Abstract
Catalytic activities can be facilitated by ordered enzymatic arrays that co-localize and orient enzymes and their substrates. The purified RNA-dependent RNA polymerase from poliovirus self-assembles to form two-dimensional lattices, possibly facilitating the assembly of viral RNA replication complexes on the cytoplasmic face of intracellular membranes. Creation of a two-dimensional lattice requires at least two different molecular contacts between polymerase molecules. One set of polymerase contacts, between the ‘thumb’ domain of one polymerase and the back of the ‘palm’ domain of another, has been previously defined. To identify the second interface needed for lattice formation and to test its function in viral RNA synthesis, a hybrid approach of both electron microscopic and biochemical evaluation of wild-type and mutant viral polymerases was used to evaluate computationally generated models of this second interface. A unique solution satisfied all constraints and predicted a two-dimensional structure formed from antiparallel arrays of polymerase fibers that use contacts from the flexible amino-terminal region of the protein. Enzymes that contained mutations in this newly defined interface did not form lattices and altered the structure of wild-type lattices. When reconstructed into virus, mutations that disrupt lattice assembly exhibited growth defects, synthetic lethality, or both, supporting the function of the oligomeric lattice in infected cells. Understanding the structure of polymerase lattices within the multimeric RNA-dependent RNA polymerase complex should faciliate antiviral drug design and provide a precedent for other positive-strand RNA viruses.
INTRODUCTION
The RNA replication complexes of positive-strand RNA viruses such as poliovirus, Dengue virus and hepatitis C virus involve multiple virus- and host-encoded proteins, assembled on the surface of cytoplasmic membranes. The lynchpins of such complexes are the RNA-dependent RNA-polymerases, which have been shown for several such viruses to form, in addition to contacts with other proteins, homo-oligomeric interactions both in solution 1; 2; 3; 4; 5; 6; 7 and in cells 6; 8; 9. For the enzyme encoded by poliovirus, termed 3D polymerase, the formation of homo-oligomeric contacts correlates with cooperative RNA binding and elongation activity 1; 10; 11; 12 and mutations that interfere with polymerase oligomerization interfere with virus growth 11; 12; 13; 14; 15. The crystal structure of full-length, monomeric 3D polymerase has been determined 16. Like other polymerases, this structure has been likened to a right hand, and the residues that contribute to each component are shown in Figure 1a.
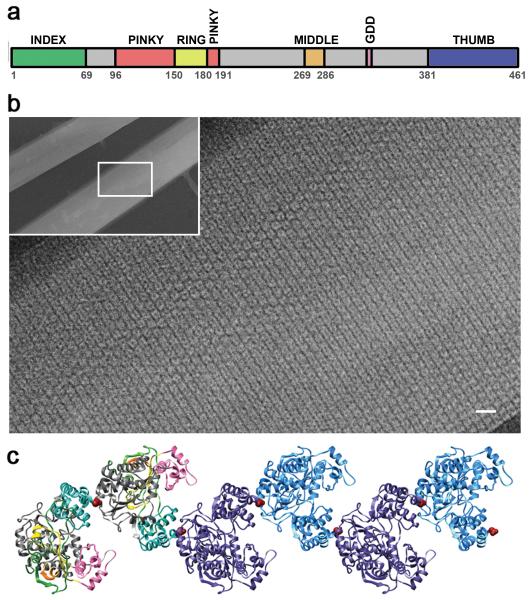
Figure 1. Structure and oligomerization of poliovirus 3D polymerase.
(a) The domain structure of the 461-amino acid poliovirus 3D polymerase is shown, with domains based on the three-dimensional structure of the full-length polymerase 16. (b) Two-dimensional lattices 12 formed by purified wild-type polymerase, as shown by negative staining followed by electron microscopy. Magnification bar: 10 nm for enlarged image, and 100 nm for inset. (c) The polymerase-polymerase interactions shown along Interface I, a set of polymerase-polymerase interaction surfaces observed in a crystal form 21 and tested for functionality by site-directed mutagenesis 11; 12; 14; 15. Domains of the first two polymerases on the left are color-coded as in panel a. Amino acid Leu446, integral to Interface I, is shown as a red space-filling model. A single polymerase molecule is approximately 5 nm in length, width and height.
Purified poliovirus polymerase forms planar, sheet-like structures that can be visualized via electron microscopy (Fig. 1b; 12; 17, reviewed in 18). To determine whether the contacts involved in lattice formation, and the lattices themselves, are important in the function of poliovirus 3D polymerase in viral RNA replication, we and others have sought to identify the contacts involved and to test their roles in the viral infectious cycle. All of the six tested mutations or sets of mutations in one set of potentially relevant polymerase-polymerase interactions, termed Interface I, have been shown to display growth defects (Table 1). Viruses that contained the single L342A mutation in Interface I showed dramatic temperature sensitivity, and those that either contained the single mutation L446A or the double mutation R455A/R456A gave rise to no viable virus 11. Viruses that contained the triple mutation D339A/S341A/D349A, initially reported to display only a small-plaque defect at 37°C 14 were found to be severely temperature-sensitive at slightly higher temperatures 15. The severity of mutational effects on viral growth was found to correlate with the extent to which oligomerization was disrupted 17, supporting the hypothesis that oligomeric contacts along Interface I are critical in viral growth.
Table 1. Polymerase-polymerase contacts in predicted Interface I and the effect of mutations on viral viability.
Interface I contacts were obtained from Hansen et al.17; all residues on the “palm” side of the interface that contain an atom with a van der Waal’s overlap of more than −0.4 of the residue on the “thumb” side are listed, and the references indicated. Residue D346, originally identified to be on the “palm” side of Interface I9is actually more distant and so not included in this table.
| Thumb residue |
Palm contacts | Mutations | Phenotype | Reference |
|---|---|---|---|---|
| Q411 | H336 | |||
| R415 | D339 | D339A/S341A/D349A | Highly temp.sens. | (14,15) |
| A444 | H336 | |||
| L446 | Y334, H336, Q337, V338 | L446A | Lethal | (11) |
| L447 | Y313 | |||
| P448 | L342 | L342A | Highly temp.sens. | (1) |
| E449 | T312 | |||
| S451 | L342, Q345 | |||
| T452 | T312, L342 | |||
| L453 | L342 | |||
| R455 | L342, Q345, S346 | R455A/R456A | Lethal | (14,17) |
| R455D | Lethal | (11) | ||
| R456 | D339, S341, L342 | R456D | Lethal | (11) |
However, the polymerase is a multifunctional protein involved in several steps in viral replication, and, of the 61 total mutations or sets of mutations that have been introduced into the poliovirus 3D coding region and tested for their viral phenotype, 47 have displayed temperature sensitive or lethal defects (Suppl. Table 1). Therefore, as has been pointed out 14; 19; 20, mutations that disrupt Interface I might also disrupt predicted interactions with other proteins in the RNA replication complex (reviewed in 18). Furthermore, although the mutational disruption of Interface I was found to reduce polymerase activity under conditions of low RNA concentration, presumably making RNA binding rate-limiting 11; 12; 17, no effect of such mutations was observed when assays were performed in template excess 14. Given the complexities of protein-protein interactions in viral RNA replication complexes, and the difficulty of determining the structure of two-dimensional protein arrays, we sought to identify additional contacts in polymerase-polymerase interactions surfaces, both to elucidate the structure of such arrays (Fig. 1b) and to identify additional targets for site-directed mutagenesis to test their function.
