Abstract
Carbon nanoparticles (CNPs) labeled with reporter molecules can serve as signaling labels in rapid diagnostic assays as an alternative to gold, colored latex, silica, quantum dots, or up-converting phosphor nanoparticles. Detailed here is the preparation of biomolecule-labeled CNPs and examples of their use as a versatile label. CNPs can be loaded with a range of biomolecules, such as DNA, antibodies, and proteins (e.g., neutravidin or a fusion protein of neutravidin with an enzyme), and the resulting conjugates can be used to detect analytes of high or low molecular mass.
Figure.
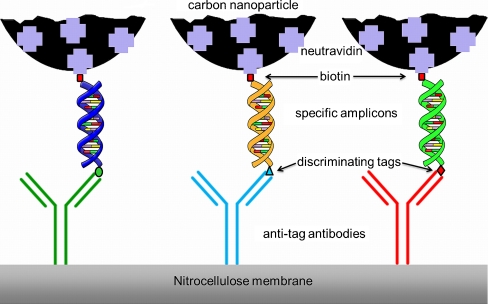
Scheme of a NALFIA-, NAMIA, or NALMIA. Neutravidin adsorbed onto CNPs detects biotin-labelled amplicons; the discriminating tag is recognized by its respective antibody, which is immobilized onto nitrocellulose membranes or pads
Keywords: Carbon nanoparticles, Lateral flow assays, Protein microarray-based assays, Nucleic acid immunoassays (DNA/RNA), Immunochemical assays, Diagnostics
Introduction
Modern analytical methods include, among others, immunochemical and DNA/RNA hybridization-based techniques. Such techniques rely on the molecular recognition of antigens by their corresponding antibodies or on the recognition of complementary DNA/RNA sequences, respectively. To visualize the interaction, a label of one kind or another must be attached to either the antigen or the antibody. Rapid assay formats have emerged at the point of care/need, and the lateral flow assay (LFA) format is very well suited to this purpose, and thus often used [1]. For this assay format, particulate labels with optical properties are obligatory for visual inspection. The use of nanoparticles as a label has been discussed in recent reviews [2, 3], but the utilization of elemental carbon as a label has not been mentioned aside from a chapter in the book Using Nanoparticles in Agricultural and Food Diagnostics [4].
Among these particulate labels, it was shown in a literature survey by the independent FIND Diagnostics organization that when CNPs are used as labels in the LFA format, they are more sensitive than gold or latex [5]. Sensitivity in the low picomolar range was reported for carbon nanoparticles (CNPs), even by visual inspection. Moreover, CNPs are the cheapest labels, and suspensions of them are very stable and easy to prepare. In addition, no activation is necessary. We will stress the difference between carbon nanoparticles—which are amorphous—and carbon nanotubes (CNTs), which have a defined structure (i.e., single- and multi-walled nanotubes). These CNTs are mainly used in pre-concentration systems, as reviewed in [6], and as labels in electrochemical systems, as reviewed in [7]. The use of CNTs in diagnostics will not be discussed here, as a general review of the use of CNTs as detection labels has recently been published [8].
Instead, we will present the preparation, functionalization, and use of carbon nanoparticles as labels in diagnostic assays such as in LFAs and in antibody microarray formats. An overview of their strengths and weaknesses is given in Table 1, and an overview of opportunities and threats associated with them is shown in Table 2.
Table 1.
SWOT analysis of CNPs: internal factors
| Strengths | Weaknesses |
|---|---|
| Inert in terms of surface charge, relatively independent of pH, does not oxidize | Low density: not really suitable for TEM/SEM |
| Amorphous carbon is suitable for physical adsorption due to hydrophobic surface characteristics | Some types of carbon require the addition of surfactants to obtain stable suspensions in aqueous media: such surfactants may interfere with test performance |
| High contrast, good sensitivity | The presence of larger, irregularly shaped carbon particles requires the use of nitrocellulose with larger pore openings; the captured ligand concentration/density is then relatively low, resulting in "relatively diffuse" signal |
| Relatively large dynamic range: at high(er) concentrations, the small(er) particles contribute to the signal, and at low(er) concentrations, the large(r) particles provide the greatest contribution | Still relatively unknown label in commercial diagnostic tests and in the literature |
| Relatively high molar extinction coefficient compared to latex; relatively strong signals offer high test sensitivities (down to low picomolar concentrations [5]) | Elemental carbon covalent linkage through the targeted use of reactive groups is problematic |
| A relatively low density, stable suspension of relatively large particles is possible (also an advantage in terms of the response) | Fixed functionality; e.g., not paramagnetic, no change of color |
| Heterogeneous particle size distribution is less prone to the hook effect | Luminescent CNPs need a more expensive readout system; visualization is not possible |
| Carbon suspensions are easy to prepare | |
| Good flow characteristics in the case of capillary migration | |
| Label is available in large to very large batches, variation in quality/features over time is negligible (especially in combination with inert materials) | |
| Label is obtainable at very low cost | |
| Scaling up production after test preparation is a relatively simple task | |
| Carbon “variants under Patents US5529901A1 [10] and US5641689A1 [11]” do not require the addition of surfactants for stable suspensions in aqueous media |
Table 2.
