Abstract
Background
Genetic susceptibility for cancer can differ substantially among families. We use trait-related covariates to identify a genetically homogeneous subset of families with the best evidence for linkage in the presence of heterogeneity.
Methods
We performed a genome-wide linkage screen in 93 families. Samples and data were collected by the familial lung cancer recruitment sites of the Genetic Epidemiology of Lung Cancer Consortium. We estimated linkage scores for each family by the Markov chain Monte Carlo procedure using SimWalk2 software. We used ordered subset analysis (OSA) to identify genetically homogenous families by ordering families based on a disease-associated covariate. We performed permutation tests to determine the relationship between the trait-related covariate and the evidence for linkage.
Results
A genome-wide screen for lung cancer loci identified strong evidence for linkage to 6q23-25 and suggestive evidence for linkage to 12q24 using OSA, with peak logarithm of odds (LOD) scores of 4.19 and 2.79, respectively. We found other chromosomes also suggestive for linkages, including 5q31-q33, 14q11, and 16q24.
Conclusions
Our OSA results support 6q as a lung cancer susceptibility locus and provide evidence for disease linkage on 12q24. This study further increased our understanding of the inheritability for lung cancer. Validation studies using larger sample size are needed to verify the presence of several other chromosomal regions suggestive of an increased risk for lung cancer and/or other cancers.
Impact
OSA can reduce genetic heterogeneity in linkage study and may assist in revealing novel susceptibility loci.
Keywords: Ordered subset analysis, lung cancer, cancer susceptibility genes, linkage analysis, standardized incidence ratio
Introduction
Lung cancer remains the leading cause of cancer-related mortality in both men (30%) and women (26%) in the United States (1). Exposures to environmental factors including tobacco smoke, radon gas, asbestos, arsenic, and some forms of silica and chromium are strongly associated with lung cancer (2, 3) . Although smoking is a contributing cause in 85% to 90% of lung cancer cases, only a small fraction of smokers actually develop lung cancer (4). This finding indicates variability in individual susceptibility to lung cancer in response to tobacco.
Epidemiologic studies have shown that lung cancer aggregates in families. Relatives of lung cancer patients are two to three times more likely to develop lung cancer than are relatives of control participants in family-based studies (5, 6). A large segregation analysis conducted by Ooi et al (7) suggested that lung cancer inheritance was compatible with Mendelian segregation of a single codominant locus that affects early onset of the disease. Xu et al (8)conducted segregation analysis and reported that a few loci contribute to lung cancer risk. Bailey-Wilson et al (9) mapped a lung cancer susceptibility locus to a region of chromosome 6q, but noted heterogeneity in evidence for linkage among families.
Lung cancer is a complex disease caused by complex interactions between multiple genes and environmental factors. Genetic susceptibility to lung cancer can differ substantially across families (9). In planning any genetic linkage analysis to map genes for lung cancer, researchers must account for this heterogeneity among the families in the model. Bailey-Wilson et al (9) performed a genome-wide linkage study to map disease susceptibility loci and used the admixture approach (10) to compute heterogeneity logarithm of odds (LOD) scores (HLODs) by assuming a single heterogeneity parameter across different families. The researchers detected significant evidence for linkage only on chromosome 6q23-25 in a subset of families with 4 or more cases, but not in the entire collection of families, indicating that of the HLOD approach did not adequately account for genetic heterogeneity in the whole dataset, while further stratification of the entire sample into subsets may define genetically more homogeneous subsets of families.
A predivided sample test may be used to assess genetic heterogeneity if families can be stratified by a priori cutoff point using a covariate(11). However, choosing the best cutoff point in defining a genetically homogeneous subset is often not trivial. Evaluating evidence for linkage according to family-specific trait-related characteristics can improve the evidence for linkage, as shown in the landmark paper by Hall et al., (1990) (12)in which evidence for linkage of breast cancer to BRCA1 was far greater among the families with an early average age of onset. However, post-hoc analysis of trait-related characteristics can be criticized as data dredging unless rigorous approaches to control the false positive rate are adopted. Hauser et al (13) proposed to use ordered subset analysis (OSA) in genetic linkage mapping of complex traits in the presence of genetic heterogeneity. They found that grouping families into subsets determined by ordering levels of family-specific trait-related covariates could identify more homogenous subsets and demonstrated significant increases in LOD scores relative to linkage scores in the entire collection of families. Simulation studies by Hauser et al (13) showed that OSA provided substantially greater statistical power than did HLODs when the covariates values in OSA were chosen from a mixture of normal distribution with means different in the linked and unlinked subsets. Based on family-specific LOD scores and trait-associated covariates such as age at onset of disease or other variables, OSA detected novel susceptibility loci for several complex diseases or traits such as Alzheimer disease (14), alcohol dependence (15), and type 2 diabetes (16) in subsets of more homogeneous families. As a part of the OSA procedure, permutation tests are implemented to assess the type I error rate.
The purpose of our current study was to re-analyze linkage scores in the previously studied families with lung cancer assessed by the National Cancer Institute funded Genetic Epidemiology of Lung Cancer Consortium (GELCC) and to evaluate the linkage evidence in subsets of genetically more homogeneous families by rank-ordering the LOD score for each family based on trait-related covariates. We were also interested in finding out whether any relationship exists between evidence for linkage to lung cancer and the risk of developing other cancers; hence, we performed further analysis ranking families by risk of other cancers.
