Abstract
While alterations in xenobiotic metabolism are considered causal in the development of bladder cancer (BCa), the precise mechanisms involved are poorly understood. In this study, we used high-throughput mass spectrometry to measure over 2,000 compounds in 58 clinical specimens, identifying 35 metabolites which exhibited significant changes in BCa. This metabolic signature distinguished both normal and benign bladder from BCa. Exploratory analyses of this metabolomic signature in urine showed promise in distinguishing BCa from controls, and also non-muscle from muscle-invasive BCa. Subsequent enrichment-based bioprocess mapping revealed alterations in phase I/II metabolism and suggested a possible role for DNA methylation in perturbing xenobiotic metabolism in BCa. In particular, we validated tumor-associated hypermethylation in the CYP1A1 and CYP1B1 promoters of BCa tissues by bisulfite sequence analysis and methylation-specific PCR, and also by in vitro treatment of T-24 BCa cell line with the DNA demethylating agent 5-aza-2′-deoxycytidine. Further, we showed that expression of CYP1A1 and CYP1B1 was reduced significantly in an independent cohort of BCa specimens compared to matched benign adjacent tissues. In summary, our findings identified candidate diagnostic and prognostic markers and highlighted mechanisms associated with the silencing of xenobiotic metabolism. The metabolomic signature we describe offers potential as a urinary biomarker for early detection and staging of BCa, highlighting the utility of evaluating metabolomic profiles of cancer to gain insights into bioprocesses perturbed during tumor development and progression.
Introduction
Bladder cancer (BCa) is the fourth most common cancer in American men and accounts for more deaths annually in women than cervical cancer (1). Diagnosis and surveillance of BCa currently consists of cystoscopy aided by cytology and biopsy. Cystoscopy identifies most papillary and sessile lesions but may have low sensitivity for high-grade superficial disease (i.e., CIS); further cystoscopy may be associated with a high psychological burden for some patients particularly when coupled with biopsy (2). While urine cytology has reasonable sensitivity and specificity for the detection of high-grade BCa, it has poor sensitivity for detecting low-grade tumors, ranging from 4% to 31% (3). There is an urgent need for noninvasive, objective, and accurate markers that are sensitive and specific for risk stratification. One method to achieve this goal is to identify and better understand the multiplex molecular events that regulate the onset and progression of this deadly disease.
The metabolomic studies on BCa published to date include profiling urine specimens to identify benign controls from BCa using mass spectral profiles with minimal compound identification (4, 5). Notably, these studies did not include external validation (6). Further, they do not provide insight into the biochemical processes that may be involved in BCa development and progression. To address these issues, we employed a unique approach to (1) identify BCa-associated metabolomic signatures, (2) nominate potential candidate biomarkers, and (3) uncover bioprocesses that could potentially alter pre-carcinogen metabolism.
Methods
Clinical Samples
Flash frozen, pathologically verified bladder tissues were obtained from tumor banks at the Winship Cancer Center at Emory and Medical College of Georgia following approval of the respective Institutional Review Boards (IRB, Clinical information in Supplementary Tables S1, S2). These included BCa as well as benign adjacent tissues (i.e., histologically confirmed benign tissue from areas adjacent to the tumor) Gold standard normal bladder tissues (from individuals having no prior/current history of BCa) were purchased from NDRI (National Disease Research Interchange, Philadelphia, PA). Clinically annotated urine samples were obtained either prior to transurethral resection of bladder tumor (TURBT) or cystectomy at the Georgia Health Sciences University and its affiliated Charlie Norwood Veteran Affairs Medical Center of Augusta (GHSU/CNVAMC, Group 1), University of Michigan (UM, Group 2), Weil Cornell Medical College (WCMC, Group 3) and University of Texas Southwestern Medical Center (UTSW, Group 4) following informed consent under IRB approved protocols (Refer Table 1 for overview of urine specimens used in this study and Supplementary Tables S3–S6 for clinical information).
Table 1.
