of TC-tagged DNA probes by UV irradiation on non-modified and agarose-coated glass surfaces [19, 20, 25], as well as non-modified PMMA [9] have been previously demonstrated. In this study, we explored the possibility of immobilizing TC-tagged probes on additional polymeric materials. Spot quality and immobilization efficiency were characterized for various non-modified plastics.
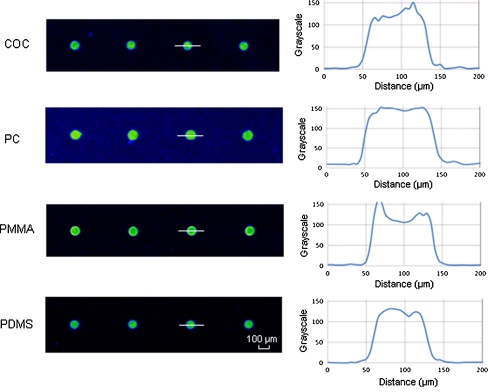
The Cy5-labeled 20-base M gene probe with a TC tag was diluted in spotting buffer to a concentration of 20 μM and spotted on untreated COC, PC, PMMA and PDMS slides. These materials were selected as they are most popular in microfluidic applications due to their wide availability in a variety of grades with high optical transmission in the visible wavelengths, good solvent and chemical compatibility, and well-characterized molding parameters [26]. Following UV irradiation and washing away unbound probes, microarrays on the four plastic surfaces were scanned (Fig. 1). DNA underwent UV-induced binding to all four plastics, demonstrating the high versatility of this immobilization method. As seen from the image profiles, highly homogenous spots were achieved. This was attributed to the addition of the proper amount of Triton X to the spotting buffer. Printing pure PBS on hydrophobic plastic surfaces led to shrinkage of the droplets during evaporation, resulting in a small spike at the center of the initial drop area. The surfactant lowers the initial contact angle of the deposited solution, facilitating homogeneous repartitioning of the immobilized probe molecules. However, too high a concentration of Triton X was not desirable either, as it would cause the pattern of coffee rings similar to those observed by Sabourin et al. when using 0.01% Triton X in the spotting buffer [9]. PBS with 0.004% Triton X was found to be the optimized spotting buffer in order to achieve uniform distribution of probes (see Electronic Supplementary Material Fig. S2). The maximum spot-to-spot variation in terms of spot size and florescence signal after immobilization was 3% and 8%, respectively. Good-quality spots with high intra- and inter-spot homogeneities were printed on the plastic substrates, which is important as spot quality is crucial to the reliability of microarray analysis.
Fig. 1.
Spot morphology of DNA probe immobilized on different native plastic surfaces. The Cy5-labeled M probe was attached with a poly(T)10-poly(C)10 tag at the 5′ end. The probe was diluted in 150 mM sodium phosphate buffer with 0.004% Triton to a concentration of 20 μM and spotted on plastic slides using a non-contact array spotter. After exposing to UV irradiation for 10 min, the slides were washed and scanned using laser scanner. Image profiles were obtained by measuring the gray scale of pixels across the center lines
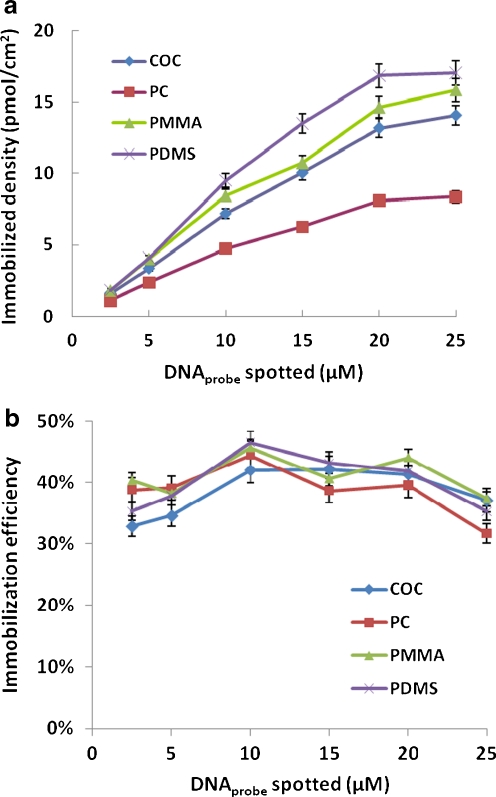
The immobilization efficiency of DNA probes achieved on different plastic surfaces was evaluated. The M probe was spotted in concentrations ranging from 2.5 to 25 μM on the four plastic substrates. The immobilization density increased linearly with the concentration of the spotting solution and reached saturation gradually (Fig. 2a). The maximum probe attachment was achieved using DNA concentrations of 20 μM or higher in the spotting buffer. The DNA immobilization efficiency was investigated by comparing the surface densities of immobilized DNA (i.e., after washing away unbound probes) and initially spotted DNA (i.e., before washing), which represented the ratio of the amount of attached probes to the actual spotted probes. Depending on materials and spotting buffer concentration, the immobilization efficiency of this simple and robust method was between 30% and 50% (Fig. 2b), fivefold higher in average when compared to DNA immobilization using amino-labeled probes on chemically aminated PMMA slides [15] or thiolated DNA probes attached on non-modified PMMA substrate [16] under the same experimental condition.