SWOT analysis of CNPs: external factors
| Opportunities | Threats |
|---|---|
| Very well suited for use in lateral flow test systems where sensitivity is associated with a relatively wide dynamic range | In the case of pollution: materials (tubing, measuring cells, devices) are not easy to clean; carbon label is less suitable for use in complex, expensive devices |
| Due to their high contrast, they are suitable for use as a label system in antibody microarray formats | |
| Because of “black on white” test results (applicable to nitrocellulose, nylon, polystyrene colored white), very suitable for quantification of results using "gray pixel" processing | Amorphous particles in the dry state during and immediately after the production phase (present in bulk quantities) can be potentially harmful to health, as they are fine particles; however, their utilization as CNPs in immunoassays does not present health problems |
| Suitable for use as a label in agglutination/precipitation assays | Finding carbon particles with the desired properties/characteristics (e.g., particles with similar properties but that are much smaller in size) is dependent on what the market can offer |
| Suitable for use in sensors that employ piezoelectric film detection/sensing |
Preparation and functionalization
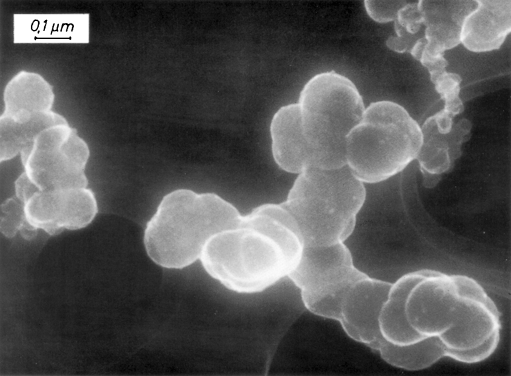
CNPs can be obtained from soot, commercially available from different sources. These preparations are intended for other uses, such as in toner or ink for printers or in automobile tires. Type SB4 (Special Black 4) from the company Degussa AG was reported as a label at first in 1993 [9]. This type receives special post-treatment in order to generate a few aldehyde groups, and does not need a stabilizer in suspension to obtain colloidally stable plain nanoparticles [10, 11]. Accordingly, the preparation of aqueous suspensions and the functionalization of the nanoparticles are easy tasks. For some applications, a shortcoming of this type is that the particles are not monodisperse in nature, as can be seen in Fig. 1, although uniform sizes can be obtained, which are called carbon nanostrings [12]. Although this heterogeneous size distribution may be a drawback, in most applications it is a clear advantage (see below).
Fig. 1.
SEM image of Flammruss 101 (comparable to SB4); images courtesy of Evonik-Degussa AG
The number of functional groups on these nanoparticles is very small, precluding the possibility of the covalent attachment of proteins or DNA. However, physical adsorption has the advantage that, in most cases, the specificity is retained. Achieving a specific orientation, which is useful for presenting binding/active domains of antibodies or enzymes to the liquid phase, can be done with intermediary biomolecules such as protein A, protein G, or a secondary antibody.
Several other carbon brands need the presence of stabilizers [10, 11]. Amongst these are preparations that lead to monodisperse particles in stable suspensions, e.g., after treatment using a ball mill [13].
Soot obtained by burning toluene was claimed to have a surface that could be covalently attached to streptavidin, antibodies, or G-protein via glutaraldehyde coupling [14]. An average size of 162 ± 52 nm was recorded, and a stability of over ten years was observed. However, the chemical reaction that facilitated the covalent binding was unclear. CNPs were prepared from fructose by hydrothermal degradation, and subsequently functionalized by the covalent attachment of enzyme-labeled anti α-fetoprotein [15].
CNPs prepared from other sources were reported to show more homogeneous dimensions. These CNPs were prepared from graphene oxide [16], carbohydrates [17–19], benzene [20], or cyclodextrins [21]. The chemical modification steps that yield nanoparticles with fluorescent, phosphorescent, or up-converting properties (luminescent CNPs) are described in [22–28].
However, these CNP preparation methods are all either rather complex or they require expensive starting materials, and they are often also made more expensive by the high energy consumption of the process. Moreover, large-scale preparation is often a serious problem. These disadvantages were sidestepped by using raw carbon as starting material and a one-step ultrasonic treatment in a hydroxide peroxide solution [24], which produced enough active functional groups for covalent attachment. This resulted in hydrophilic nanoparticles with fluorescent, phosphorescent, and up-converting properties that were 5–10 nm in size.