Overview of the specimens used to evaluate biomarker potential of bladder-associated metabolites in urine
| Group Number | Sample collection site | Total number of specimens | Controls | BCa | Training/Validation |
|---|---|---|---|---|---|
| Group 1 | Georgia Health Science University/VA(GHSU) | 26 | 13 | 13 | Validation |
| Group 2 | University of Michigan (UM) | 44 | 16 | 28 | Training/Validation |
| Group 3 | Weil Cornell Medical College (WCMC) | 45 | 11 | 34 | Validation |
| Group 4 | University of Texas Southwestern Medical Center (UTSW) | 19 | 11 | 8 | Validation |
Tissue metabolome extraction
Following harvest, bladder tissues were stored at −140°C in liquid nitrogen until analysis. For extraction of the metabolome, 25 mg of tissue was homogenized in 1:4 ice cold water:methanol mixture containing an equimolar mixture of 11 standard compounds (refer to Supplementary Table S7). This was followed by metabolic extraction using sequential application of ice cold organic and aqueous solvents (water:methanol:chloroform:water; ratio 1:4:3:1), deproteinization and drying of the extract. The latter was resuspended in injection solvent, and analyzed by liquid chromatography-coupled to mass spectrometry (LC-MS). Detailed procedural description of tissue metabolome extraction is given in the Supplementary Methods.
Sample preparation for urine
Following collection, urine specimens were spun and the supernatant was stored at −70 °C until analysis. Osmolarity of all urine samples was measured using MULTI-OSMETTE™ 2430 osmometer (Precision Systems Inc, Natick, MA), and values calibrated using standards as per the manufacturer’s instruction. The osmolarity of the urine samples examined was restricted to 140–400 milliosmoles/liter, which was achieved by measuring out a defined volume of the urine prior to extraction. This was followed by the introduction of an equimolar mixture of 4 standard compounds ([15N] Tryptophan, [D4] Thymine, [15N] N-Acetyl Aspartic acid and [D5] Glutamic acid) into the specimen and the mixture was dried under vacuum (Genevac EZ-2plus, Gardiner NY). Prior to analysis, all samples were resuspended in an identical volume of water: acetonitrile injection solvent (98:2) consisting of 0.1% formic acid prior to LC-MS analysis.
Liquid Chromatography/Mass Spectrometry (LC/MS)
The chromatographic separation of metabolites was performed using either reverse phase (RP) separation or aqueous normal phase (ANP) separation online with Quadrupole-Time-Of-Flight (QTOF)/Triple Quadrupole (QQQ) mass spectrometers (both Agilent Technologies, Santa Clara, CA) as published in (7) and described in detail in Supplementary Methods. As controls to monitor the profiling process, an equimolar mixture of 11 standard compounds (described under Tissue metabolome extraction) and a characterized pool of mouse liver tissue were extracted and analyzed in multiple times tandem with the clinical samples. Two blank runs were interspersed between clinical samples to prevent carry-over of metabolites. Parameters used to operate the mass spectrometers for unbiased (Q-TOF) and targeted analysis (QQQ, SRM transitions in Supplementary Table S8) of metabolites and associated data pre-processing methods were identical to our earlier study (7) and are described in Supplementary Methods.
Metabolites were identified from the pre-processed mass spectral data using a 1000-compound metabolomic library, METLIN (Agilent Technologies), using both mass and retention time information, as described in (7). In addition a subset of compounds associated with BCa was identified using targeted MS/MS analysis.
Metabolomic Data Analysis
Metabolites in the profiling data, with more than 75% missing values across samples were removed from the analysis. The remaining metabolites included 1509, 510 and 55 compounds in unbiased positive (+) and negative (−) ionization and SRM data, respectively, resulting in 1905 unique compounds (listed in Supplementary Table S9) across platforms from which 99 metabolites were named. For included metabolites, the distribution of proportion of missing values in bladder cancer and benign samples were examined, and missing measures in metabolites were imputed according to the adaptive procedure described in the Supplementary Methods. Imputed data from positive and negative ionization were log2 transformed and quantile normalized per sample using the R-package “limma” (8). To avoid bias due to the small number of compounds measured by SRM, median centering was employed instead of quantile normalization. Data obtained from different platforms were z-transformed and compared across samples. The data from different platforms were combined by averaging over duplicate samples for each compound. Hierarchical clustering was performed using complete linkage with Pearson’s correlation and heat maps were drawn using R-packages “gplot” (9). A permutation-based (n=10,000) two-sided t-test coupled to false discovery rate correction using procedure implemented in R-package “fdrtool” (10) (for all metabolites) or that described in (11) (for named metabolites), with FDR threshold of q*=0.2, was used to assess the association of each metabolite with the cancer status of the sample.