The residues originally thought to form a second oligomeric interface involve N-terminal residues, making contact with a surface on the thumb domain of the adjacent polymerase in the original crystal structure 21. This intermolecular contact was formed at an oblique angle to the interface I fibers via the intermolecular donation of amino-terminal residues, leading to a lateral association that would not be able to form planar lattices. Furthermore, subsequent structures of viral RNA-dependent RNA polymerases, beginning with NS5B of hepatitis C virus 22 and including the structure of full-length poliovirus polymerase 16 showed exclusively intramolecular protein folding, making the domain-swap model for Interface II in poliovirus polymerase unlikely. Thus, the nature of the contacts required for the second oligomeric interface, in solution or in infected cells, is an open question.
Several lines of evidence argue that the poliovirus polymerase can change conformation upon forming oligomers and modifying substrates. In the original three-dimensional structure, determined from crystals of wild-type polymerase, almost 100 amino acids of the amino terminus were disordered, and residues from this “fingers” domain were unresolved due to conformational mobility 21. The three-dimensional structure of the full-length polymerase, in which the fingers domain is resolved, was solved from crystals of polymerase that contained mutations designed to prevent formation of Interface I 16. The N-terminal residues of this polymerase still show very labile folding at physiological temperatures 23. In addition, a conformational change in the polymerase is rate-limiting during nucleotide addition in some RNA polymerase assays 24, allosteric effects of substrate and template binding have been reported 25; 26, and the lattices formed by polymerase homo-oligomerization differ morphologically in the presence and absence of RNA 12, supporting conformational flexibility of 3D polymerase.
Successful computational methods for prediction of protein-protein interactions involve exhaustive geometric searches combined with free energy calculations 27. Modeling polymerase oligomers involves several additional challenges to ensure that candidate oligomers be extendable and approximately planar. We have used computational modeling to generate plausible hypotheses that could be distinguished experimentally. A scanning wavelength turbidity assay, developed originally for microtubules28, was used to analyze polymerase oligomerization in solution. Electron microscopy was used as an independent means to examine the formation of oligomer lattices. Introduction of mutations into infectious cDNAs showed the viruses with defects in the predicted lattices showed reduced viability and synthetic lethality. These data have provided experimental support for a unique prediction for Interface II, which we propose links fibers of Interface I to form two-dimensional polymerase lattices.
RESULTS
Formation of a two-dimensional lattice depends on contacts along Interface I
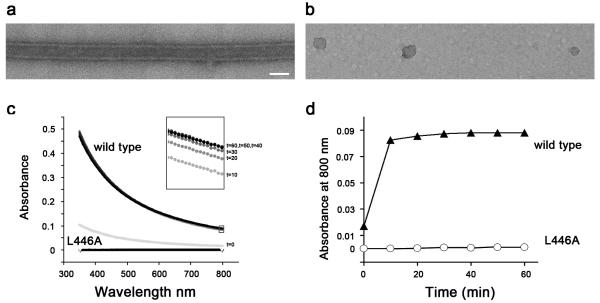
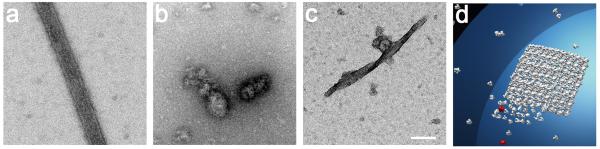
The poliovirus polymerase 3D, when purified, assembles into planar lattices of flat or curved sheets (Fig. 1b, 12; 17), which can also roll into tubular structures. We hypothesize that one of the polymerase-polymerase contacts required to form such arrays is the previously studied Interface I, observed in the first three-dimensional crystal structure of the poliovirus polymerase 11; 21. To study the effect of a single Interface I mutation on the formation of the two-dimensional lattices, we tested the effect of the lethal Leu446A point mutation (Fig. 1c). Electron microscopy of purified preparations of wild-type and L446A mutant polymerase revealed that two-dimensional arrays did not form in the presence of the mutation (Fig. 2a,b). To employ another assay to quantify the formation of large arrays in solution, we noted that wavelength-scanning turbidity assays have been used to monitor the oligomerization of tubulin29, as well as coagulin30, desmin31, thrombin and fibrinogen32, fibrinogen Longmont33, and fibrin34. Turbidity of protein oligomers in solution is determined by their size relative to the wavelength of the incident light; oligomers that are much larger than the wavelength of the light will scatter the beam. Thus, very large oligomers scatter light at both shorter wavelengths and longer wavelengths, whereas small oligomers only scatter light at shorter wavelengths 29. As shown in Figure 2c, the turbidity of both wild-type and L446A poliovirus polymerase across a range of 350-800 nm over a 60-minute time course revealed that the wild-type polymerase developed significant turbidity at both shorter and longer wavelengths. No increase in turbidity was observed at any wavelength for the purified L446A mutant polymerase, reinforcing the role of Leu446 in the polymerase-polymerase interactions (Fig. 1c) that give rise to oligomerization in solution. Lattice formation (Fig. 2a,b) correlated with turbidity at short and long wavelengths (Fig. 2c). Turbidity experiments to determine the effects of mutations at either Interface I or putative Interface II alone were subsequently performed at the longest wavelength tested, 800 nm, to avoid any background from the formation of small aggregates or fibers that used only the non-mutated interface.
Figure 2. Lattice formation and wavelength-dependent turbidity of L446A mutant polymerase.
Electron microscopy of purified wild-type (a) and L446A (b) polymerase following incubation for 24 hours and negative staining. Magnification Bar 500 Å. (c) Time courses of the acquisition of turbidity as a function of wavelength in preparations of wild-type and L446A mutant polymerase, which contains a mutation lethal to the virus and predicted to disrupt Interface I (Fig. 1c), following dilution from high-salt, high-glycerol storage buffer to a concentration of 6 μM. All times from 0 to 60 minutes were super-imposable for the L446A mutant polymerase, whereas the 0-minute time point (grey line) could be differentiated from the later time points for the wild-type polymerase. (d) The time course of the acquisition of turbidity by wild-type and L446A mutant polymerase at a single wavelength, 800 nm.