Applications to lateral flow assays (LFA)
Sandwich-type lateral flow immunoassays (LFIA)
In this kind of assay, the analyte is sandwiched between the capture antibody (which has been sprayed at the test line of the nitrocellulose strip) and the detection antibody (which has been labeled for visualization). The analyte is usually a molecule with a relatively high molecular mass, such as a protein. The detection antibody can be immobilized onto the CNPs by physical adsorption, thus generating an immunolabel. As in other immunoassays, such as ELISAs, the signal is proportional to the analyte concentration. Applications include pregnancy test kits based on human chorionic gonadotropin [29] (working range 5-300 mIU/mL); verotoxin-producing Escherichia coli detection [30, 31] (specificity in ring tests: 95–100%); the diagnosis of schistosomiasis using circulating cathodic antigen in urine [32] (specificity 90%), and the detection of fungal amylase in the workplace [33, 34] (in a field test using wipes, 100% agreement with an ELISA at allergen levels of >5 ng/mL was observed). The results of these tests are mainly interpreted visually, sometimes by comparison with a standard set. The use of a flatbed scanner to quantify the response was first mentioned in 2001 [35] for immunoglobulin E detection. This assay showed enhanced sensitivity and yielded semi-quantitative or even quantitative results.
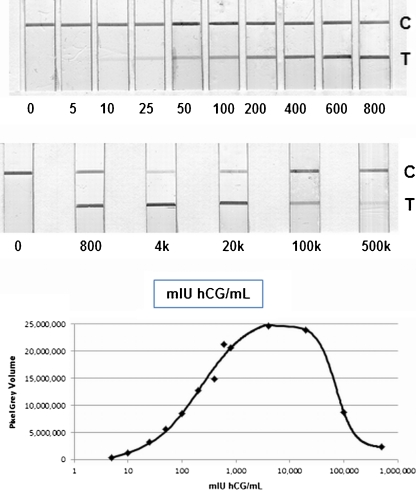
In a survey performed by the Foundation for Innovative New Diagnostics (FIND) in which the sensitivities of immunolabels used in lateral flow assays were investigated, CNPs were ranked in the top five, well above more conventional labels such as gold and latex [5]. The sensitivities of assays with CNPs were in the low pM range, whether the results were interpreted visually or using a flatbed scanner or digital camera. In this kind of assay, the heterogeneity in particle size in the carbon suspension is an advantage, as the smaller particles reach the test line more rapidly than the larger particles. The hook effect—which often affects immunochemical sandwich type assays [36]—is clearly reduced, and the dynamic range of the lateral flow assay is substantially increased by up to four orders of magnitude, as shown in Fig. 2 (unpublished results).
Fig. 2.
Large dynamic range of a sandwich-type lateral flow assay that shows a diminished hook effect. hCG levels in pregnant women may reach concentrations of up to 50,000 mIU/mL at 8 weeks since the last menstruation. Even at a concentration ten times higher than this (i.e., 500,000 mIU/mL), there is still a positive response. Upper panel: LFIA strips (C control line, T test line). Lower panel: graph based on pixel gray volumes obtained by flatbed scanning and digitization
The detection of p24 antigen from HIV-infected infants was reported using an unspecified carbon black preparation from Evonik Degussa Corporation (Parsippany, NJ, USA) [37]. The sensitivity of this assay with pure recombinant antigen was reported to be 50 pg/mL (2 pM). In infant plasma, an overall sensitivity and a specificity of 90% and 100%, respectively, were achieved using a flatbed scanner and a homemade MATLAB program to evaluate the results.
Isotyping of transferrin in serum was achieved using a lateral flow test and CNPs [38]. An integrated setup combining a separation membrane and a lateral flow strip was used to detect carbohydrate-deficient transferrin. Measured isoforms constituted a minimal part (0.3%) of the total amount of transferrin. The measuring time was about 10 min, and the correlation coefficient with an established, commercial, two-step procedure, which takes 4–5 h to perform, was 0.99 [38]. This group later used carbon nanostrings to isotype transferrin isotypes in a lab-on-a-chip setup with a lateral flow strip (pI differences of 0.1 pH units could be successfully discriminated) [39]. Carbon nanostrings were also used to detect erythropoietin in urine [12].
CNPs prepared from soot by burning toluene have been used to develop several rapid assays [14]. Protein G covalently attached to these particles was the intermediate in an assay that was specific for HIV antigens [14], whereas anti-human chorionic gonadotropin antibodies were used for a pregnancy hormone-specific assay [14]. Covalent attachment using CNPs prepared hydrothermally in a closed system in the presence of fructose was reported for the detection of α-fetoprotein using HRP-labeled anti α-fetoprotein [15]. Very good sensitivity was obtained in an electrical readout system using sheets of anti α-fetoprotein-functionalized graphene on a graphite electrode.