Data from urine samples were first normalized with respect to the osmolarity level of the sample, and then median centered and normalized using the inter-quartile range (IQR). Partial Least Square Discriminant Analysis (PLS-DA) classification models with two principal components (R-packages “pls” (12) and “caret”(13)) were used to classify the benign and BCa samples in different datasets, as well as Muscle-Invasive and Non-Muscle-Invasive samples in the UM data set. Cross validation (CV) coupled with repeated random splitting (n = 1000) as described in Supplementary Methods was used to obtain reliable estimates of classification errors in the training data.
Cell Culture & 5-aza-2′-deoxycytidine (Aza) Treatment
Bladder cancer cell line (T-24) was obtained from American Type Culture Collection (ATCC, Manassas, VA) tested per standards described by the ATCC Genuine Cultures® by the vendor and grown per vendor’s instructions. All experiments were carried out within 6 months of procurement of these cells. The cells were seeded at a density of 1×105 cells/well in 6-well plates and exposed to culture media containing either DMSO (≤ 0.05%, controls) or 5 μM Aza 24 hours after attachment, for a total duration of 2 days (48h). AZA was replenished at 24h intervals. At the end of the treatment period, both AZA-treated or control cells were trypsinized, rinsed with phosphate-buffered saline (PBS), pelleted and stored at −80°C until further analyses.
Real Time PCR (Q-PCR) Analysis
Total RNA and cDNA were made from frozen bladder-derived tissues (~50 mg) or cell line pellets using methods described in (14). QPCR for enzymes involved in the phase I and II metabolic pathway were performed as described (14) using genespecific oligonucleotide primers (Supplementary Table S10). The −ΔΔCt method (15) was used to calculate relative changes in mRNA levels measured by the qRT-PCR experiments, after normalizing the transcript levels of each gene by the levels of Glyceraldehyde-control (11) were used to assess the changes of expressions in different experimental conditions.
Bisulfite Treatment and DNA Sequencing
Genomic DNA was extracted from tissues and/or cell lines using DNA extraction kit (Qiagen Inc., Valencia, CA) and bisulfite treated using EZDNA methylation gold kit (Zymo Research Inc., Orange, CA), both per manufacturer’s instructions. Regions containing CpG islands on CYP1A1 and CYP1B1 promoters were PCR amplified using specific primers (synthesized by Integrated DNA Technologies, Coralville, IA) 5′-AAACTAACCCTTTAAAAACCCC-3′(sense) and 5′-TTTTTAGGGGGTAGAGGTTAGG-3′ (anti-sense) for CYP1A1 and 5′-TTTTATTTGGTGAAGAGGAGGAGTA-3′(sense) and 5′-CAACAACTTCATCCTAAACAAATTCT-3′(anti-sense) for CYP1B1. The PCR product (191 bp of the CYP1A1 promoter and 246 bp of the CYP1B1 promoter) covered 9 and 14 CpG sites respectively, located ~ 1000 bp upstream of the transcription start site. The PCR conditions that were employed were as follows: 95°C for 5 min, 40 cycles of 95°C for 15 secs, 55°C for 30 secs, 72°C for 30 secs and a final extension at 72°C for 7 min. PCR products were gel eluted and sub cloned into T/A cloning vector pCR4 (Invitrogen, Carlsbad, CA) using TOPO TA Cloning Kit as per manufacturer’s instructions. The cloned products were sequenced by Molecular Cloning laboratories (MC Labs, San Francisco, CA) using M13 reverse primer. The bisulfite sequences were analyzed using BiQ Analyzer (16).