Normal mode analysis of poliovirus polymerase yields many thermodynamically plausible conformations
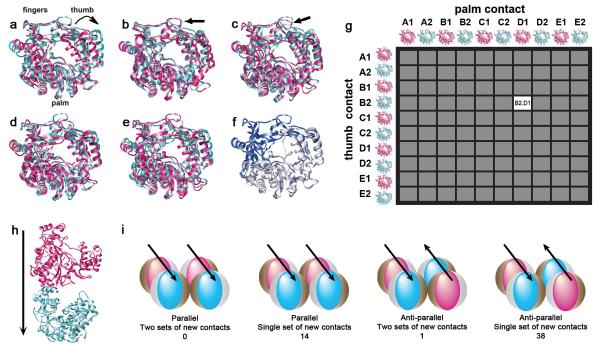
As a first step to modeling the potential interactions between fibers of Interface I that might constitute Interface II, normal mode analysis was used to sample a range of potential polymerase conformations computationally (Fig. 3a-e). Although several three-dimensional structures of the poliovirus polymerase as determined by X-ray crystallography have been published, they do not sample all the conformational space available to such a protein and do not provide a usable framework from which to model the lattice formation. The first crystal structure21 of the polymerase clearly showed Interface I, but left the fingers domain unresolved. The subsequent crystallography study produced a full-length structure 16 but introduced mutations in the polymerase at Interface I that result in a non-viable virus (Suppl. Table 1). Here, the initial step in modeling lattice formation was to explore the putative conformations of wild-type polymerase computationally (Fig 3a-e). Figure 3f compares the first two solved structures: the structure in which the N-terminal domain is disordered, but the contacts at Interface I are intact17 is shown in white, and the full-length structure in which the residues required for Interface I formation were mutagenized16 is shown in blue. To sample a range of potential conformations of 3D polymerase, the full-length structure was used as a starting point and the introduced mutations were restored to their wild-type identity. The structure was computationally modeled using normal mode analysis 35 to find a low-energy structure for the wild-type polymerase. In normal mode analysis, each atom in the system is modeled as a mass on a spring. Conformations are identified that arise from motion in which all masses in the system oscillate at the same frequency. It has been postulated that all classic motions of a system can be made from combinations of normal modes (reviewed in 36). Though many normal modes exist for any given structure, the five lowest-frequency and highest-amplitude normal modes, termed A-E, were selected to sample diverse polymerase states. Within each mode, the two conformations (1 and 2) that showed the maximum differences between them were selected as putative conformational states of the polymerase, yielding ten candidate structures (Fig. 3a-e). The modes differ qualitatively and quantitatively in the relative motion of the various domains of the polymerase. For example, in the pair of conformations shown in mode A (Fig. 3a), the thumb domain shows a hinging motion, while the fingers move laterally. In mode D (Fig. 3d), the anti-parallel beta sheets in the middle of the palm domain shift laterally. The RMSD values between states 1 and 2 of each normal mode are provided in Supplemental Table 2. It is clear that a range of potential conformations of poliovirus polymerase, as with other proteins, can be captured by normal mode analysis 35.
Figure 3. Computational modeling of polymerase conformations and of docking of two-polymerase fibers with each other.
(a-e) Normal mode analysis yielded several different modes of conformational change for the poliovirus polymerase, based on the known full-length structure of a polymerase monomer 16. The two states in contrasting colors in panels a-e are those within each of the five different normal modes that yielded the highest RMSD when compared with each other (Suppl. Table 2). (f) Two different three-dimensional structures of the poliovirus polymerase determined by X-ray crystallography are superimposed for comparison. The white structure 21 includes all but 100 N-terminal residues of the 3D polymerase, and leaves Interface I intact. The blue structure shows the full-length monomeric poliovirus 3D polymerase 16. (g) One hundred different two-polymerase Interface I fibers were formed, including the two most different structures from each of the five normal modes. (h) One of the 100 different two-polymerase fibers, with conformation B2 in the ‘thumb contact’ position in blue and conformation D1 in the ‘palm contact’ position in pink. (i) Symmetries capable of self-propagation in a parsimonious manner that were found in the 200,000 structures identified by surface convolution. The direction of the arrow signifies the palm-to-thumb alignment along Interface I. The number of potential solutions that were identified from the 200,000 candidates in each symmetry group is indicated in parentheses.
Computational generation of hypotheses for Interface II contacts
To explore potential interactions between fibers of polymerase formed by Interface I contacts, we first modeled the shortest possible polymerase fiber, two polymerases in length. Two-polymerase fibers were generated by superimposing the full-length structure of the polymerase monomer 16 onto the structure of the more incomplete crystal structure 21 that nevertheless contained the Interface I contacts. As shown schematically in Figure 3g, one hundred such two-polymerase fibers were generated computationally, so that each of the ten potential conformations could be found in the position of the ‘thumb contact’, the ‘palm contact’, or both. These 100 different two-polymerase fibers are named, as shown in Figure 3g, by the name of the conformation used in the blue ‘thumb contact’ position and the conformation used in the pink ‘palm contact’ position; the two-polymerase fiber B2.D1 is shown in Figure 3h.
To generate specific hypotheses for Interface II, the set of protein-protein interactions that would align Interface I fibers into a two-dimensional lattice, the surface correlations between each of the 100 sets of identical two-polymerase fibers were calculated 37. The use of identical fibers was based on the assumption that, while we may not know the conformation of the polymerase in the lattice, any conformation used would be likely to repeat in the iterative array. The only constraint placed on the initial space complementarity was that the residues at Interface I should not be used. All dimer-dimer complexes generated were screened based on an energetic scoring function 38; 39. The 200 lowest free energy complexes from each of the 100 sets of identical two-polymerase fibers were retained for further analysis, resulting in 20,000 candidate complexes.
To sort these potential complexes computationally, the requirements for the propagation of two-dimensional arrays were encoded to automate the selection. For example, potential complexes in which the two fibers were aligned in “T” or “L” shapes could not form two-dimensional arrays without invoking additional interfaces and therefore were excluded from further consideration due to lack of parsimony.
The remaining complexes contained only parallel or anti-parallel fibers, related by different symmetries such that one new set of protein-protein interactions would be required, or that two new interfaces were represented (Fig. 3i). One symmetry, with parallel fibers aligned so that two new interfaces would be formed, was not found in any of the 20,000 potential solutions. However, 14 parallel solutions that created one new interface, one anti-parallel solution that required two new interfaces, and 38 solutions that required one new protein-protein interaction surface were found (Fig. 3i). These potential solutions sampled all ten different predicted conformations of the polymerase (Suppl. Table 3).
Site-directed mutagenesis to test contacts in predicted Interface II
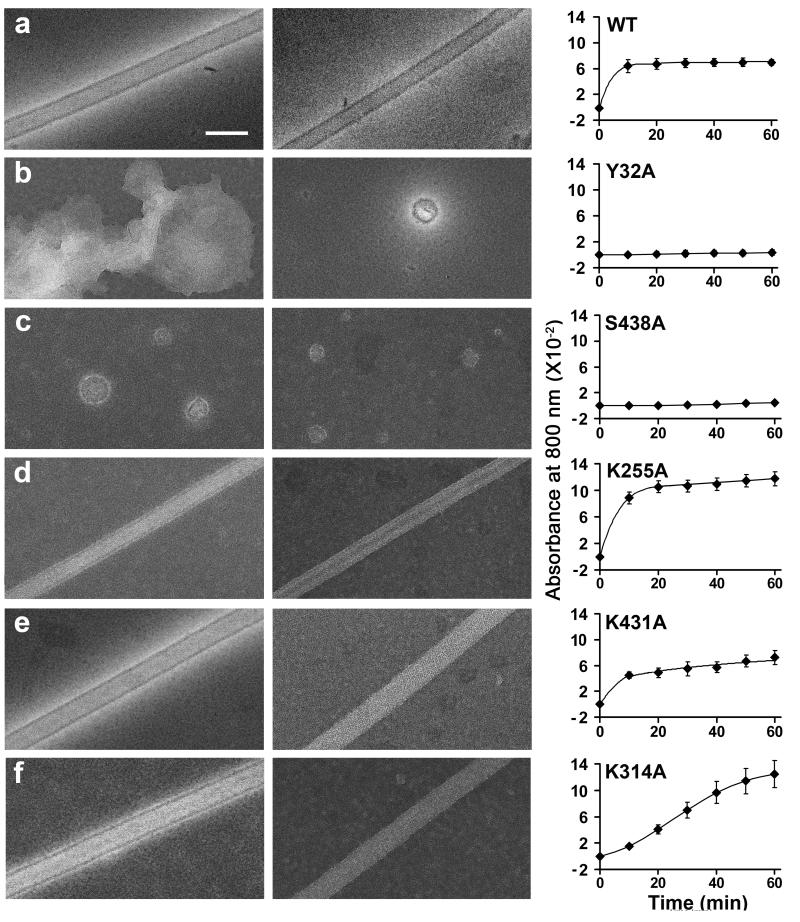
To determine whether any of the hypothesized polymerase-polymerase contacts were critical for polymerase oligomerization and lattice formation, several different site-directed mutations were made in the polymerase coding region, and the mutant polymerases were expressed and purified. To choose which residues to mutate, all of the parallel and anti-parallel solutions were examined for the most highly used residues, and for combinations of residues that would provide additional constraints on the putative lattice contacts, regardless of their effect on oligomerization as tested in our assays. After developing a candidate list of residues by looking at utilization patterns, residues residing on the surface were selected for further consideration, using the criterion of greater than 10% solvent exposure. These residues were tested for proximity within 6Å of surface residues on the adjacent polymerase to define potential contact sites. Based on this analysis, residues Tyr32 and Ser438 were chosen for mutation because these were the contacts most frequently observed in the potential structures. Residues Lys255, Lys431 and Lys314 were chosen because they were predicted to be involved in some but not all of the proposed solutions, and thus could provide discrimination between the 53 potential structures. Alanine substitutions Y32A, S438A, K255A, K431A and K314A were each introduced into an expression clone for poliovirus 3D polymerase, expressed in E. coli, and purified.