Fig. 2.
a Immobilized densities of TC-tagged M probe on four non-modified plastic slides with spotting buffer concentrations ranging from 2.5 to 25 μM. Fluorescence signals were quantified and compared to standard curves to obtain the probe densities on the surfaces (see “Experimental” section). b Immobilization efficiency achieved on four plastics as functions of spotted probe concentrations. The immobilization efficiency was calculated from the ratio of surface densities of immobilized DNA and initially spotted DNA. Each experiment was repeated three times and error bars represent standard deviations
The one-step, cost-effective microarray fabrication method is applicable to various non-modified plastics and results in both high spot quality and high immobilization efficiency. As such, the method significantly simplifies microarray fabrication procedures and permits great flexibility with respect to material selection.
DNA hybridization
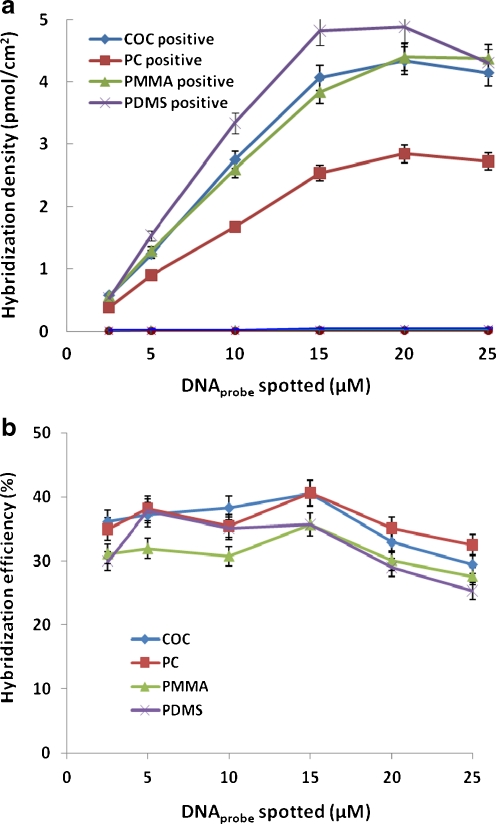
Retaining accessibility and selectivity of the surface-bound probes for complementary targets during hybridization was a concern in this work as UV light and radiation can damage DNA. To assess the specificity of the immobilization method, increasing concentrations of TC-tagged M probes ranging from 2.5 to 25 μM were directly attached to the four non-modified plastics by UV. The DNA probes were then hybridizing with 1 μM Cy3-labeled complementary and non-complementary DNA targets. The surface densities of hybridized DNA at different probe concentrations were quantified (Fig. 3a). Unspecific hybridization did not occur as negligible signals were obtained with the non-complementary fluorescent DNA. The signal-to-noise ratio (SNR) observed between the signal from the specific and unspecific binding was 120 in average, showing the functionality and selectivity of DNA oligonucleotide probes were well maintained on the plastic surfaces despite of UV irradiation.
Fig. 3.