Methylation specific PCR (MSP)
Methyl specific PCR (MSP) was performed on bisulfite treated genomic DNA (described above) using the CYP1A1 promoter-specific primers 5′-CGTGTGGTTTTTGTTTGC-3′ (sense) and 5′-GAAACAACGTCGAAAACA-3′ (anti-sense), to obtain a 123 bp product. The PCR conditions were: 95°C for 5 min, 33 cycles of 95°C for 15 secs, 48.8°C for 30 secs, 72°C for 25 secs and a final extension at 72°C for 7 min followed by 4°C. Human methylated and un-methylated DNA purchased from Zymo Research Corp. (Orange, CA) served as positive and negative controls respectively. Protocols published in (17) and described in Supplementary Methods were used to carry out immunoblot analysis of CYP1A1 and CYP1B1 in bladder tissues.
Results
Metabolomic Profiling of BCa
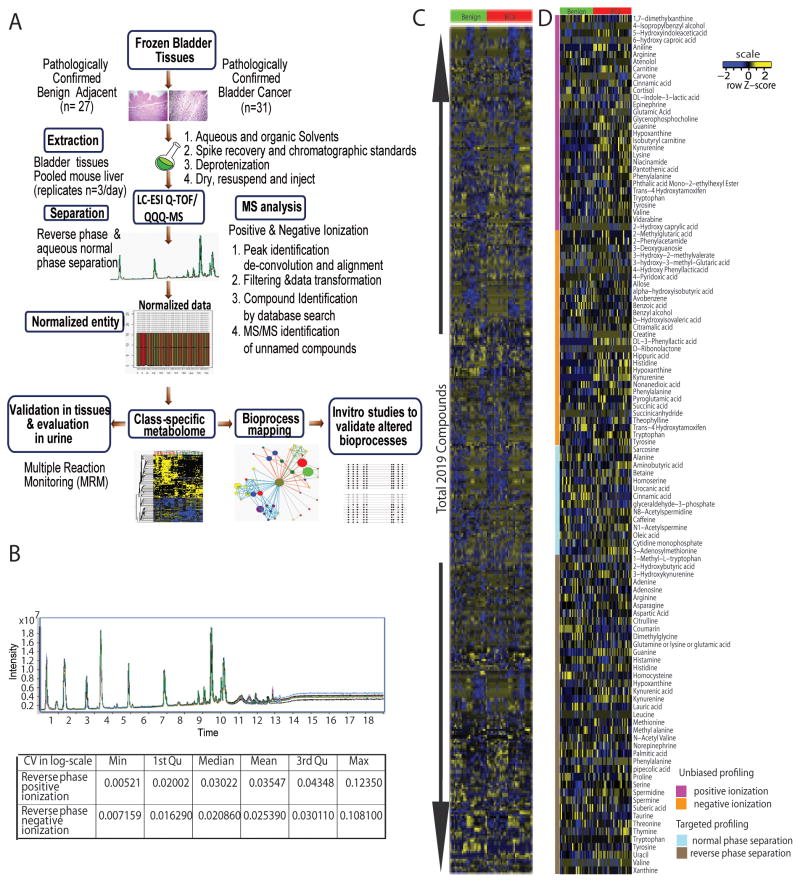
A total of 58 pathologically evaluated bladder tissues (benign adjacent n = 27 and BCa n = 31; matched pairs n = 25, power 87%, Supplementary Table 1) were examined for their metabolomic profiles using liquid chromatography-mass spectrometry. The metabolome was separated using both reverse phase (RP) and aqueous normal phase (ANP) chromatography and examined using Quadrupole-Time-Of-Flight (Q-TOF) and Triple Quadrupole (QQQ) mass spectrometers employing positive (+) and negative (−) electrospray ionization (ESI). The Q-TOF analysis involved unbiased profiling while the QQQ was used to examine a focused set of 55 compounds using Single Reaction Monitoring (SRM, refer Supplementary Table S8 for a list of SRM transitions). Fig. 1A and Supplementary Fig. 1 provide an overview of the metabolomics profiling and associated data analysis strategy employed in this study. Both the chromatographic separation of metabolites and their analysis by mass spectrometry were monitored for drifts using internal standards (refer Methods) and biological replicates of pooled extract of mouse liver and were highly reproducible (CV ≤5%, Fig. 1B).