To determine whether any of the mutations interfered with lattice formation, wild-type and mutant polymerases were visualized by electron microscopy after 24 hours of incubation in the presence or absence of U24 RNA; the presence of low concentrations of RNA is known to stabilize the lattice structures 12. As shown in Figure 4, only the Y32A and S438A mutations disrupted lattice formation, whereas the K255A, K431A and K314A mutations did not. To test the formation of large oligomers of polymerase by another method, wild-type and mutant polymerases were each diluted from high-salt storage buffer into a buffer in which the polymerases were active, and the acquisition of turbidity at 800 nm was monitored as in Figure 1. Neither the Y32A nor the S438A mutant polymerases showed oligomerization during the 60-minute time period shown or at longer times. Conversely, the K255A, K431A and K314A polymerases all formed high-order oligomers upon incubation, although the detailed rates and extents of oligomerization differed from each other and from wild-type. For each of these polymerases, the ability to oligomerize in solution is consistent with the observation of lattices by electron microscopy (Fig. 4). We conclude that Tyr32 and Ser438 are likely to be directly involved in lattice formation, whereas Lys255, Lys314 and Lys431 are not. Five different candidate complexes, shown in bold in Supplemental Table 3, fit this mutational profile and thus seemed equally likely candidates for Interface II by this criterion.
Figure 4. Effect of selected mutations on lattice formation and turbidity acquisition by poliovirus 3D polymerase.
Electron microscopy of indicated polymerases incubated for 24 hours in the absence (left) or presence (middle) of 4 μM U24 RNA are shown, as is the absorbance at 800 nm as a function of time after dilution to 6 μM for each polymerase indicated (right). (a) wild-type, (b) Y32A, (c) S438A, (d) K255A, (e) K431A, and (f) K314A polymerases. White magnification bar, 100 nm.
For a set of contacts to form a repeatable, two-dimensional lattice such as that shown in Figure 1b, the mini-lattices, comprising four polymerases each, should be further extendable in a two-dimensional plane. Of the five solutions in Suppl. Table 3 supported by the mutagenesis data, only the anti-parallel complex A2.C1.142 displayed this property, in which a two-dimensional lattice could be extended. After approximately 16 polymerases in the A2.C1.142 conformation were encompassed, however, the addition of further subunits led to steric hindrance. To explore the conformational space around complex A2.C1.142 further, a second round of space complementarity was performed, using all 200 two-polymerase fibers from Figure 3g, and specifying that residues Tyr32 and Ser438, but not Lys255, Lys431 or Lys314, should be within 5Å of the protein-protein interface. Of ten predicted antiparallel complexes that resulted from this computational search, one was extendable into an approximately flat lattice using unlimited numbers of polymerases. This complex, termed the F2 lattice, was further refined to minimize its Gibbs free energy, and became our leading candidate (Fig. 5a); atomic coordinates of this tetramer and of a 30-mer array are included as Supplementary Information, Files #1 and #2, respectively.
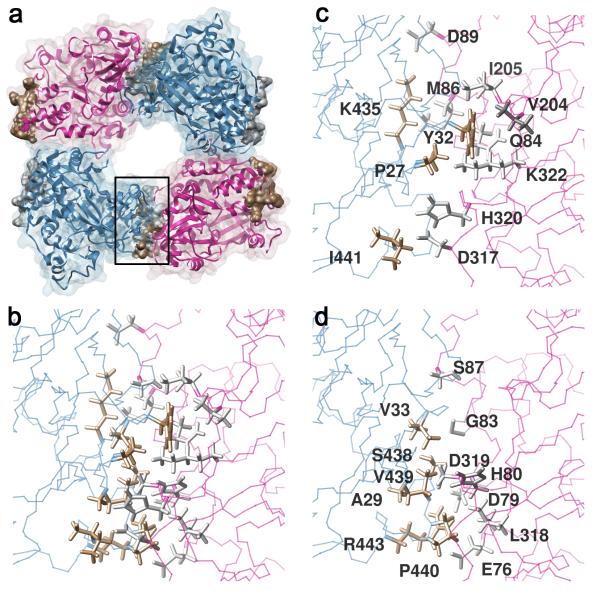
Figure 5. Characteristics of predicted polymerase complexes with Interface II interactions in complex F2.
(a) Ribbon diagram of four polymerase molecules, interacting along two interfaces to form the building block for a two-dimensional lattice. Previously characterized Interface I links the pink and blue polymerases vertically, as in Figure 3h. Computationally predicted Interface II links the blue and pink polymerases from left to right as in the “Anti-Parallel, Single Set of New Contacts” example in Figure 3i. Contact residues on the S438-containing surface are shown in gold, and contact residues on the S87-containing surface are shown in silver. (b) Close-up of proposed Interface II shows all the contacts in gold on the blue, Ser 438-containing, interface and in silver on the pink, Ser87-containing interface. All residues that contain an atom with a van der Waal’s overlap of more than −0.4 of an atom are shown. (c,d) The individual amino acids and their interactions can be seen more clearly when separated into two diagrams.