Methods
Our methods for sample collection were summarized by Bailey-Wilson et al (9) and Amos et al (17). Samples and data were collected by the familial lung cancer recruitment sites of the GELCC, which included the University of Cincinnati, University of Colorado Health Science Center, Karmanos Cancer Center, Louisiana State University, Mayo Foundation and Clinic, Medical University of Ohio, Johns Hopkins University, and Saccomanno Research Institute. Of the 28,085 lung cancer patients screened at the GELCC sites for inclusion in this report, 23.7% had at least one first-degree relative with lung cancer (17). Families were identified from the Mayo Clinic and Karmanos Cancer Center as part of an ongoing case series based in these hospitals. All other sites accrued patients by physician referral; in addition, some patients were self-referred to the Johns Hopkins University, Karmanos Cancer Center, and University of Cincinnati sites. All sites accrued participants into institutional review board (IRB)-approved protocols and obtained informed consent from each participant. The site conducting statistical analyses (The University of Texas MD Anderson Cancer Center) also had an IRB-approved protocol for data analysis.
The pedigree development process began at all GELCC sites by screening lung cancer patients by family history (focusing on the number of first-degree relatives affected with lung cancer). Supplementary Fig. S1 presented the process for recruitment of lung cancer cases as described by Amos et al(17). After the initial screening process, we collected additional data from 3,827 willing probands or their family representatives about additional cancer-affected persons in the extended family, vital status of cancer-affected individuals, availability of archival tissue, and willingness of family members to participate in the study. We then initiated full pedigree development and biospecimen collection among 871 families, most of which had three or more affected relatives.
The majority of families did not meet our inclusion criteria for further study because they did not contain enough family members with lung cancer from whom blood samples or nontumor tissue samples could be obtained for genotyping or, if the affected family members were deceased, they did not have children willing to participate in the study from whom the genotype of the affected parent could be deduced. To date, 93 families with genetic information for at least two lung cancer–affected relatives have been genotyped with microsatellite markers, representing 0.3% of the cases we screened and 2.4% of the potential families identified (17).
The data on cancers in the families were obtained by requesting pathology reports, death certificates, and original tumor blocks and slides, when available. When tumor blocks or slides could be obtained, they were transmitted to the tumor pathology core of the GELCC, which is headed by Adi Gazdar at The University of Texas Southwestern Medical Center. Otherwise, tumor histology was assessed according to the pathology report or death certificate. Cancer diagnoses could not be verified for 72 of 489 subjects who were reported by relatives to have had cancers. Although all lung and throat cancers were verified by medical records or death certificates, few cancers at the other sites were verified.
Blood, buccal cells, and archival biospecimens were used as sources of DNA for genotyping family members of the lung cancer patients. Sample preparation, genotyping, integration of genotype data across platforms and quality control procedures were described by Amos et al (17).
Our primary analytic approach to the data from the GELCC assumed a model with 10% penetrance in carriers and 1% penetrance in the noncarriers. This analytic approach weights information primarily from the affected subjects (18)and so provides an essentially model-free analysis. To obtain linkage results, we used SimWalk2 (19) and calculated HLOD scores (10) from the output using Perl scripts we have developed. In this analysis, we estimated the evidence for linkage from each family separately using the Markov chain Monte Carlo (MCMC) method provided by SimWalk2. The MCMC analysis was used to estimate LOD scores because the pedigrees were too large to permit exact multipoint computation of the likelihood of the data. We performed all analyses separately within each genotyping batch and within each racial group to avoid any issues that might arise if marker alleles were not faithfully mapped among studies.
OSA assesses the linkage evidence in subsets of potentially more homogenous families by ordering families according to a trait-related covariate and successively summing LOD scores in each family to find a subset with maximal linkage to a given chromosome (13). Specifically, families were rank-ordered by their family-specific covariate values and the multiple-point LOD score for each family was added successively at each position on the chromosome. We had to store the maximum LOD score and its map position for each ordered subsets of families. By repeating this procedure and adding the families one at a time in order until all families were included in the final subset, the optimal subset of families with the maximum LOD score was determined for each ranking covariate. The reduced heterogeneity in identified subsets may increase the statistical power of detecting evidence for linkage and may refine gene localization. The identified ordered subset may also be used to examine genetic variants on disease-related traits through high-density sequencing. In this study, age at onset (minimum, maximum, average and range)( early onset of complex disease being more likely due to heritability factors), number of affected individuals (large number indicating familial aggregation of inheritable disease), proportion of smokers and maximum pack-years in the family(lung cancer being smoking-related) were investigated to determine if these covariates significantly influence the evidence for linkage. Cancer risk and person-year incidence rate at other tumor sites in each family were estimated using the CAGE program (20). We also performed OSA with families ranked by relative risks and incidence rates of diverse other cancers(as an exploratory study, we used available data for all other cancers). We used permutation tests to determine the empirical significance of the increase in linkage achieved with covariate-determined subsets relative to the overall sample. Permutation tests were performed by randomly ordering families and identifying the maximum LOD score for each permutation. We used the Besag-Clifford sequential stopping rule to determine how many permutation tests were needed to compute empirical P-values. The rule stopped permutations after 20 random orderings had subset scores greater than or equal to the maximum ordered subset score obtained when ordering by family covariate values. The P-value was then 20 divided by the number of replicate permutations that were required. OSA was performed by applying the FLOSS software, which was originally developed for analyzing weighted nonparametric multipoint linkage Z-scores in the Merlin(21), but was modified by the author (SF) to input LOD scores without weighting. To assess the significance of the maximum LOD score allowing for the multiple tests that we performed when considering orderings by multiple covariates, we applied the adjustment suggested by Ott (22) in which the critical LOD score was adjusted upward by the log10 of the number of covariates evaluated. Applying this very conservative approach and using an initial critical value of 3.3 for genome-wide significance, we required a maximum LOD score of 4.15 for genome-wide significance after allowing for 7 different covariates.