Inhibition-type lateral flow immunoassays (LFIA)
In this kind of assay, the reagents consist of a specific antibody and a carrier protein to which small analyte molecules (with a low molecular mass; so-called haptens) have been covalently bound. Both reagents can be used as either the detection or the capture ligand, although in most cases the carrier protein–analyte (hapten) conjugate is sprayed onto the nitrocellulose membrane at the test line. The free analyte in the sample competes with the carrier protein bound analytes for antibody binding sites at the test line. Both the detection antibody and the analyte–carrier protein conjugate are either immobilized onto the nitrocellulose membrane or labeled onto the CNPs by physical adsorption.
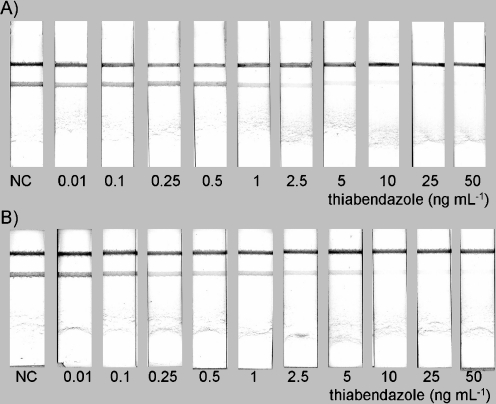
In another approach, a secondary antibody raised against the animal species of the specific detection antibody is coupled to the CNPs. This means that titration of the specific antibody will give a 100% signal that can be easily scored by visual inspection. This approach allows the sensitivity of the inhibition-type immunoassay to be greatly increased compared to tests in which the specific antibody has been coupled onto the nanoparticles. Also, in this case the response is negatively correlated to the amount of analyte. Applications include the detection of sulfamethazine in urine [40], progesterone in buffer [41], methiocarb in surface water [42], and carbaryl and thiabendazole residues in fruit juices [43, 44]. A typical example is depicted in Fig. 3.
Fig. 3A–B.
Typical example of an inhibition-type lateral flow assay: A analyte in buffer, B analyte in spiked fruit juice. Reproduced from [43] with permission from Elsevier
Nucleic acid lateral flow (immuno)assays (NALF(I)A)
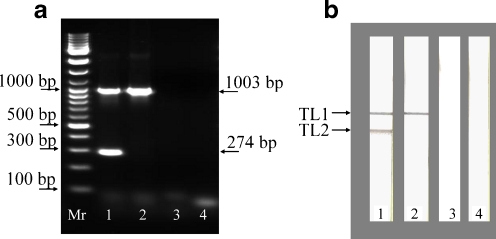
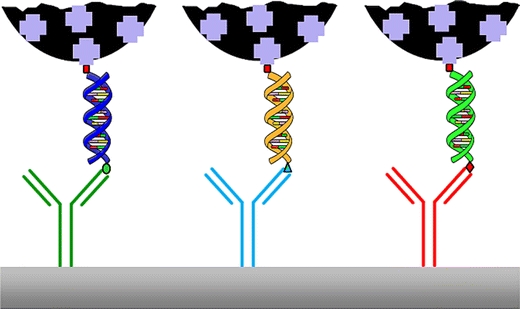
This technique was presented in the journal IVD Technology [45] by the company Xtrana. The method relies on the amplification of specific DNA or RNA sequences using labeled primers. Nucleic acid lateral flow immunoassays using carbon nanoparticles were first reported in 2005 for the detection of Shiga toxin producing Escherichia coli [46]. One primer was labeled with a discriminating tag, and the other primer with a biotin moiety. Anti-discriminating tag antibodies were immobilized onto the lateral flow strip and neutravidin was adsorbed onto CNPs. After applying a fast amplification protocol to the specific nucleic acid sequences (30 min), a small amount of amplicon was added to the CNP–neutravidin suspension, and it was then run on the prepared NALFIA. Several applications show the versatility of this procedure. Plasmodium falciparum species were detected in sera of patients suffering from malaria [47]; also reported were the presence/absence of Listeria bacteria in food (see Fig. 4) [48], and the detection of Cronobacter spp. in baby food [49], and the presence of virulence factors in Shiga-toxin producing Escherichia coli [50].
Fig. 4a–b.