Effect of mutations in predicted Interface II contacts on viral viability
If the predicted Interface II contacts in the F2 lattice are important for two-dimensional array formation, and if two-dimensional arrays are involved in some aspect of the poliovirus replicative cycle, then mutations that disrupt these contacts in the F2 lattice should have deleterious effects on viral growth. The polymerase-polymerase interactions that give rise to the Interface II contacts in the F2 lattice are shown in Figure 5b-d and summarized in Table 2. Ser438 is found in the center of this interface, contacting residues Asp79, His80 and Gly83 of the opposing polymerase. To test the importance of the identity of Ser438 in viral viability, mutations were introduced into the infectious poliovirus cDNA to substitute a Trp residue at this position. RNA from the mutagenized cDNA was transfected into cells and the resulting virus (Materials and Methods) was recovered. Compared to that of wild-type virus recovered in the same way, the mutant virus was found to have a strong temperature-sensitive defect (Fig. 6). Therefore, substitution of Ser438 with a bulky residue disrupted viral viability. To attempt to identify residues on the opposite side of Interface II from Ser438, mutant virus 3D-S438W virus was propagated for six hours at 39.5°C in three different pools to select phenotypic revertants of the temperature-sensitive phenotype. Each individual isolate sequenced from each of the three pools was found to be a direct revertant, encoding only Ser at position 438, although different mutations were instituted to achieve this codon change (data not shown). Even when Ser438 was changed to a similarly sized Ala residue, a greater than ten-fold decrease in yield was observed during a single-cycle infection at 39.5°C (Fig. 6b) even though no difference in the plaque phenotype was observed (Fig. 6a). Therefore, the identity of Ser438, predicted to be central to the protein-protein contacts at Interface II, is important for viral viability.
Table 2. Polymerase-polymerase contacts in complex F2.
Potential Interface II contacts were determined from the predicted structures of complex F2. The two polymerases are denoted as the polymerase on the “Ser87” side, the pink one in Figure 5, with silver intermolecular contacts, and the “Ser438 side”, the blue one in Figure 5, with gold intermolecular contacts. All residues on the “Ser87” polymerase that contain an atom with a van der Waal’s overlap of more than −0.4 of an atom on the “Ser438” polymerase are listed, and their partners indicated. Any known mutations that contain these residues and have been tested for their phenotype in viruses are indicated and the results summarized. The phenotypes of mutant viruses reported in the present manuscript (Fig. 6) are reported in parentheses.
| Ser87 side silver contacts |
Ser438 side gold contacts |
Known mutations | Phenotypes | Reference |
|---|---|---|---|---|
| E76 | P440, R443 | K75A/ E76A | Temperature sensitive | (17) |
| D79 | S438, R443 | D79A/ H80A | Temperature sensitive | (17) |
| H80 | S438, V439, P440 | |||
| G83 | V33, S438 | V33A | Lethal | (11) |
| Q84 | Y32, V33 | |||
| M86 | K435 | |||
| S87 | Y32, V33, F34, E35 | F34L | Temperature sensitive | (48) |
| D89 | E35 | D89A/ E93A | No defect | (17) |
| V204 | Y32 | |||
| I205 | Y32 | |||
| D317 | I441 | |||
| L318 | P440 | |||
| D319 | S28, A29, F30, V439 | F30S | Lethal | (48) |
| H320 | E26, P27 | |||
| K322 | P27, Y32 | |||
| S438A | (Slightly temp. sens.) | (this work) | ||
| S438W | (Severely temp. sens.) | (this work) | ||
| D89A | (No defect) | (this work) | ||
| D89A/S438A | (Severely temp. sens.) | (this work) |
Figure 6. Growth phenotypes of viruses that contain mutations that affect the stability of Interface II of the predicted F2 lattice.
(a) Plaque phenotypes of viruses at 32.5°C and 39.5°C on HeLa cells. (b) Time courses of viral yields at various times after infection of HeLa cells with wild-type and mutant viruses as indicated, at multiplicities of infection of ten plaque-forming units per cell. The values for the final yields are given next to the relevant curve.
To investigate further the effects of mutation on the formation of proposed Interface II, we inquired whether any the 61 mutations or sets of mutations introduced into poliovirus 3D polymerase whose phenotypes have been tested in viruses (Suppl. Table 1) were found in the proposed interface. Six mutations, or sets of mutations, were predicted to affect Interface II as defined by the F2 lattice. Two of these mutations were lethal, and three gave rise to temperature-sensitive viruses. However, one of them, comprising mutations D89A and E93A, gave rise to virus with no apparent growth defect. To reconcile this observation with the predicted participation of Asp89 in Interface II, virus that contained the single mutation D89A was constructed. The recovered virus was also found to display no growth defect in plaque assays (Fig. 6a) and only a slight decrease in yield during a single-cycle infection at 39.5°C (Fig. 6b). Therefore, if the interaction along predicted Interface II of the F2 lattice shown in Figure 5 is important during viral infection, it must be sufficiently robust that it can withstand the D89A mutation. In support of this idea, when the D89A mutation was combined with the S438A mutation, the resulting D89A/S438A virus was highly temperature sensitive in plaque assays (Fig. 6a), and showed a greater than 200-fold decrease in yield in a single-cycle infection at 39.5°C (Fig. 6b). Thus, the D89A and S438A mutations display synthetic lethality at 39.5°C. Although synthetic lethality between two weak alleles can occur when the alleles are in different processes, it most often indicates that they are involved in the same process or pathway 40.
If the Interface II contacts made by S438A polymerase are weakened, but those in Interface I can still form, then the addition of S438A mutant polymerase to wild-type polymerase might disrupt regular lattice formation by wild-type polymerase. As can be seen in Figure 7, the presence of S438A polymerase in a solution of wild-type polymerase altered the resulting wild-type lattice structure. Thus, although S438A polymerase alone does not form stable lattices (Fig. 4), the residual interactions at Interface I, Interface II, or both, allow it to interact with wild-type polymerase and thereby disrupt wild-type lattice formation.
Figure 7. Effect of mixing S438A mutant polymerase with wild-type polymerase on lattice formation.
Electron microscopy of structures formed by incubating 2 μM wild-type polymerase for 24 hours, followed by negative staining. (b) Structures formed by incubating 2 μM wild-type polymerase in the presence of 2 μM S438A polymerase for 24 hours. (c) Scaled diagrammatic model of polymerase lattice being formed on a membranous vescicle 200 nm in diameter, and being disrupted by the presence of mutant polymerase. Polymerases from the F2 lattice were placed using Chimera software 52. Magnification Bar 500 Å.
DISCUSSION
Conformational flexibility of individual subunits has led to a unique prediction for lattice formation by poliovirus 3D polymerase into an oligomeric array. Interface I contacts were previously described, and are confirmed here by turbidity and electron microscopic data obtained with L446A mutant polymerase. The second interface was posited by a combination of normal-mode analysis, shape complementarity and energy minimization. Hypotheses were screened computationally for their ability to form approximately planar arrays.
Potential complexes were then tested experimentally by site-directed mutagenesis of putative contact residues, with results that support the reliability of the computational approach. Computational models were eliminated when mutation of a residue that appeared to be essential caused no change in the oligomerization properties of the expressed protein. Mutations that resulted in significant changes in the ability of the protein to oligomerize were considered potential contact points and investigated further. We conclude that residues K255, K314 and K431 are not essential for lattice formation, whereas Y32 and S438 are important for the polymerase-polymerase interactions that lead to 3D polymerase oligomerization. Our analysis led to one independent solution for placement of polymerase subunits in a two-dimensional lattice; this “F2 lattice” was further optimized by energy minimization.
The proposed lattice features polymerase-polymerase interactions that include, on both sides of Interface II, residues from the very flexible amino terminus of the protein in addition to more stably folded residues. A loop formed by residues 29-34 from the index finger contacts an alpha helical region in the palm, residues 82-88. A coiled region, residues 62-65 from the index finger, contacts residues 239-242 from the palm, shown in the crystal structure to be a beta strand/ alpha helix transition.