Results
The 93 families that we studied included 474 persons with lung cancer with an average age of onset of 61.6±11.3 years, of whom 35 were unrelated and 439 were related to other affected family members and were informative for linkage analysis. From these families, we collected 1,156 blood samples, 24 buccal cell samples, 58 sputum samples, and 274 archival blocks containing normal tissue. Archival tumor blocks from lung cancer–affected subjects were collected from 186 persons, along with 88 blocks from other tissues. Among patients who did not have lung cancer, 54.4% reported smoking, with an average of 34.1±25.8 pack-years among smokers. Among lung cancer patients, 92% reported smoking, with an average of 51.0±32.5 pack-years among smokers.
OSA Using the Lung Cancer-Related Covariates Age of Onset, Smoking, and Number of Affected Individuals per Family
Several chromosomal regions revealed strong or suggestive evidence for linkage using OSA (Table 1). On chromosome 6q23-25, OSA yielded a maximum LOD score of 4.19, exceeding the empirically defined genome-wide LOD score significance threshold of 3.3 (23) and the critical value of 4.15 after allowing for 7 covariates. Another locus on chromosome 12q24 demonstrated a borderline significant LOD score of 2.79. Chromosomes 5q31-q33, 14q11 and 16q24 also showed suggestive evidence for linkage (>2.0) in trait-determined subsets using OSA. However, a statistically significant increase in LOD scores was only observed for two loci—12q24 and 16q24 when a corrected significance of 0.00357 was used, following conservative Bonferronni adjustment for the study of 7 different covariates in both descending and ascending orders (P ≤0.05/14=0.00357).
Table 1.
Subsets with LOD > 2.0 or nominally significant increases in LOD scores (P<0.05) associated with lung cancer related covariates, as identified by OSA. “Cut points ” denote the cut points for the covariate in the selected subsets with maximum LOD. P-values in bold font were significant for a change in LOD score when modeling the covariate after imposing a Bonferronni correction
| Chromosome and nearest marker |
cM | LOD score |
Change from baseline |
P | One-LOD interval |
Number of Families in the Subset |
Covariate | Order | Cut points |
|---|---|---|---|---|---|---|---|---|---|
| 5q31-q33 D5S816 |
139 | 2.45 | 2.85 | 0.004 | 132.7-153.5 | 6 | Range of age at onset | Descending | ≥40 years |
| 6p21 D6S1017 |
63 | 3.35 | 2.39 | 0.118 | 52.8-69.0 | 12 | Minimum age at onset | Ascending | ≤39 years |
| 6q23-25 D6S2436 |
158 | 3.66 | 2.70 | 0.0476 | 150.5-164.0 | 51 | Number of affected individuals | Descending | ≥5 |
| 6q23-25 D6S1035 |
168.2 | 4.19 | 3.23 | 0.0216 | 160-177.2 | 53 | Proportion of smokers | Ascending | ≤0.567 |
| 12q24.2 D12S2070 |
123.4 | 2.79 | 2.95 | 0.002 | 110.6-143.5 | 30 | Maximum age at onset | Descending | ≥78 years |
| 14q11.2 D14S306 |
44 | 2.26 | 1.87 | 0.0358 | 14.8-51.2 | 14 | Maximum pack-years | Descending | ≥141 pack-years |
| 16q24 D16S539 |
125 | 2.33 | 2.27 | 0.0008 | 119.4-140.0 | 44 | Range of age at onset | Ascending | ≤20 years |
| 20p12 D20S604 |
34.8 | 2.20 | 2.37 | 0.160 | 21.1-39.9 | 49 | Average age at onset | Descending | ≥61.7 years |
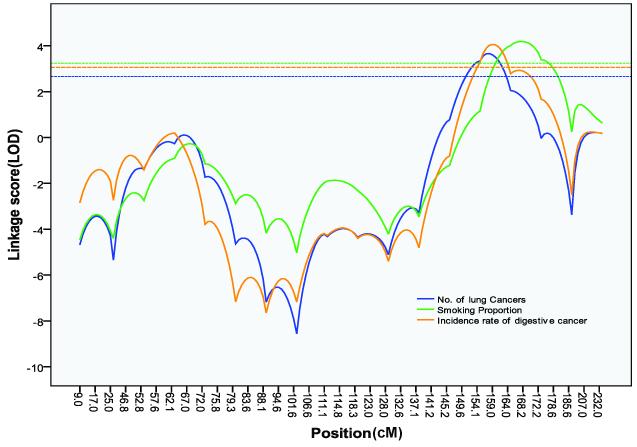
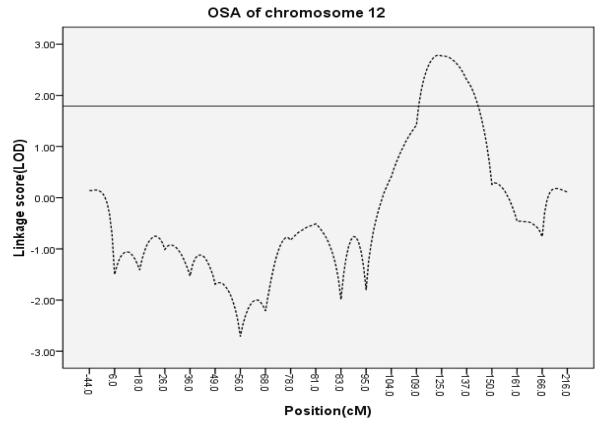
When creating subsets of families according to the number of affected individuals, the 51 families with the greatest number of lung cancer cases (more than 4 per family) had a maximum LOD score of 3.66 (Fig. 1) at 158 cM (1-LOD support interval =[150.5-164.0 cM]) on chromosome 6q(D6S2436). This was a significant increase from the baseline LOD score of 0.96(P=0.0476). We found a significant increase (P=0.0216) in the LOD score to 4.19 at 168.2 cM (D6S1035) among the 53 families with the smallest proportion of smokers (1-LOD-unit support interval = [160.0 - 177.2 cM]). The 1-LOD support interval for subsets of families defined by having the most family members with lung cancer and the interval for those identified by having the smallest proportion of smokers overlapped (Fig. 1). Thirty of the 51 families with the largest number of affected cases were among the 53 families with the smallest proportion of smokers. On chromosome 12q, a subset of families defined by a decreasing maximum age at onset yielded a maximum LOD score of 2.79 at 123.4 cM (D12S2070; P=0.002, 1-LOD support interval = [110.6 -143.5 cM]; Table 1, Fig. 2).