Typical results from the simultaneous detection of L. monocytogenes and generic Listeria spp. amplicons by agarose gel electrophoresis (a) and NALFIA (b) after duplex PCR. For both a and b, chromosomal DNA of L. monocytogenes (lane 1), L. innocua (lane 2), and Enterobacter cloacae (lane 3) were used for the duplex PCR. The negative control (lane 4) is a primer control (PCR without template DNA). Mr DNA size marker, TL1 test line specific to all species from the genus Listeria (line with anti-FITC antibody), TL2 test line specific to L. monocytogenes (line with anti-DIG antibody). Reproduced from [47]
In another NALF approach, carbon nanostrings onto which BSA had been physically adsorbed were applied. The BSA molecules had been functionalized through the covalent coupling of DNA oligonucleotide probes [51]. As a proof of principle, the detection of the maize invertase gene was reported. This gene can function as the reference gene in maize when looking for genetically modified maize. Also reported was the detection of a single nucleotide polymorphism in the toll-like receptor 4 gene as a pathogen recognition scheme, and the activation of innate immunity.
Applications of antibody microarray immunoassays (MIA)
As well as creating assays in the LFA format, it is also possible to print capture antibodies as very small spots in a microarray format and use a comparable assay format to that outlined for NALFIA [46]. We suggest that such a format where antibodies are used to capture labeled amplicons should be called a nucleic acid microarray immunoassay (NAMIA). In this format, the number of discrete targets can be easily expanded up to 10–15 different targets. The results of this approach, a 30 min amplification followed by a one-step incubation on the microarray for 1 h, can be obtained by automated data processing following the digitization of the spots by flatbed scanning or digital photography (10 min of processing time in total). A scheme showing the principle of a NALFIA, NALMIA, or NAMIA is depicted in Fig. 5.
Fig. 5.
Scheme for a NALFIA, NAMIA, or NALMIA. Neutravidin adsorbed onto CNPs detects biotin-labeled amplicons; the discriminating tag is recognized by its respective antibody, which is immobilized onto nitrocellulose membranes or pads
Applications of lateral flow microarray immunoassay formats (LMIA)
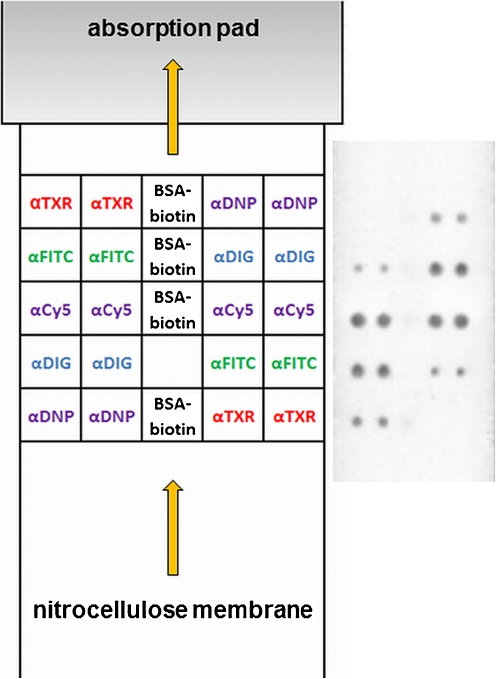
A new approach is to print an antibody microarray onto a lateral flow strip, thus combining the advantages of nucleic acid lateral flow immunoassays (NALFIA) and nucleic acid microarray immunoassays (NAMIA) [52, 53]. Again, when antibodies are used to capture tagged amplicons, the format is referred to as a nucleic acid lateral flow microarray immunoassay (NALMIA). In this format, more antibodies can be spotted than on a “classical” LFA strip, so more target molecules/microorganisms can be detected simultaneously. Since the reaction time is similar to that needed for the conventional lateral flow assay (5–10 min), this format is much quicker than the general antibody microarray format on slides, which may take 2–4 h to yield the final results. A recent example in which specific amplicons of virulence factors of Shiga toxin-producing Escherichia coli were applied is shown in Fig. 6 (unpublished results).
Fig. 6.
A lateral flow microarray immunoassay (LMIA) test showing double-labeled amplicons specific for virulence factors of Shiga toxin producing Escherichia coli. Left panel: layout of the LMIA. Right panel: typical example of a double-labeled (tag/biotin) amplicon solution, and detection using neutravidin-coated carbon nanoparticles
Applications in other formats
Mouse immunoglobulin isotyping, the detection of human serum albumin (HSA), and the determination of Kunitz-type trypsin inhibitor from soy (KSTI) were reported in [9] using a nitrocellulose strip with spots of specific antibodies or antigens and incubation of the nitrocellulose strips in 1 mL volumes.
Upon adsorbing carboxyfluorescein (FAM)-labeled ssDNA onto CNPs, the fluorescence is quenched. This phenomenon was used to develop an assay format in which the hybridization of DNA strands complementary to the FAM-labeled ssDNA released the FAM label from the CNPs, thus restoring fluorescence [17]. As a proof of principle, sequences from HIV were labeled with FAM. Binding to CNPs quenched the fluorescence. Following hybridization with the complementary sequence, the fluorescence was restored.