a The surface densities of hybridized DNA on four plastic substrates. TC-tagged M probes ranging from 2.5 to 25 μM were directly attached to the slides by UV. The DNA probes were then hybridizing with 1 μM Cy3-labeled complementary and non-complementary DNA targets for 1 h. Fluorescence signal of spots were quantified and the amount of hybridized DNA was extracted from the calibration curve. b Hybridization efficiency as functions spotted DNA probe concentrations. The hybridization efficiency is defined as ratio of surface densities of hybridized and immobilized DNA. Each experiment was repeated three times and error bars represent standard deviations
Relatively high hybridization densities were achieved on all of the four substrates. For all the four substrates studied, the hybridization signals increased linearly with the concentration of the spotted probe solution until 15 μM. The binding capacity of the substrate surfaces was nearly saturated at the spotting solution concentration of 20 μM and started to decline above that, showing that high surface coverage of DNA probes did not consequently lead to the formation of more hybrids. This might be due to the repulsive electrostatic and steric interactions which were increasing with probe density [27]. Hybridization efficiency, calculated from the ratio of hybridized target DNA density and immobilized probe density, was used to determine the percentage of attached probes that participated in hybridization (Fig. 3b). In average, 30% to 40% of immobilized probes were involved in hybridization, fourfold higher in average than use of aminated or thiolated DNA probes [15, 16]. It is unclear why higher hybridization efficiency was obtained here as compared to previous results. Possible explanations are different oligo sequences and hybridization buffers employed of a spacer effect of the TC tag. Molecular spacers lifting the oligo up in solution is a well-known method to increase signal by avoiding surface effects [28, 29].
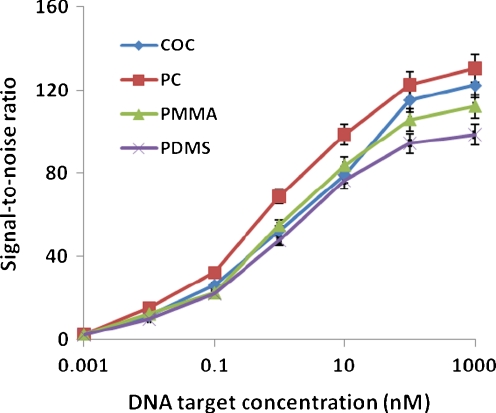
To quantify the target to probe hybridization sensitivity, 20 μM TC-tagged M probe was spotted on all of the four plastic substrates and these arrays were hybridized with increasing concentration of a Cy3-labeled complementary target ranging from 0.001 to 1,000 nM. As shown in Fig. 4, the fluorescent signals showed a trend to reach plateau at a target concentration of 100 nM for all substrates. The lowest detection limit was 0.01 nM where the signal-to-noise ratio was around 12 in average. The linear response range was from 0.1 to 100 nM. These results showed that the microarrays fabricated on non-modified plastics using TC-tagged probes could provide equally good sensitivity as the arrays on commercial aldehyde-modified plastic slides functionalized with amino-labeled probes [8] and commercial glass slides [30]
Fig. 4.
Sensitivity and dynamic range of hybridization on four polymeric materials. TC-tagged M probe at a concentration of 20 μM was spotted on different substrates. Cy3-labeled complementary oligo DNA target was hybridized at concentrations ranging from 0.001 to 1,000 nM. The points represent the average values obtained from three experiments, and the error bars indicate standard deviations
Hybridization in microfluidic biochip
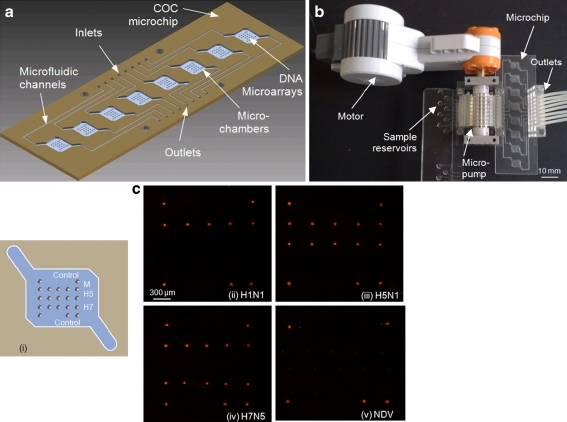
Using the TC-tag microarray fabrication method, a COC microfluidic biochip containing eight microfluidic chambers each with a microarray was made to demonstrate parallel and rapid hybridization. DNA microarray containing probes specific to the M gene of AIV type A as well as H5 and H7 subtypes was immobilized in each microchamber (Fig. 5a). Eight sample reservoirs were connected to the COC chip through the micropump (Fig. 5b). PCR products of one NDV viral strain and three AIV viral strains, H1N1, H5N1 and H7N5, were run in parallel through the microchambers at a flow rate of 1 μL/min (one sample repeated in two chambers) and hybridized with the DNA microarrays at 37 °C.