Figure 1.
OSA results on chromosome 6q. The blue curve represents OSA results using the largest number of affected individuals in the family; the green curve represents OSA results using the lowest proportion of smokers in the family; and the yellow curve represents OSA results using the smallest incidence rate of digestive cancer in the family. For each horizontal dash line, the part under the curve with the same color represents its corresponding1-LOD-unit support interval. Three intervals overlap over one region of 160.0-164.0 cM.
Figure 2.
OSA results on chromosome 12. The horizontal line represents 1-LOD-unit support interval(110.6-143.5 cM)
The six families identified through OSA as having the largest range of age at onset were observed to have a maximum LOD score of 2.45 at 139 cM (D5S816; 1-LOD support interval = [132.7- 153.5 cM]) on chromosome 5q. This was a significant increase from the baseline LOD score of −0.4 (P=0.004). These results need to be confirmed by testing for linkage in larger datasets because our sample size was small.
Ranking families by decreasing maximum pack-years in a family yielded significant improvements in linkage evidence (P=0.0358), with LOD scores of 2.26 at 44 cM (D14S306; 1-LOD support interval = [14.8-51.2 cM]) on chromosome 14q. On chromosome 16q, the peak LOD score of 2.33 at 125cM (D16S539; P=0.0008, 1-LOD interval = [119.4-140.0 cM]) was obtained with families ordered increasingly by the range of age at onset.
Two chromosome regions showed evidence for linkage with non-significant increases of LOD scores in a subset of families(Table 1). Although the 12 families with the youngest minimum age at onset were identified as having a maximum LOD score of 3.35 at 63 cM on chromosome 6p(D6S1017), this LOD score was not significantly different from the baseline LOD score of 0.96 (P=0.118). The 1-LOD-unit down support interval extended from 52.8 cM to 69.0 cM (Supplementary Fig. S2). Similarly, ordering families by descending average age at onset resulted in a maximum LOD score of 2.20 at 34.8 cM on chromosome 20p in the subset of 49 families with the greatest average age at onset; however, this score was not significantly different from the baseline LOD score in all families (P=0.160).
OSA Using Other Cancer Risk in the Family
To search for loci or environmental factors that enhance risk for diverse cancers besides lung cancer, we estimated standardized cancer incidence ratio and incidence rate per 100 person-years for the other 14 major cancer sites in our study population (Supplementary Table S1). Compared with the baseline populations in cancer registries in the United States, our study population were 1.71 (1.59-1.83) times more likely to develop any type of cancer, but this increased propensity probably reflected selection bias, as our probands had been selected because their families had 2 or more lung cancer cases. There was also a preferential selection of families that had throat cancers, and therefore the inflated frequency of these cancers in our population is understandable. For all malignancies excluding lung and throat cancer, we found that family members showed a standardized incidence ratio (SIR) of 0.86 (95% CI, 0.77-0.96), indicating they had less or similar risk of developing other cancers with respect to the population in the cancer registries. In contrast, significantly decreased risks were observed for non-Hodgkin lymphoma, with a SIR of 0.43 (95% CI, 0.20-0.82), and leukemia, with a SIR of 0.52 (95% CI, 0.24-0.98), and a non-significant decreased risk was observed for Hodgkin lymphoma, with a SIR of 0.47 (95% CI, 0.09-1.37). For cancer incidence rates at 13 other cancer sites, no significant differences were reported between our study and standardized populations in SEER cancer registries.