Another approach was described in [18], where a fluorescently labeled DNA sequence does not bind to CNPs in the presence of Hg2+ions, because the induced hairpin structure formed by thymidine–Hg2+–thymidine prevents binding. The fluorescent properties of the DNA are retained. In the absence of Hg2+ ions, the DNA binds to the CNPs, which results in the quenching of the fluorescence. The same principle was used to detect Ag+ ions in solution [54]. In all cases, the fluorescence was measured using a fluorometer. The LODs were very low (10 nM to 500 pM), showing good sensitivity and reproducibility.
Although the preparation of fluorescent, phosphorescent, or up-converting carbon nanoparticles has been reported, none of these methods have yet been used as detection labels in diagnostic assays. The question remains as to whether it will be possible to functionalize such CNPs with biomolecules while retaining their fluorescent, chemiluminescent, up-converting, or phosphorescent properties.
Commercialization
SB4-derived CNPs are being applied and commercialized by the company Genzyme Diagnostics (Cambridge, MA, USA; http://www.genzyme.com/), which was recently acquired by Sekisui Diagnostics (West Malling, UK; http://www.sekisuidiagnostics.com/), and by the company Oxoid (Cambridge, UK; http://www.oxoid.com/uk/blue/index.asp) for their syphilis agglutination test.
The company Vivacta (http://www.vivacta.com/) is applying and commercializing carbon nanoparticles in a format using a piezoelectric film for signal readout [55].
The company Maiia is commercializing carbon nanostrings (http://www.maiiadiagnostics.com/products/carbon_black_nano-strings/).
Conclusions
CNPs prepared from soot—amorphous carbon produced in bulk quantities, with the properties outlined in patents US5529901A1 [10] and US5641689A1 [11], e.g., SB4—can be used satisfactorily as versatile labels in lateral flow and microarray immunoassay test formats. They provide a less expensive yet more sensitive and stable alternative to other labels. The signal can be read easily by visual inspection (black on a white background), and can be quantified using a flatbed scanner and image analysis software.
CNPs not obtained from soot intended for bulk applications, such as in ink, toner or tires, are often prepared on the small scale in laboratories. Only a few applications with these “homemade” nanoparticles have been reported at the time of writing. Hence, their applicability is still questionable. Luminescent CNPs need more sophisticated (and often more expensive) equipment to visualize and quantify the test results.
Acknowledgments
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- CNPs
Carbon nanoparticles
- CNTs
Carbon nanotubes
- LFIA
Lateral flow immunoassay
- NALFIA
Nucleic acid lateral flow immunoassay
- NALMIA
Nucleic acid lateral flow microarray immunoassay
- NAMIA
Nucleic acid microarray immunoassay
References
- 1.Posthuma-Trumpie GA, Korf J, van Amerongen A. Anal Bioanal Chem. 2009;393:569–582. doi: 10.1007/s00216-008-2287-2. [DOI] [PubMed] [Google Scholar]
- 2.El-Ansary A, Faddah LM (2010) Nanotechnol Sci Appl 3:65–76 [DOI] [PMC free article] [PubMed]
- 3.Perez-Lopez B, Merkoci A. Anal Bioanal Chem. 2011;399:1577–1590. doi: 10.1007/s00216-010-4566-y. [DOI] [PubMed] [Google Scholar]
- 4.Posthuma-Trumpie GA, Van Amerongen A (2011) In: Frewer LJ, Norde W, Fisher AR, Kampers FW (eds) Nanotechnology in the agri-food sector: implications for the future. Wiley-VCH, Weinheim
- 5.Gordon J, Michel G. Clin Chem. 2008;54:1250–1251. doi: 10.1373/clinchem.2007.102491. [DOI] [PubMed] [Google Scholar]
- 6.Hussain C, Mitra S. Anal Bioanal Chem. 2011;399:75–89. doi: 10.1007/s00216-010-4194-6. [DOI] [PubMed] [Google Scholar]