How might such a highly oligomeric structure facilitate RNA replication? First, the porous nature of the polymerase lattice is likely to allow the participation of other viral and cellular proteins, which are known to be present in the viral RNA replication complex. In imagining the organization of an RNA replication complex associated with a two-dimensional membrane surface, it is interesting to remember that, for most DNA-dependent DNA or RNA synthesis, the proteins are immobilized and the DNA template moves through a static DNA or RNA polymerase complex (reviewed in 41; 42). For RNA synthesis of double-stranded RNA viruses, for which the polymerases are embedded into virion structures 43, it must certainly be the case that the template RNA strand transits through an immobilized polymerase; we propose that this is the case for the membrane-associated poliovirus polymerase complex as well and implies the formation of a large, immobile complex on the membrane surface. The possible generality of a planar array of polymerases is noted by tests that superposed polymerases from other RNA viruses onto the poliovirus polymerase lattice. The superposition of the conserved catalytic domain of Dengue virus polymerase (DENV pdb 2j7w) onto the corresponding domain of poliovirus polymerase in the F2 lattice (Fig. 5) showed only minor steric hindrance. For the polymerase of hepatitis C virus (HCV pdb 1csj), overlaps of loop regions and of a single α-helical region could both be relieved by rotation of the helix and small conformational changes in the loops. Therefore, the predicted interactions at Interface II in poliovirus can already be used for structure-function analysis of similar interactions in other RNA-dependent polymerases.
During infection, polymerases that are catalytically inactive can rescue infections with viruses whose polymerases have other defects 17, arguing that 3D polymerase has multiple functions during infection, at least one of which is structural. We suggest that the polymerase lattice creates channels of aligned enzymes, in which active sites and other RNA-binding residues are positioned in spaced rows so that RNA elongation, which is likely to take place at a single active site in copying any individual template 17, is supported by aligned RNA-binding surfaces. Thus, the functional oligomer is robust and tolerates polymerases that are deficient in RNA polymerization because only a single functional active site is needed for elongation; non-elongating enzymes still play a functional role in supporting the RNA strand as it is passed through the lattice. Cooperativity and processivity are predicted advantages of such an intracellular structure. Furthermore, we consider it highly likely that the genetic dominance of certain classes of defective polymerases 44 results from the formation of mixed polymerase lattices whose defective members poison the functions of other polymerases in the lattice. Investigations are currently ongoing to identify which polymerase functions, when present in the lattice, dominantly inhibit wild-type polymerase function.
How do flat two-dimensional sheets form on a curved membrane surface, tethered by contacts with viral protein 3AB 45; 46; 47, affinity for phosphatidyl inositol 4-phosphate 48, or both? A lattice that coats a spherical vesicle would bend to follow the natural curvature of the vesicle. As illustrated in Figure 7c, a polymerase lattice that completely coated a 200-nm membranous vesicle would cover the 125,000 square nanometers of surface area with approximately 5,000 polymerases. Although sufficient polymerase is made within infected cells to fulfill this scenario, it seems more likely that polymerase lattices are found in smaller patches on the membrane surface.
Many proteins form two-dimensional sheet-like lattices that can be visualized by electron microscopy, including FtsZ, the bacterial homolog of tubulin, and P1, the polymerase-associated protein of phage Φ29 49. For FtsZ, crystal structures are available and a single protein-protein interaction site yielding a fiber is known 50. The problem of how the subunits are geometrically arranged to produce the overall structure is almost identical to the question of lateral association of polymerase fibers discussed here. The general features of these problems are similar enough that the same computational methods of surface convolution and selection based on parsimony and repeatable structure may also be employed to produce directly testable, plausible orientations of the fibers within such lattices , which might then be testable using new tubular lattices comprising SepF rings and FtsZ fibers 51.
MATERIALS AND METHODS
Visualization of the Interface I fiber and polymerase lattice
The model polymerase fibers along Interface I (Fig 1c) were made with structural superpositioning of the exact contacts of Interface I, defined in an asymmetric dimer of the incomplete structure 21 with the full-length structure of polymerase that was crystallized using a mutant polymerase unable to form Interface I contacts 16. The full-length polymerase structure was superimposed onto the two incomplete structures of wild-type polymerase to create a two-unit fiber using the Deep View structural alignment program 53. Additional polymerases were added to either end of the fiber by superimposing the asymmetric dimer onto the respective thumb-contacting and palm-contacting polymerases. Superposition beyond four polymerases was not straightforward with Deep View, and was done using Chimera 52; 54. Polymerases were added to produce an approximately planar lattice using the superposition of the polymerase from the F2 tetramer that extended the array with minimized steric interference from adjacent polymerases.
Modeling potential conformations of polymerases in an asymmetric dimer
Potential polymerase conformations were sampled using normal mode analysis 35. Coordinates for a full-length wild-type polymerase were obtained by substituting the wild-type residues Arg455 and Leu446 for the aspartic acid residues used in the mutant polymerase used to solve the crystal structure 16. The resulting structure was subjected to repeated rounds of energetic minimization with the Deep View program. Normal modes were calculated with the elastic network model server EL NEMO 35. Within each of the five lowest energy modes, the two conformations with the highest RMSD, were selected (Suppl. Table 2). The RMSD between each normal mode conformation and the full-length crystal structure 16 is approximately 1.5 Å, and approximately 3 Å between the two conformations taken from the same normal mode (Suppl. Table 2).
Finding energetically favorable complexes by docking
One hundred different asymmetric dimers were created by superpositioning different polymerase conformations, each made by normal mode analysis, onto an Interface I fiber. Each asymmetric dimer comprised one polymerase in the thumb-contacting position, sampled from the ten possible conformations, and the other in the palm-contacting position, also sampled from the ten possible conformations (Fig 3a). For each of these one hundred asymmetric dimers, energy minimization was performed using Deep View 53 to eliminate any steric clashes created during the superpositioning.
To search for possible Interface II contacts, each asymmetric dimer was docked to an identical fiber using ZDOCK 38; 55 and the GROMOS forcefield 56. During the docking calculation, the residues involved in Interface I, on both thumb and palm surfaces of the protein, were excluded from use as contact surfaces during the docking. The required search-space was reduced by limiting the composition of each lattice to two polymerase conformations. The 200 lowest energy conformations were kept for further analysis, yielding 20,000 complexes.
Clustering of polymerase conformations
Pairs of asymmetric dimers generated in the docking step were categorized initially by hierarchical clustering. To compute the similarity in contact residues, a list of contact residues with C-α carbons within 7Å was computed for each polymerase in the asymmetric dimer pair. A quantitative comparison between any two of the 20,000 asymmetric dimer pairs of the polymerase-polymerase contacts was expressed as the ratio of shared contacts to total contacts, with a weighting factor to adjust for the number of contacts involved. An initial clustering approach classified geometrically similar complexes into distinct sets. To relax the initial clustering and get a course-grained yet geometrically similar classification scheme, the merge operation was modified to take the union of intersecting residues in the pair of complexes, as opposed to the intersection.