We also performed OSA with families ranked by relative risks or incidence rates for 14 other types of cancer. Analysis conducted with families ordered by levels of risk of several cancers presented significantly increased LOD scores for lung cancer on chromosomes 1q23, 2p11, 6q23-25, 13p12, and 17p11(Table 2), but none of these increases reached significance if a Bonferronni correction was applied, allowing for the number of cancer sites used for covariate analysis. The above results showed that there exists a weakly positive relationship between risk of developing other cancers and evidence for linkage to lung cancer on several chromosomes; but, there also exists a negative relationship between risk for one cancer and that for lung cancer. Families with the smallest digestive cancer incidence rates were observed to have a peak LOD score of 4.06 at 159 cM on chromosome 6q (D6S2436; P = 0.0284). The 1-LOD support interval overlapped with OSA results using the covariates of the number of lung cancer or proportion of smokers in a family at one region (160-164 cM; Fig. 1).
Table 2.
Significant increases in LOD score for lung cancer associated with other cancer risk, as identified by OSA. “Cut point “ denotes the cutoff value for the covariate in the selected subsets with maximum LOD. “*” represents the unit of incidence rate that is measured in” per 100 person-years”. None of increases in LOD score were significant after correction for testing multiple covariates on the same chromosome (P≤0.05/54=0.0009).
| Chromosome and peak marker |
CM | LOD score |
Change from baseline |
P-value | 1-LOD interval |
Number of families in the subset |
Covariate | Order | Cut point |
|---|---|---|---|---|---|---|---|---|---|
| 1q23 D1S1677 |
176 | 2.24 | 2.80 | 0.0287 | 168.9-189.6 | 18 | Breast cancer incidence rate |
Descending | ≥0.158* |
| 2p11.2 D2S1790 |
103 | 1.78 | 1.93 | 0.0389 | 99.4-112.9 | 13 | Bladder cancer incidence rate |
Descending | ≥0.035* |
| 6q23-25 D6S2436 |
159 | 4.06 | 3.10 | 0.0284 | 154.1-164 | 61 | Digestive cancer incidence rate |
Ascending | ≤0.051* |
| 13p12 D13S217 |
19.7 | 1.13 | 1.59 | 0.0417 | 11.4-42.5 | 8 | Relative risk for leukemia |
Descending | ≥2.48 |
| 19.7 | 1.13 | 1.59 | 0.0352 | 11.4-42.5 | 8 | Leukemia incidence rate |
Descending | ≥0.017* | |
| 17p11.2 D17S799 |
35.9 | 1.34 | 1.40 | 0.0375 | 25.6-45.6 | 12 | Relative risk for breast cancer |
Descending | ≥2.57 |
| 37.2 | 1.34 | 1.39 | 0.0412 | 24.8-45.6 | 11 | Breast cancer incidence rate |
Descending | ≥0.186 * |
Discussion
Our study included an additional 41 families since our first report in 2004 (9), and we performed OSA to examine evidence for linkage on each chromosome. Our results further confirm previous evidence for linkage on 6q with genome-wide significance and provide suggestive evidence for linkage on 12q in an identified subset of families with age at onset ≥78. We also observed several other suggestive linkages (LOD > 2.0) on chromosomes 5q, 14q, and 16q in subsets of families who were etiologically more homogeneous as defined by trait-related covariates. There was weak evidence for linkage (LOD > 1.0) on chromosomes 1q, 2p, 6q, 13p, and 17p if we sorted on risks for breast cancer, bladder cancer, digestive cancer or leukemia, but the exact mechanism of the relationship between diverse cancers and lung cancer remains to be determined in the future study. Of note, the region on chromosome 17p that showed the greatest evidence for linkage in families with the highest incidence of breast cancer includes p53 gene, suggesting that perhaps a subset of the families contains p53 variants. Two families that contain multiple primary cancers (family #21 had 6 lung, 5 breast, 2 colon, 1 lymphoma and 1 brain cancer; family #104 had 1 synovial sarcoma, 1 brain, 1 melanoma, 7 lung, 1 tonsil and 1 uterine cancers) previously underwent sequencing of exons 4-9 of the p53 gene and no variants were identified.
A genome-wide linkage study performed by Bailey-Wilson et al (9) successfully mapped a major susceptibility locus to chromosome 6q23-25 in families with four or more individuals affected with lung cancer. The HLOD score in 38 families that included four or more affected cases in two or more generations increased from 3.47 to 4.26 in the 23 families with five or more affected members. A follow-up study by Amos et al (17) that included 50 families with five or more affected individuals found a HLOD score of 4.69 on chromosome 6q at 158cM. These findings provided further evidence that a region of chromosome 6q was associated with lung cancer risk.