- 7.Yanez-Sedeno P, Pingarron JM, Riu J, Rius FX (2010) Trend Anal Chem 29:939–953
- 8.Scida K, Stege PW, Haby G, Messina GA, García CD. Anal Chim Acta. 2011;691:6–17. doi: 10.1016/j.aca.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Amerongen A, Wichers JH, Berendsen LBJM, Timmermans AJM, Keizer GD, van Doorn AWJ, Bantjes A, van Gelder WMJ. J Biotechnol. 1993;30:185–195. doi: 10.1016/0168-1656(93)90112-Z. [DOI] [PubMed] [Google Scholar]
- 10.van Doorn AWJ, Wichers JH, van Gelder WMJ (1996) Method for determining the presence of analyte using a stable colloidal carbon sol. Int Patent 5,529,901
- 11.van Doorn AWJ, Wichers JH, van Gelder WMJ (1997) Stable aqueous carbon sol composition for determining analyte. Int Patent 5641689
- 12.Lonnberg M, Drevin M, Carlsson J. J Immunol Methods. 2008;339:236–244. doi: 10.1016/j.jim.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 13.Wang S-J, Chang W-F, Chen C-I, Hsu F-S, Hsiung K-P, Liu Y-C. J Biotechnol. 2008;136:S193. doi: 10.1016/j.jbiotec.2008.07.1833. [DOI] [Google Scholar]
- 14.Rayev M, Shmagel K. J Immunol Methods. 2008;336:9–15. doi: 10.1016/j.jim.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Du D, Zou Z, Shin Y, Wang J, Wu H, Engelhard MH, Liu J, Aksay IA, Lin Y. Anal Chem. 2010;82:2989–2995. doi: 10.1021/ac100036p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S, Song H, Guo P, Zhou J, Chen X. Carbon. 2010;48:4211–4214. doi: 10.1016/j.carbon.2010.07.025. [DOI] [Google Scholar]
- 17.Li H, Zhang Y, Wang L, Tian J, Sun X. Chem Commun. 2011;47:961–963. doi: 10.1039/c0cc04326e. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Zhai J, Tian J, Luo Y, Sun X (2011) Biosens Bioelectron (in press). doi:10.1016/j.bios.2011.03.026 [DOI] [PubMed]
- 19.Yao C, Shin Y, Wang L-Q, Windisch CF, Samuels WD, Arey BW, Wang C, Risen WM, Exarhos GJ. J Phys Chem C. 2007;111:15141–15145. doi: 10.1021/jp074188l. [DOI] [Google Scholar]
- 20.Wesolowski MJ, Kuzmin S, Moores B, Wales B, Karimi R, Zaidi AA, Leonenko Z, Sanderson JH, Duley WW. Carbon. 2011;49:625–630. doi: 10.1016/j.carbon.2010.10.008. [DOI] [Google Scholar]
- 21.Shin Y, Wang L-Q, Bae I-T, Arey BW, Exarhos GJ. J Phys Chem C. 2008;112:14236–14240. doi: 10.1021/jp801343y. [DOI] [Google Scholar]
- 22.Gonçalves H, Esteves da Silva J. J Fluoresc. 2010;20:1023–1028. doi: 10.1007/s10895-010-0652-y. [DOI] [PubMed] [Google Scholar]
- 23.Hu S-L, Niu K-Y, Sun J, Yang J, Zhao N-Q, Du X-W. J Mater Chem. 2009;19:484–488. doi: 10.1039/b812943f. [DOI] [Google Scholar]
- 24.Li H, He X, Liu Y, Yu H, Kang Z, Lee S-T. Mater Res Bull. 2011;46:147–151. doi: 10.1016/j.materresbull.2010.10.013. [DOI] [Google Scholar]
- 25.Li H, He X, Liu Y, Huang H, Lian S, Lee S-T, Kang Z. Carbon. 2011;49:605–609. doi: 10.1016/j.carbon.2010.10.004. [DOI] [Google Scholar]
- 26.Liu H, Ye T, Mao C. Angew Chem Int Edit. 2007;46:6473–6475. doi: 10.1002/anie.200701271. [DOI] [PubMed] [Google Scholar]
- 27.Zhang B, Liu C-y, Liu Y (2010) Eur J Inorg Chem 2010:4411–4414
- 28.Zhu H, Wang X, Li Y, Wang Z, Yang F, Yang X (2009) Chem Commun 5118–5120 [DOI] [PubMed]
- 29.van Amerongen A, van Loon D, Berendsen LBJM, Wichers JH. Clin Chim Acta. 1994;229:67–75. doi: 10.1016/0009-8981(94)90229-1. [DOI] [PubMed] [Google Scholar]
- 30.Aldus CF, van Amerongen A, Ariens RMC, Peck MW, Wichers JH, Wyatt GM. J Appl Microbiol. 2003;95:380–389. doi: 10.1046/j.1365-2672.2003.01989.x. [DOI] [PubMed] [Google Scholar]
- 31.Capps KL, McLaughlin EM, Murray AWA, Aldus CF, Wyatt GM, Peck MW, van Amerongen A, Ariëns RMC, Wichers JH, Baylis CL, Wareing DRA, Bolton FJ. J AOAC Int. 2004;87:68–77. [PubMed] [Google Scholar]