Fig. 5.
a Schematic diagram of the eight-chamber microfluidic biochip. Ten-microliter microchambers and inlet/outlet channels were milled on a COC slide. DNA microarrays containing 20 μM TC-tagged probes specific to the M gene of AIV type A as well as H5 and H7 subtypes were immobilized in each microchamber. b System setup for flow-through hybridization. Sample reservoirs were connected the microfluidic device through an eight-channel peristaltic micropump. The Cy3-labeled PCR products were pumped through the chambers at a flow rate of 1 μL/min for 10 min at 37 °C. c Results of DNA hybridization using the microfluidic biochip. (i) Microarray layout. The DNA probes for M gene, H5 and H7 are represented as gray circles. Circles at four corners are Cy3-labeled M gene probe used for control. (ii–v) Fluorescent images of the microchambers after hybridization with Cy3-labeled PCR products of four viral strains: (ii) AIV H1N1, (iii) AIV H5N1, (iv) AIV H7N5, and (v) NDV
Fluorescent images of the microchambers after 10-min hybridization showed that AIV viruses and their subtypes were unambiguously identified by distinct patterns (Fig. 5c). No positive signal was obtained for the NDV and unspecific hybridization was almost negligible, showing the immobilized capture probes were all specific to their respective target viruses even after UV irradiation and high-temperature bonding (120 °C). Despite the short hybridization time, SNR of 90 was achieved for flow-through hybridization, while SNR was only 5 for plastic slides with cover slips after 10 min passive hybridization. The hybridization speeds were accelerated by microfluidic addressing of the arrays due to reduced diffusion distances and enhanced mass transport of the target molecules. The results demonstrate the feasibility of producing DNA microarrays on non-modified plastics using TC-tagged probes and its potential for fabrication of inexpensive and robust diagnostic devices.
Conclusion
In this paper, we demonstrated that TC-tagged DNA oligonucleotide probes could be reliably immobilized onto various non-modified plastic surfaces via simple UV exposure. Compared to surface-modified slides with aminated or thiolated DNA probes, the technique described here provided in average, five- and fourfold improvement in immobilization and hybridization efficiency, respectively. The immobilization method is rapid (10-min UV irradiation), efficient (yielding high immobilization and hybridization densities), versatile (working on various surface types), robust (can withstand high-temperature) and economical (cost only 30% of amino-modified probes). Highly selective hybridization was demonstrated on the portable COC microarray device with eight addressable microfluidic channels for parallel and rapid identification of AIV samples. The direct attachment of DNA by UV irradiation addresses the challenge of developing inexpensive microarray tools and is ideal for the integration with microfluidic devices. In the future, the selectivity and specificity of the probe attachment technique will be further evaluated by performing single nucleotide polymorphism genotyping of real human clinical samples, and the thermal stability will also be explored for the development of devices for PCR-based bioassays.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 438 kb)
Acknowledgments
This work is co-supported by Forskningsrådet Teknologi og Produktion (FTP), Sagsnr. 09066477, DTU Food Pathogen Project No. 8, Grant No. 150627 and EU FP-7 IP LABONFOIL project, Grant No. 224306.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Yoo SM, Jong HC, Lee SY, Yoo NC (2009) J Microbio Biotech 19
- 2.West J, Becker M, Tombrink S, Manz A. Anal Chem. 2008;80:4403–4419. doi: 10.1021/ac800680j. [DOI] [PubMed] [Google Scholar]
- 3.Lien KY, Lee GB. Analyst. 2010;135:1499–1518. doi: 10.1039/c000037j. [DOI] [PubMed] [Google Scholar]
- 4.Kimura N, Oda R, Inaki Y, Suzuki O (2004) Nucleic Acids Res 32 [DOI] [PMC free article] [PubMed]