Figure 1. Metabolomic Profiling of BCa.
A. Overview of the strategy used to profile and characterize the metabolome of bladder cancer B. Chromatographic reproducibility of a mixture of 11 metabolite standards over ten technical replicates identified on the Q-TOF using positive ionization (top panel) and a table showing the coefficient of variation (CV) for detection of 142 metabolites across 23 biological replicates of pooled liver extract. C. Heat map of hierarchical clustering of 2,019 metabolites detected across 58 bladder-related samples. Columns represent individual tissue samples and rows refer to distinct metabolites. Shades of yellow represent elevation of a metabolite and shades of blue represent a decrease of a metabolite relative to the median metabolite levels (see scale). D. Same as C, but for 99 named compounds identified across 58 bladder-related tissue specimens.
A total of 2,019 entities were detected using this robust platform, across the 58 biospecimens (Fig. 1C), of which 1,905 compounds were unique (Supplementary Table S9). Among these 99 compounds were identified using a combination of database search and MS/MS (Figure. 1D).
BCa-associated Metabolic Profiles
A total of 459/1,905 metabolites were differential between BCa and benign adjacent bladder tissue after multiple comparison adjustment at an FDR level of 20%. These included 50 named compounds that were altered in BCa compared to adjacent benign tissues (Fig. 2A). Among the perturbed metabolites were elevated levels of aliphatic amino acids, namely serine, asparagine, and valine or their aromatic counterparts, namely tryptophan, phenylalanine, and histidine. There were also hydroxylated metabolites like 3-hydroxy-kyneurenine, 4-hydroxy phenyl lactic acid and 5-hydroxy indoleacetic acid in the BCa-associated compendia. Furthermore, levels of Sadenosyl methionine (SAM) were also elevated in BCa tissues. Apart from these metabolites, BCa tissues also had higher levels of aniline, a xenobiotic compound known to be involved in bladder carcinogenesis (18, 19) while, levels of palmitic, lauric, and oleic acids were decreased in BCa compared to adjacent benign tissues.
Figure 2. Metabolomic Alteration in Bladder Cancer.
A. Heat map showing 50 named differential metabolites in BCa relative to benign samples. B. Heat map showing unsupervised hierarchical classification of an independent set of 50 bladder-derived tissues using 35 BCa-derived significant compounds nominated by metabolic profiling studies. The color scheme for the heat maps is the same as in Fig. 1C.
We examined 50-named BCa-associated metabolites (refer Fig. 2A) in a distinct set of 50 pathologically confirmed bladder tissues (n = 13 matched BCa and benign adjacent pairs and n = 24 normal bladder tissues, Supplementary Table S2), using a targeted SRM-based approach. A total of 39/50 BCa-associated metabolites were measured with certainty in these independent tissues, with alterations in 35/39 compounds remaining significant in BCa, compared to normal controls (Fig. 2B). Among these 35 compounds, levels of 31 corroborated the results in the prior profiling study (Fig. 2A). The remaining four compounds, namely hippuric acid, oleic acid, 4-pyridoxic acid, and pipecolic acid, which were modestly elevated in BCa compared to adjacent benign tissue (Fig 2A, profiling data), were found to be significantly reduced in tumors compared to normal bladder tissues (Fig 2B). This opposing trend in expression observed when normal bladder specimens are used as controls, indicate the existence of a subtle “field effect” (20) in the benign adjacent tissues that were used in the profiling study. Therefore in light of this, our metabolic signature for BCa consists of 35 named compounds from the “discovery profile” (Fig. 2A) that were also significant in the comparison between normal bladder and BCa tissues (Fig. 2B). Notably, unsupervised hierarchical clustering of bladder tissues using these metabolites distinguished BCa from benign adjacent and normal bladder controls (Fig 2B) as well as delineated benign adjacent tissue from normal bladder controls with the mis-assignment of only four normal bladder tissues (Fig 2B). Furthermore, when examined in prostate cancer specimens (10 benign-cancer pairs), only 5/35 BCa-associated compounds had nominal p-values below 0.05 (Supplementary Figure 2), and only Glyceraldehyde-3-phosphate, was found significant after multiple comparison adjustment at 20% FDR level.