- 32.van Dam GJ, Wichers JH, Ferreira TMF, Ghati D, van Amerongen A, Deelder AM. J Clin Microbiol. 2004;42:5458–5461. doi: 10.1128/JCM.42.12.5458-5461.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogdanovic J, Koets M, Sander I, Wouters I, Meijster T, Heederik D, van Amerongen A, Doekes G. J Allergy Clin Immun. 2006;118:1157–1163. doi: 10.1016/j.jaci.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Koets M, Sander I, Bogdanovic J, Doekes G, van Amerongen A. J Environ Monitor. 2006;8:942–946. doi: 10.1039/b605389k. [DOI] [PubMed] [Google Scholar]
- 35.Lonnberg M, Carlsson J. Anal Biochem. 2001;293:224–231. doi: 10.1006/abio.2001.5130. [DOI] [PubMed] [Google Scholar]
- 36.Amarasiri Fernando S, Wilson GS (1992) J Immunol Methods 151:47–66 [DOI] [PubMed]
- 37.Parpia ZA, Elghanian R, Nabatiyan A, Hardie DR, Kelso DM. JAIDS. 2010;55:413–419. doi: 10.1097/QAI.0b013e3181f1afbc. [DOI] [PubMed] [Google Scholar]
- 38.Lonnberg M, Carlsson J. J Immunol Methods. 2000;246:25–36. doi: 10.1016/S0022-1759(00)00287-8. [DOI] [PubMed] [Google Scholar]
- 39.Lonnberg M, Carlsson J. J Chromatogr A. 2006;1127:175–182. doi: 10.1016/j.chroma.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 40.O'Keeffe M, Crabbe P, Salden M, Wichers J, van Peteghem C, Kohen F, Pieraccini G, Moneti G. J Immunol Methods. 2003;278:117–126. doi: 10.1016/S0022-1759(03)00207-2. [DOI] [PubMed] [Google Scholar]
- 41.Posthuma-Trumpie GA, Korf J, van Amerongen A. Anal Bioanal Chem. 2008;392:1215–1223. doi: 10.1007/s00216-008-2362-8. [DOI] [PubMed] [Google Scholar]
- 42.Blazkova M, Mickova-Holubova B, Rauch P, Fukal L. Biosens Bioelectron. 2009;25:753–758. doi: 10.1016/j.bios.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 43.Blazkova M, Rauch P, Fukal L. Biosens Bioelectron. 2010;25:2122–2128. doi: 10.1016/j.bios.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 44.Holubova-Mickova B, Blazkova M, Fukal L, Rauch P. Eur Food Res Technol. 2010;231:467–473. doi: 10.1007/s00217-010-1301-z. [DOI] [Google Scholar]
- 45.Mondesire RR, Kozwich D, Johansen KA, Gerdes JC, Beard SE (2000). Solid-phase nucleic acid extraction, amplification, and detection. IVD Technology 1 May 2000. http://www.ivdtechnology.com/article/solid-phase-nucleic-acid-extraction-amplification-and-detection
- 46.van Amerongen A, Koets M (2005) In: van Amerongen A, Barug D, Lauwaars M (eds) Rapid methods for biological and chemical contaminants in food and feed. Wageningen Academic, Wageningen
- 47.Mens PF, van Amerongen A, Sawa P, Kager PA, Schallig HDFH. Diagn Micr Infec Dis. 2008;61:421–427. doi: 10.1016/j.diagmicrobio.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 48.Blazkova M, Koets M, Rauch P, van Amerongen A. Eur Food Res Technol. 2009;229:867–874. doi: 10.1007/s00217-009-1115-z. [DOI] [Google Scholar]
- 49.Blazkova M, Javurkova B, Fukal L, Rauch P. Biosens Bioelectron. 2011;26:2828–2834. doi: 10.1016/j.bios.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 50.Noguera P, Posthuma-Trumpie G, van Tuil M, van der Wal F, de Boer A, Moers A, van Amerongen A. Anal Bioanal Chem. 2011;399:831–838. doi: 10.1007/s00216-010-4334-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalogianni D, Boutsika L, Kouremenou P, Christopoulos T, Ioannou P. Anal Bioanal Chem. 2011;400:1145–1152. doi: 10.1007/s00216-011-4845-2. [DOI] [PubMed] [Google Scholar]
- 52.Carter DJ, Cary RB. Nucleic Acids Res. 2007;35:1–11. doi: 10.1093/nar/gkm207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gantelius J, Hamsten C, Neiman M, Schwenk JM, Persson A, Andersson-Svahn H. J Microbiol Meth. 2010;82:11–18. doi: 10.1016/j.mimet.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 54.Li H, Zhai J, Sun X. Langmuir. 2011;27:4305–4308. doi: 10.1021/la200052t. [DOI] [PubMed] [Google Scholar]
- 55.Ross S (2008) Developing an optically stimulated piezofilm immunoassay. IVD Technology 1 June 2008. http://www.ivdtechnology.com/article/developing-optically-stimulated-piezofilm-immunoassay