Selection of repeatable parallel and anti-parallel tetramers from all possible parallel and anti-parallel asymmetric dimer pairs (tetramers)
Repeatable anti-parallel and parallel tetramers have structural properties that allow for parsimonious propagation. Vector representations for the 100 different protein fibers (Fig. 3g,h) were implemented in Python to allow for automated encoding and selection of repeatable tetramers. A vector from the active site of the thumb-contacting polymerase to the active site of the palm-contacting polymerase was defined for both of the asymmetric dimers in a candidate tetrameric complex. The cosine of the angle between the two vectors was calculated; parallel fibers had cosines close to 1.0 while anti-parallel fibers had cosines close to −1.0 .
A vector was defined from the active site of the thumb-contacting polymerase to the atom closest to the weighted average position (x, y, z coordinates) of the contact surface with the polymerase contacting it through the predicted interface II. The identical vector was defined in the adjacent polymerase and the cosine of the angle between the vectors was calculated as before. Values near −1.0 indicated rotational symmetry whereas values near +1.0 indicated translational symmetry for the two asymmetric dimers.
Site-directed mutagenesis, purification of poliovirus polymerase and scanning wavelength turbidity assays
Wild-type and Y32A, Y73A, K255A, K314A, K431A, S438A and L446A mutant poliovirus polymerases were purified as described 17. Turbidity assays were performed with 6μM polymerase in 13.5mM Tris (pH 7.5) at a total volume of 50μL. The wavelength-dependent turbidity was measured over the 60 minute time course with a Perkin-Elmer Lambda 35 UV/vis.
Electron microscopy of higher-order structures of poliovirus polymerase
Reaction mixes containing 2μM polymerase, 10mM Tris-HCl (pH 7.5), 1mM EDTA, 30mM NaCl and 0.01% n-octyl glucoside were incubated at 30° C for 30 min, followed by 10 min on ice and overnight at 4° C. Eight μL of each reaction were placed on carbon-coated copper grids and incubated at room temperature for 10 min. The grids were washed with 10mM Tris-HCl pH 7.4, 1mM EDTA, 30mM NaCl and negatively stained with 1% uranyl acetate. Images were recorded at minimal electron dose using a Philips CM-12 electron microscope. The negatives were digitized using a Nikon scanner at a step size corresponding to a final pixel size of 1.4 Angstroms.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Douglas Brutlag and Russ Altman for ongoing discussions of computational approaches, Christopher Akey for helpful discussions, and Peter Sarnow and Stephen Floor for critical reading of the manuscript. This work was supported by NIH training grants to Stanford University (for A.T., E.J.T. and J.W.), an NIH Director’s Pioneer award (to K.K.), and Boston University School of Medicine (for E.B. and J.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Pata JD, Schultz SC, Kirkegaard K. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA. 1995;1:466–77. [PMC free article] [PubMed] [Google Scholar]
- 2.Wang QM, Hockman MA, Staschke K, Johnson RB, Case KA, Lu J, Parsons S, Zhang F, Rathnachalam R, Kirkegaard K, Colacino JM. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J Virol. 2002;76:3865–72. doi: 10.1128/JVI.76.8.3865-3872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin W, Luo H, Nomura T, Hayashi N, Yamashita T, Murakami S. Oligomeric interaction of hepatitis C virus NS5B is critical for catalytic activity of RNA-dependent RNA polymerase. J Biol Chem. 2002;277:2132–7. doi: 10.1074/jbc.M106880200. [DOI] [PubMed] [Google Scholar]
- 4.O’Reilly EK, Paul JD, Kao CC. Analysis of the interaction of viral RNA replication proteins by using the yeast two-hybrid assay. J Virol. 1997;71:7526–32. doi: 10.1128/jvi.71.10.7526-7532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goregaoker SP, Culver JN. Oligomerization and activity of the helicase domain of the tobacco mosaic virus 126- and 183-kilodalton replicase proteins. J Virol. 2003;77:3549–56. doi: 10.1128/JVI.77.6.3549-3556.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dye BT, Miller DJ, Ahlquist P. In vivo self-interaction of nodavirus RNA replicase protein a revealed by fluorescence resonance energy transfer. J Virol. 2005;79:8909–19. doi: 10.1128/JVI.79.14.8909-8919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinnaswamy S, Murali A, Li P, Fujisaki K, Kao CC. Regulation of de novo-initiated RNA synthesis in hepatitis C virus RNA-dependent RNA polymerase by intermolecular interactions. J Virol. 84:5923–35. doi: 10.1128/JVI.02446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellon-Echeverria I, Lopez-Jimenez AJ, Clemente-Casares P, Mas A. Monitoring hepatitis C virus (HCV) RNA-dependent RNA polymerase oligomerization by a FRET-based in vitro system. Antiviral Res. 2010;87:57–66. doi: 10.1016/j.antiviral.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Hogbom M, Jager K, Robel I, Unge T, Rohayem J. The active form of the norovirus RNA-dependent RNA polymerase is a homodimer with cooperative activity. J Gen Virol. 2009;90:281–91. doi: 10.1099/vir.0.005629-0. [DOI] [PubMed] [Google Scholar]
- 10.Beckman MT, Kirkegaard K. Site size of cooperative single-stranded RNA binding by poliovirus RNA-dependent RNA polymerase. J Biol Chem. 1998;273:6724–30. doi: 10.1074/jbc.273.12.6724. [DOI] [PubMed] [Google Scholar]
- 11.Hobson SD, Rosenblum ES, Richards OC, Richmond K, Kirkegaard K, Schultz SC. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 2001;20:1153–63. doi: 10.1093/emboj/20.5.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyle JM, Bullitt E, Bienz K, Kirkegaard K. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science. 2002;296:2218–22. doi: 10.1126/science.1070585. [DOI] [PubMed] [Google Scholar]
- 13.Paul AV, Mugavero J, Yin J, Hobson S, Schultz S, van Boom JH, Wimmer E. Studies on the attenuation phenotype of polio vaccines: poliovirus RNA polymerase derived from Sabin type 1 sequence is temperature sensitive in the uridylylation of VPg. Virology. 2000;272:72–84. doi: 10.1006/viro.2000.0354. [DOI] [PubMed] [Google Scholar]
- 14.Pathak HB, Ghosh SK, Roberts AW, Sharma SD, Yoder JD, Arnold JJ, Gohara DW, Barton DJ, Paul AV, Cameron CE. Structure-function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol). A surface of the primary oligomerization domain functions in capsid precursor processing and VPg uridylylation. J Biol Chem. 2002;277:31551–62. doi: 10.1074/jbc.M204408200. [DOI] [PubMed] [Google Scholar]
- 15.Burgon TB, Jenkins JA, Deitz SB, Spagnolo JF, Kirkegaard K. Bypass suppression of small-plaque phenotypes by a mutation in poliovirus 2A that enhances apoptosis. J Virol. 2009;83:10129–39. doi: 10.1128/JVI.00642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson AA, Peersen OB. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004;23:3462–71. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spagnolo JF, Rossignol E, Bullitt E, Kirkegaard K. Enzymatic and non-enzymatic functions of viral RNA-dependent RNA polymerases within oligomeric arrays. RNA. 2010;16:382–93. doi: 10.1261/rna.1955410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirkegaard K, Semler B. RNA replication: the process. In: Ehrenfeld E, Domingo E, Roos R, editors. The Picornaviruses. A.S.M.; Herndon, VA: 2010. [Google Scholar]