Evaluation of Biomarker Potential of BCa-associated Metabolites in Urine
The classification power of BCa-associated metabolites in tissues motivated us to investigate their biomarker potential in urine. We used 134 urine specimens collected independently at four institutions as described under Methods. In Group 1 (GHSU/CNVAMC), specimens were collected from BCa patients (n=13) immediately prior to radical cystectomy (n=7) or transurethral resection of bladder tumor (TURBT) (n=6), with or without prior intravesical immuno/chemotherapy and, age-matched control individuals (n=13) with no history of cancer (e.g., neurogenic bladder (n=2) or removal of kidney stones (n=10), Supplementary Table S3). In Groups 2 (UM) and 3 (WCMC), specimens were collected from BCa patients with or without any prior immuno/chemotherapy followed by cystoscopy, with urine collection before cystoscopy (Supplementary Tables S4 and S5). Here, patients were categorized as benign (responders to therapy or pT0) or BCa (non-responders to therapy). Individuals with suspicious cystoscopy findings were evaluated with transurethral resection of the bladder tumor (TURBT). All histology slides from cystectomy specimens were reviewed and staged by a genitourinary pathologist without knowledge of the metabolomic data. In Group 4 (UTSW), specimens were collected from BCa patients (n=9) prior to TURBT or cystectomy and controls were from age-matched control individuals (n=11) with no history of BCa (Supplementary Table S6). Specifically, the controls for our urine study contained benign controls in Groups 2 and 3 who had a prior history of BCa but were considered disease-free or benign after treatment, while the controls in the Groups 1 and 4 had no prior history of BCa.
Using a SRM-based approach (Supplementary Table S8 for SRM transitions), we could measure 25/35 tissue-derived BCa-associated metabolites in urine specimens collected at the four institutions. These 25 metabolites were altered in urine of BCa patients compared to control urine from individuals having no history of BCa (Supplementary Figure 3). The reproducibility of these 25-metabolite measurements in urine was ascertained by independent replicate analysis of 10 specimens, which gave a Pearson Correlation of 0.965–0.995 (Supplementary Fig. 4). We then examined the ability of these 25 metabolites to distinguish benign from BCa in urine specimens using a partial least squares discriminant analysis (PLS-DA) with two principal components as described in Methods and illustrated in Fig. 3A. Supplementary Fig. 5 shows, the 25 BCa-associated metabolites, contributed differently to the two principal components that together defined the classificatory power of the model. The Receiver Operator Characteristic curve (ROC) for this classifier in training data from Group 2 had an Area Under the Curve (AUC) of 0.67 (90% CI: 0.52, 0.83) (Fig. 3B). When applied to independent test samples from Group 1 (n = 26), Group 3 (n = 45) and Group 4 (n=19) the classifier gave an AUC of 0.79, 0.62 and 0.76 respectively in delineating benign and BCa (Fig. 3B). Additionally, a two-stage model (refer Fig. 3A), successively combining urine cytology with the metabolic panel showed an improved AUC of 0.76 and 0.7 in classifying benign and BCa specimens in Groups 2 and 3, respectively (Fig. 3C). Further, in a pilot study, an independent cross-validated PLS-DA model (refer Supplementary Fig 6 for relative contribution of the metabolites), the 25-metabolite signature was able to distinguish muscle-invasive (n=8) from non-invasive BCa (n=20) in Group 2 specimens, with an AUC of 0.84 (90% CI: 0.63, 0.94, Fig. 3D).
Figure 3. Biomarker Potential of Bladder Cancer-associated Metabolites in Urine.