You et al (24) identified RGS17 as a candidate familial lung cancer susceptibility gene for the locus at 158 cM on chromosome 6q through epidemiologic and biologic studies. RGS17 was found to have opioid receptor function and act as a potential oncoprotein, promoting tumor cell growth. In our study, OSA identified a peak LOD of 3.66 at this region in the 51 families with more than 4 affected lung cancer cases in each family. We also observed a maximum LOD of 4.19 at 168.2cM on 6q in 53 families with lowest proportion of smokers. This finding is consistent with findings from the study of Amos et al (17) showing high risks for lung cancer in never and light smokers from families that link to chromosome 6q. The 1-LOD down support intervals of the regions for the subsets of families identified by these two covariates overlapped, suggesting that those two regions might share the same lung cancer susceptibility locus.
Our results on chromosome 12q showed a nearly significant linkage at the genome-wide significance level of linkage score 3.3 (23) in 30 families with the highest maximum age of onset, and the increase of linkage relative to the overall sample was significant after correction for multiple comparisons. This was the largest linkage score of lung cancer on 12q yet reported (9, 17), and to some degree our OSA suggested linkage between the late onset of lung cancer and chromosome 12q. The support interval under this peak encompasses ~33 cM, in which one candidate gene - insulin-like growth factor-1(IGF-1) at 114.24 cM was found to be involved with lung cancer because high plasma levels of IGF-I were associated with an increased risk of lung cancer (OR = 2.06) and a significant lower survival among patients(25, 26). IGF-1 signaling is important for cancer development and progression because it is involved in cell proliferation, differentiation, migration and death (27). IGF-1 helps cells to pass the G1-S checkpoint in the cell cycle, and effects of overexpression of IGF-1 receptors are also important in tumorigenesis (25). Raised concentrations of IGF-1 were also reported to increase the risk of several other cancers such as prostate cancer, premenopausal breast cancer and colorectal cancer (28-30).
On three chromosomes—5q, 14q, and 16q—OSA produced a suggestive linkage score of greater than 2.0 in ordered subsets and the increase of LOD scores was significant compared with the whole sample. Such a suggestive linkage was previously reported only at 14q by Bailey-Wilson et al (9) in a subset of families with five or more affected cases, but no candidate genes have been identified on this chromosome thus far. For the peak linkage region on 16q24.2-q24.3, previous evidence for linkage has not been demonstrated. Our results show an increased LOD score among families with early onset lung cancer. A candidate gene in this region is cadherin13 (CDH13), which encompasses the marker D16S3091 at 111cM. CDH13 was reported to be inactivated in lung cancer by Sato et al (31), who found that chromosomal deletion accompanied by hypermethylation inactivated the CDH13 gene in a considerable number of lung cancer specimens. The gene locus was also observed to be hypermethylated or deleted in breast cancer (32) and ovarian cancer (33). This particular cadherin is a putative regulator of cell-to-cell interaction in the heart and may function as a negative guiding molecule in neural cell growth (34). The CDH13 gene was mapped to chromosome 16q, in which allelic loss in patients with lung cancer was also reported (35).
Although the OSA yielded subsets of families with significant evidence for linkage on chromosome 6p (3.35 at 63 cM) and suggestive evidence for linkage on chromosome 20p (2.20 at 34.8 cM), neither of them were significantly increased from the overall baseline(P>0.05). Evidence suggestive of linkage at these two regions has been previously noted by our colleagues (9, 17), but no candidate genes have been demonstrated to be associated with lung cancer on these chromosomes thus far.
A strong evidence for linkage to lung cancer was observed at 159cM on 6q in 61 families with the lowest digestive cancer incidence rate. Allelic losses were reported on bands 6q16, 6q21-22, and 6q27 in gastric carcinoma (36) and on 6q25 in lung cancer (37), but this observation can’t explain why the subset of families with low digestive cancer risk had high linkage to lung cancer. The roles of some other genes or environmental factors in the development of both digestive and lung cancers need to be further verified.
The OSA with breast-cancer risk-defined subsets produced a significant increase in LOD scores on chromosomes 1q23 (LOD = 2.24). On chromosome 1q21, the MUC1 gene was more strongly expressed in neoplastic lung tissues than in normal counterparts (38), and it was highly overexpressed in breast carcinomas (39, 40).
In summary, our OSA results strongly support the previous evidence for linkage on 6q and provide nearly significant evidence for linkage on 12q in a subset of families defined by trait-related covariates. Several other regions suggestive of linkage with LOD scores greater than 2.0 were detected in the OSA. Genetic variants at one region that confer inherited susceptibility to lung cancer could be risk factors for other cancers. But we should also notice that common environmental factors may play in the development of both lung cancer and other cancers, for which an elevated LOD score for lung cancer can also be obtained when using other cancer risk as a covariate in OSA. Although our results provide a clue of utilizing disease-related covariates to identify potential linkage heterogeneity in genomic scans of lung cancer, confirmation of the linkage in another study with a larger sample size may be necessary to further reinforce our findings.
Supplementary Material
Acknowledgments
Grant Support: This work was supported in part by the National Institutes of Health grants UO1CA076293, P30ES06096, P30CA016772, R01CA133996, RO1CA060691, RO1CA87895, P30ES007789, P50CA70907, and NO1PC35145, the intramural programs of the National Cancer Institute, and the National Human Genome Research Institute. This publication was made possible by grant P30ES007784 from the National Institute of Environmental Health Sciences.