- 19.Shen M, Reitman ZJ, Zhao Y, Moustafa I, Wang Q, Arnold JJ, Pathak HB, Cameron CE. Picornavirus genome replication. Identification of the surface of the poliovirus (PV) 3C dimer that interacts with PV 3Dpol during VPg uridylylation and construction of a structural model for the PV 3C2-3Dpol complex. J Biol Chem. 2008;283:875–88. doi: 10.1074/jbc.M707907200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcotte LL, Wass AB, Gohara DW, Pathak HB, Arnold JJ, Filman DJ, Cameron CE, Hogle JM. Crystal structure of poliovirus 3CD protein: virally encoded protease and precursor to the RNA-dependent RNA polymerase. J Virol. 2007;81:3583–96. doi: 10.1128/JVI.02306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen JL, Long AM, Schultz SC. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–22. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 22.Bressanelli S, Tomei L, Roussel A, Incitti I, Vitale RL, Mathieu M, De Francesco R, Rey FA. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc Natl Acad Sci U S A. 1999;96:13034–9. doi: 10.1073/pnas.96.23.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson AA, Albertini RA, Peersen OB. Stabilization of poliovirus polymerase by NTP binding and fingers-thumb interactions. J Mol Biol. 2007;366:1459–74. doi: 10.1016/j.jmb.2006.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnold JJ, Gohara DW, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+ Biochemistry. 2004;43:5138–48. doi: 10.1021/bi035213q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boerner JE, Lyle JM, Daijogo S, Semler BL, Schultz SC, Kirkegaard K, Richards OC. Allosteric effects of ligands and mutations on poliovirus RNA-dependent RNA polymerase. J Virol. 2005;79:7803–11. doi: 10.1128/JVI.79.12.7803-7811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richards OC, Spagnolo JF, Lyle JM, Vleck SE, Kuchta RD, Kirkegaard K. Intramolecular and intermolecular uridylylation by poliovirus RNA-dependent RNA polymerase. J Virol. 2006;80:7405–15. doi: 10.1128/JVI.02533-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sousa SF, Fernandes PA, Ramos MJ. Protein-ligand docking: current status and future challenges. Proteins. 2006;65:15–26. doi: 10.1002/prot.21082. [DOI] [PubMed] [Google Scholar]
- 28.Gaskin F, Cantor CR, Shelanski ML. Biochemical studies on the in vitro assembly and disassembly of microtubules. Ann N Y Acad Sci. 1975;253:133–46. doi: 10.1111/j.1749-6632.1975.tb19197.x. [DOI] [PubMed] [Google Scholar]
- 29.Hall D, Minton AP. Turbidity as a probe of tubulin polymerization kinetics: a theoretical and experimental re-examination. Anal Biochem. 2005;345:198–213. doi: 10.1016/j.ab.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Moody TP, Donovan MA, Laue TM. Turbidimetric studies of Limulus coagulin gel formation. Biophys J. 1996;71:2012–21. doi: 10.1016/S0006-3495(96)79399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou RG, Stromer MH, Robson RM, Huiatt TW. Determination of the critical concentration required for desmin assembly. Biochem J. 1990;272:139–45. doi: 10.1042/bj2720139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blomback B, Carlsson K, Hessel B, Liljeborg A, Procyk R, Aslund N. Native fibrin gel networks observed by 3D microscopy, permeation and turbidity. Biochim Biophys Acta. 1989;997:96–110. doi: 10.1016/0167-4838(89)90140-4. [DOI] [PubMed] [Google Scholar]
- 33.Lounes KC, Lefkowitz JB, Henschen-Edman AH, Coates AI, Hantgan RR, Lord ST. The impaired polymerization of fibrinogen Longmont (Bbeta166Arg-->Cys) is not improved by removal of disulfide-linked dimers from a mixture of dimers and cysteine-linked monomers. Blood. 2001;98:661–6. doi: 10.1182/blood.v98.3.661. [DOI] [PubMed] [Google Scholar]
- 34.Bale MD, Mosher DF. Effects of thrombospondin on fibrin polymerization and structure. J Biol Chem. 1986;261:862–8. [PubMed] [Google Scholar]
- 35.Suhre K, Sanejouand YH. On the potential of normal-mode analysis for solving difficult molecular-replacement problems. Acta Crystallogr D Biol Crystallogr. 2004;60:796–9. doi: 10.1107/S0907444904001982. [DOI] [PubMed] [Google Scholar]
- 36.Bahar I, Lezon TR, Yang LW, Eyal E. Global dynamics of proteins: bridging between structure and function. Annu Rev Biophys. 39:23–42. doi: 10.1146/annurev.biophys.093008.131258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, Vakser IA. Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc Natl Acad Sci U S A. 1992;89:2195–9. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiehe K, Pierce B, Mintseris J, Tong WW, Anderson R, Chen R, Weng Z. ZDOCK and RDOCK performance in CAPRI rounds 3, 4, and 5. Proteins. 2005;60:207–13. doi: 10.1002/prot.20559. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Chen R, Weng Z. RDOCK: refinement of rigid-body protein docking predictions. Proteins. 2003;53:693–707. doi: 10.1002/prot.10460. [DOI] [PubMed] [Google Scholar]
- 40.Hartman J. L. t., Garvik B, Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291:1001–4. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- 41.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–40. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 42.Giaever GN, Snyder L, Wang JC. DNA supercoiling in vivo. Biophys Chem. 1988;29:7–15. doi: 10.1016/0301-4622(88)87020-0. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 A. Nat Struct Biol. 2003;10:1011–8. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crowder S, Kirkegaard K. Trans-dominant inhibition of RNA viral replication can slow growth of drug-resistant viruses. Nat Genet. 2005;37:701–9. doi: 10.1038/ng1583. [DOI] [PubMed] [Google Scholar]
- 45.Lama J, Paul AV, Harris KS, Wimmer E. Properties of purified recombinant poliovirus protein 3aB as substrate for viral proteinases and as co-factor for RNA polymerase 3Dpol. J Biol Chem. 1994;269:66–70. [PubMed] [Google Scholar]
- 46.Towner JS, Ho TV, Semler BL. Determinants of membrane association for poliovirus protein 3AB. J Biol Chem. 1996;271:26810–8. doi: 10.1074/jbc.271.43.26810. [DOI] [PubMed] [Google Scholar]
- 47.Lyle JM, Clewell A, Richmond K, Richards OC, Hope DA, Schultz SC, Kirkegaard K. Similar structural basis for membrane localization and protein priming by an RNA-dependent RNA polymerase. J Biol Chem. 2002;277:16324–31. doi: 10.1074/jbc.M112429200. [DOI] [PubMed] [Google Scholar]
- 48.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bravo A, Salas M. Polymerization of bacteriophage phi 29 replication protein p1 into protofilament sheets. EMBO J. 1998;17:6096–105. doi: 10.1093/emboj/17.20.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliva MA, Cordell SC, Lowe J. Structural insights into FtsZ protofilament formation. Nat Struct Mol Biol. 2004;11:1243–50. doi: 10.1038/nsmb855. [DOI] [PubMed] [Google Scholar]
- 51.Gündoğdu ME, Kawai Y, Pavlendova N, Ogasawara N, Errington J, Scheffers DJ, Hamoen LW. Large ring polymers align FtsZ polymers for normal septum formation. EMBO J. 2011;30:617–26. doi: 10.1038/emboj.2010.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–7. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 53.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–92. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005;13:473–82. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 55.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–7. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 56.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–18. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.