- 5.Misra A, Dwivedi P. Anal Biochem. 2007;369:248–255. doi: 10.1016/j.ab.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 6.Situma C, Hassimoto M, Soper SA. Biochem Eng J. 2006;23:213–231. doi: 10.1016/j.bioeng.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Z, Peytavi R, Diaz-Quijada GA, Picard FJ, Huletsky A, Leblanc E, Frenette J, Boivin G, Veres T, Dumoulin MM, Bergeron MG. J Clin Microbiol. 2008;46:3752–3758. doi: 10.1128/JCM.00377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinoshita K, Fujimoto K, Yakabe T, Saito S, Hamaguchi Y, Kikuchi T, Nonaka K, Murata S, Masuda D, Takada W, Funaoka S, Arai S, Nakanishi H, Yokoyama K, Fujiwara K, Matsubara K. Nucleic Acids Res. 2006;35:e3. doi: 10.1093/nar/gkl939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabourin D, Petersen J, Snakenborg D, Brivio M, Gudnadson H, Wolff A, Dufva M. Biomed Microdevices. 2010;12:673–681. doi: 10.1007/s10544-010-9420-7. [DOI] [PubMed] [Google Scholar]
- 10.Diaz-Quijada GA, Peytavi R, Nantel A, Roy E, Bergeron MG, Dumoulin MM, Veres T. Lab Chip. 2007;7:856–862. doi: 10.1039/b700322f. [DOI] [PubMed] [Google Scholar]
- 11.Soper SA, Hashimoto M, Situma C, Murphy MC, McCarley RL, Cheng YW, Barany F. Methods. 2005;37:103–113. doi: 10.1016/j.ymeth.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Liu D, Perdue RK, Sun L. Langmuir. 2004;20:5905–5910. doi: 10.1021/la049605p. [DOI] [PubMed] [Google Scholar]
- 13.Li YC, Wang Z, Qu LML, Yu HZ. Anal Chem. 2007;79:426–433. doi: 10.1021/ac061134j. [DOI] [PubMed] [Google Scholar]
- 14.Kimura N. Biochem Biophys Res Commun. 2006;347:477–484. doi: 10.1016/j.bbrc.2006.06.130. [DOI] [PubMed] [Google Scholar]
- 15.Fixe F, Dufva M, Telleman P, Christensen CBV. Nucleic Acids Res. 2004;32:e9. doi: 10.1093/nar/gng157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fixe F, Dufva M, Telleman T, Christensen CBV. Lab Chip. 2004;4:191–195. doi: 10.1039/b316616c. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Rauch CB. Anal Biochem. 2003;317:76–84. doi: 10.1016/S0003-2697(03)00051-4. [DOI] [PubMed] [Google Scholar]
- 18.Sun Y, Dhumpab R, Bang DD, Handberg K, Wolff A. Diagn Microbiol Infect Dis. 2011;69:432–439. doi: 10.1016/j.diagmicrobio.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Gudnason H, Dufva M, Bang DD, Wolff A. Biotechniques. 2008;45:261–271. doi: 10.2144/000112905. [DOI] [PubMed] [Google Scholar]
- 20.Dufva M, Petersen J, Stoltenborg M, Birgens H, Christensen CBV. Analytical Biochem. 2006;352:188–197. doi: 10.1016/j.ab.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Starick E, Romer-Oberdorfer A, Werner O. J Vet Med Ser B Infect Dis Vet Public Health. 2000;47:295–301. doi: 10.1046/j.1439-0450.2000.00386.x. [DOI] [PubMed] [Google Scholar]
- 22.Sabourin D, Snakenborg D, Skafte-Pedersen P, Kutter JP, Dufva M (2010) Fast and simple: reconfigurable elements and solutions for creating and driving fluidic networks, 14th International Conference on Miniaturized Systems for Chemistry and Life Science, Gronnigen, The Netherlands. pp. 1433–1435
- 23.Skafte-Pedersen P, Sabourin D, Dufva M, Snakenborg D. Lab Chip. 2009;9:3003–3006. doi: 10.1039/b906156h. [DOI] [PubMed] [Google Scholar]
- 24.Sabourin D, Dufva M, Jensen T, Kutter J, Snakenborg D. J Micromech Microeng. 2010;20:037001. doi: 10.1088/0960-1317/20/3/037001. [DOI] [Google Scholar]
- 25.Sun Y, Dhumpab R, Bang DD, Høgberg J, Handberg K, Wolff A. Lab Chip. 2011;11:1457–1463. doi: 10.1039/c0lc00528b. [DOI] [PubMed] [Google Scholar]
- 26.Tsao CW, DeVoe DL. Microfluid Nanofluid. 2009;6:1–16. doi: 10.1007/s10404-008-0361-x. [DOI] [Google Scholar]
- 27.Peterson AW, Heaton RJ, Georgiadis RM. Nucleic Acids Res. 2001;29:5163–5168. doi: 10.1093/nar/29.24.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shchepinov MS, CaseGreen SC, Southern EM. Nucleic Acids Res. 1997;25:1155–1161. doi: 10.1093/nar/25.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poulsen L, Soe MJ, Snakenborg D, Møller L, Dufva M. Nucleic Acids Res. 2008;36:e132. doi: 10.1093/nar/gkn600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berre VL, Trevisiol E, Dagkessamanskaia A, Sokol S, Caminade AM, Majoral JP, Meunier B, Francois J. Nucleic Acids Res. 2003;31:e88. doi: 10.1093/nar/gng088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 438 kb)