Footnotes
Disclaimers: The author(s) indicated no potential conflicts of interest
Reference List
- (1).American Cancer Society . Cancer Statistics 2009 Presentation. American Cancer Society, Inc; Atlanta, GA: 2009. [Google Scholar]
- (2).International Agency for Research on Cancer (IARC) IARC Monographs on the Evaluation of Carcinogenic Risks to Humans and their Supplements: A complete list. 83 ed. World Health Organization; 2002. Available from: http://monographs.iarc.fr/ENG/Monographs/vol83/mono83-6B-4.pdf. [Google Scholar]
- (3).Samet JM. Epidemiology of lung cancer. M. Dekker; New York: 1994. [Google Scholar]
- (4).Mattson ME, Pollack ES, Cullen JW. What Are the Odds That Smoking Will Kill You. American Journal of Public Health. 1987;77(4):425–31. doi: 10.2105/ajph.77.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Etzel CJ, Amos CI, Spitz MR. Risk for smoking-related cancer among relatives of lung cancer patients. Cancer Res. 2003;63(23):8531–5. [PubMed] [Google Scholar]
- (6).Jonsson S. Familial risk of lung carcinoma in the Icelandic population (vol 292, pg 2977, 2004) JAMA. 2005;293(2):163. doi: 10.1001/jama.292.24.2977. [DOI] [PubMed] [Google Scholar]
- (7).Ooi WL, Elston RC, Chen VW, Baileywilson JE, Rothschild H. Increased Familial Risk for Lung-Cancer. Journal of the National Cancer Institute. 1986;76(2):217–22. [PubMed] [Google Scholar]
- (8).Xu HY, Spitz MR, Amos CI, Shete S. Complex segregation analysis reveals a multigene model for lung cancer. Human Genetics. 2005;116(1-2):121–7. doi: 10.1007/s00439-004-1212-9. [DOI] [PubMed] [Google Scholar]
- (9).Bailey-Wilson JE, Amos CI, Pinney SM, et al. A major lung cancer susceptibility locus maps to chromosome 6q23-25. American Journal of Human Genetics. 2004;75(3):460–74. doi: 10.1086/423857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ott J. Linkage Analysis and Family Classification Under Heterogeneity. Annals of Human Genetics. 1983 Oct;47:311–20. doi: 10.1111/j.1469-1809.1983.tb01001.x. [DOI] [PubMed] [Google Scholar]
- (11).Morton N. The detection and estimation of linkage between the genes for elliptocytosis and the Rh blood type. American Journal of Human Genetics. 1956;8(2):80–96. [PMC free article] [PubMed] [Google Scholar]
- (12).Hall JM, Lee MK, Newman B, et al. Linkage of Early-Onset Familial Breast-Cancer to Chromosome-17Q21. Science. 1990;250(4988):1684–9. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- (13).Hauser ER, Watanabe RM, Duren WL, et al. Ordered subset analysis in genetic linkage mapping of complex traits. Genetic Epidemiology. 2004;27(1):53–63. doi: 10.1002/gepi.20000. [DOI] [PubMed] [Google Scholar]
- (14).Scott WK, Hauser ER, Schmechel DE, et al. Ordered-subsets linkage analysis detects novel Alzheimer disease loci on chromosomes 2q34 and 15q22. American Journal of Human Genetics. 2003;73(5):1041–51. doi: 10.1086/379083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Reck BH, Mukhopadhyay N, Tsai HJ, Weeks DE. Analysis of alcohol dependence phenotype in the COGA families using covariates to detect linkage. BMC Genetics. 2005:6. doi: 10.1186/1471-2156-6-S1-S143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Sale MM, Lu LY, Spruill IJ, et al. Genome-Wide Linkage Scan in Gullah-Speaking African American Families With Type 2 Diabetes The Sea Islands Genetic African American Registry (Project SuGAR) Diabetes. 2009;58(1):260–7. doi: 10.2337/db08-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Amos CI, Pinney SM, Li Y, et al. A Susceptibility Locus on Chromosome 6q Greatly Increases Lung Cancer Risk among Light and Never Smokers. Cancer Res. 2010;70(6):2359–67. doi: 10.1158/0008-5472.CAN-09-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Speer MC. Use of LINKAGE Programs for Linkage Analysis. Current Protocols in Human Genetics. 2006 doi: 10.1002/0471142905.hg0107s48. p. Unit1.7. [DOI] [PubMed] [Google Scholar]
- (19).Weeks DE, Sobel E, Oconnell JR, Lange K. Computer-Programs for Multilocus Haplotyping of General Pedigrees. American Journal of Human Genetics. 1995;56(6):1506–7. [PMC free article] [PubMed] [Google Scholar]
- (20).McLaughlin L. CAGE program. Philadelphia,PA: 1997. [Google Scholar]
- (21).Browning BL. FLOSS: flexible ordered subset analysis for linkage mapping of complex traits. Bioinformatics. 2006;22(4):512–3. doi: 10.1093/bioinformatics/btk012. [DOI] [PubMed] [Google Scholar]
- (22).Jurg Ott. Analysis of Human Genetic Linkage. Revised ed The Johns Hopkins University Press; Baltimore: 1991. pp. 74–6. [Google Scholar]
- (23).Lander E, Kruglyak L. Genetic Dissection of Complex Traits -Guidelines for Interpreting and Reporting Linkage Results. Nature Genetics. 1995;11(3):241–7. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- (24).You M, Wang DL, Liu PY, et al. Fine Mapping of Chromosome 6q23-25 Region in Familial Lung Cancer Families Reveals RGS17 as a Likely Candidate Gene. Clinical Cancer Research. 2009;15(8):2666–74. doi: 10.1158/1078-0432.CCR-08-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Rajski M, Zanetti-Dallenbach R, Vogel B, et al. IGF-I induced genes in stromal fibroblasts predict the clinical outcome of breast and lung cancer patients. BMC Med. 2010;8:1. doi: 10.1186/1741-7015-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Yu H, Spitz MR, Mistry J, et al. Plasma Levels of Insulin-Like Growth Factor-I and Lung Cancer Risk: a Case-Control Analysis. J Natl Cancer Inst. 1999;91(2):151–6. doi: 10.1093/jnci/91.2.151. [DOI] [PubMed] [Google Scholar]
- (27).Jones JI, Clemmons DR. Insulin-like growth factors and their binding proteins: biological actions. Endocr Rev. 1995;16(1):3–34. doi: 10.1210/edrv-16-1-3. [DOI] [PubMed] [Google Scholar]
- (28).Chan JM, Stampfer MJ, Giovannucci E, et al. Plasma insulin-like growth factor-I and prostate cancer risk: a prospective study. Science. 1998;279(5350):563–6. doi: 10.1126/science.279.5350.563. [DOI] [PubMed] [Google Scholar]
- (29).Hakam A, Yeatman TJ, Lu L, et al. Expression of insulin-like growth factor-1 receptor in human colorectal cancer. Human Pathology. 1999;30(10):1128–33. doi: 10.1016/s0046-8177(99)90027-8. [DOI] [PubMed] [Google Scholar]
- (30).Hankinson SE, Willett WC, Colditz GA, et al. Circulating concentrations of insulin-like growth factor-I and risk of breast cancer. Lancet. 1998;351(9113):1393–6. doi: 10.1016/S0140-6736(97)10384-1. [DOI] [PubMed] [Google Scholar]
- (31).Sato M, Mori Y, Sakurada A, Fujimura S, Horii A. The H-cadherin (CDH13) gene is inactivated in human lung cancer. Human Genetics. 1998;103(1):96–101. doi: 10.1007/s004390050790. [DOI] [PubMed] [Google Scholar]
- (32).Lee SW. H-cadherin, a novel cadherin with growth inhibitory functions and diminished expression in human breast cancer. Nature Medicine. 1996;2(7):776–82. doi: 10.1038/nm0796-776. [DOI] [PubMed] [Google Scholar]
- (33).Kawakami M, Staub J, Cliby W, et al. Involvement of H-cadherin (CDH13) on 16q in the region of frequent deletion in ovarian cancer. International Journal of Oncology. 1999;15(4):715–20. doi: 10.3892/ijo.15.4.715. [DOI] [PubMed] [Google Scholar]
- (34).Takeuchi T, Misaki A, Liang SB, et al. Expression of T-cadherin (CDH13, H-cadherin) in human brain and its characteristics as a negative growth regulator of epidermal growth factor in neuroblastoma cells. Journal of Neurochemistry. 2000;74(4):1489–97. doi: 10.1046/j.1471-4159.2000.0741489.x. [DOI] [PubMed] [Google Scholar]
- (35).Sato M, Mori Y, Sakurada A, et al. Identification of a 910-Kb region of common allelic loss in chromosome bands 16q24.1-q24.2 in human lung cancer. Genes Chromosomes & Cancer. 1998;22(1):1–8. [PubMed] [Google Scholar]
- (36).Li BCY, Chan WY, Li CYS, et al. Allelic loss of chromosome 6q in gastric carcinoma. Diagnostic Molecular Pathology. 2003;12(4):193–200. doi: 10.1097/00019606-200312000-00003. [DOI] [PubMed] [Google Scholar]
- (37).Virmani AK, Fong KM, Kodagoda D, et al. Allelotyping demonstrates common and distinct patterns of chromosomal loss in human lung cancer types. Genes Chromosomes & Cancer. 1998;21(4):308–19. doi: 10.1002/(sici)1098-2264(199804)21:4<308::aid-gcc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- (38).Seregni E, Botti C, Lombardo C, et al. Pattern of mucin gene expression in normal and neoplastic lung tissues. Anticancer Research. 1996;16(4B):2209–13. [PubMed] [Google Scholar]
- (39).Boyce B, Rakha E, El-Rehim D Abd, et al. The expression of mucins (MUC1, MUC2, MUC3, MUC4, MUC5AC and MUC6) and their prognostic significance in human breast cancer. Journal of Pathology. 2005;205:5. doi: 10.1038/modpathol.3800445. [DOI] [PubMed] [Google Scholar]
- (40).Rakha EA, Boyce RWG, El-Rehim D Abd, et al. Expression of mucins (MUC1, MUC2, MUC3, MUC4, MUC5AC and MUC6) and their prognostic significance in human breast cancer. Modern Pathology. 2005;18(10):1295–304. doi: 10.1038/modpathol.3800445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.