A. Overview of the strategy used to develop and validate the Partial Least Square-Discriminate Analysis (PLS-DA)-based metabolomic model for delineating benign and BCa in urine. B. Receiver Operator Characteristic (ROC) Curve describing the ability of PLS-DA model using 25 BCa-associated metabolites (M) to delineate benign and BCa in urine C. Same as in B, but for combination of BCa-associated metabolites (M) and urine cytology (UC) in delineating benign and BCa in urine. D. Same as in B, but for muscle-invasive (n=8) and non-muscle invasive (n=20) BCa in Group 2 (UM cohort).
Integrative Molecular Concept Modeling of BCa Progression
We examined whether BCa-associated metabolites might be associated with altered biochemical processes fundamental to BCa development and progression using Oncomine Concept Maps (OCM), as described earlier (21). OCM compares the information within the BCa-associated metabolome with different biologic concepts represented by molecular signatures a.k.a. molecular concepts (i.e., lists of genes, proteins etc) (21). Here, OCM analysis of the BCa-associated metabolic signature was carried out using genes associated with metabolic pathways in Homo sapiens and listed in KEGG# as the null set and following the method described in (7) and outlined in Supplementary Methods. A FDR threshold of 10% was used to select concepts of potential interest, all of which are listed in Supplementary Table S11 with a subset displayed in Fig. 4A.
Figure 4. Cytochrome-driven Xenobiotic Metabolism is altered in Bladder Cancer.
A. Network view of the molecular concept analysis for the metabolomic profiles of our “BCa associated metabolic signature” (black node). Each node represents a molecular concept or a set of biologically related genes. The node size is proportional to the number of genes in the concept. Each edge represents a statistically significant enrichment (FDR q-value < 0.10). Enriched concepts describing cytochrome P450-driven xenobiotic metabolism and methylation in BCa indicated by yellow and blue edges respectively. B. Box plots for real time PCR data showing relative transcript levels for enzymes in phase I/II metabolic pathway in matched BCa (red) and adjacent benign tissues (green). C. Immunoblot of CYP1A1 and CYP1B1 in bladder tumors compared to their matched benign adjacent tissues.
The results of the OCM analysis validated our earlier prediction of altered amino acid metabolism in these specimens described in Fig. 2A. The enriched concepts (Fig. 4A) included those that described altered utilization of amino acids (i.e., methionine) and their aromatic counterparts (i.e., tyrosine and tryptophan), as well as metabolism of fatty acids, lactic acid, intermediates of tricarboxylic acid cycle (TCA) and electron transport. In addition to these, the BCa-associated metabolomic profile also enriched for two additional biological processes describing cytochrome P450-dependent xenobiotic metabolism and methylation. In our earlier study on prostate cancer (PCa), we described elevated methylation potential as a hallmark of aggressive prostate tumors (14). It is important to note that at least 39% of tumors we examined in our profiling experiments were muscle-invasive and hence categorized as high-grade tumors (Supplementary Table S1). Furthermore, similar to our observation in PCa (14), enriched methylation in BCa may be a result of significantly elevated levels of the methyl donor S-adenosyl methionine (SAM, p = 1.71E-05, FDR q-value < 2%), as well as methylated metabolites in tumor specimens compared to adjacent benign tissue (Fig. 2A). Notably, this enrichment of methylation potential in BCa was described by multiple interconnected concepts describing DNA and histone methylation, chromatin modification, and involvement of SET domain containing proteins (Fig. 4A, blue bridges). Although this finding of perturbed methylation in BCa tumors is consistent with other studies (22, 23), its co-enrichment with the bioprocess describing altered cytochrome P40-mediated xenobiotic metabolism is intriguing (Fig. 4A, yellow bridges). Alteration in xenobiotic metabolism in BCa, reflected in our data by elevated levels of aniline (Fig. 2A, B), has been attributed to be one of the causal factors for BCa development (24–26), resulting from mutations in the genes programming phase I/II metabolic enzymes (25, 27–30). However, its co-enrichment with methylaton leads us to hypothesize that epigenetic modification could also regulate the expression of phase I/II metabolic genes in BCa.
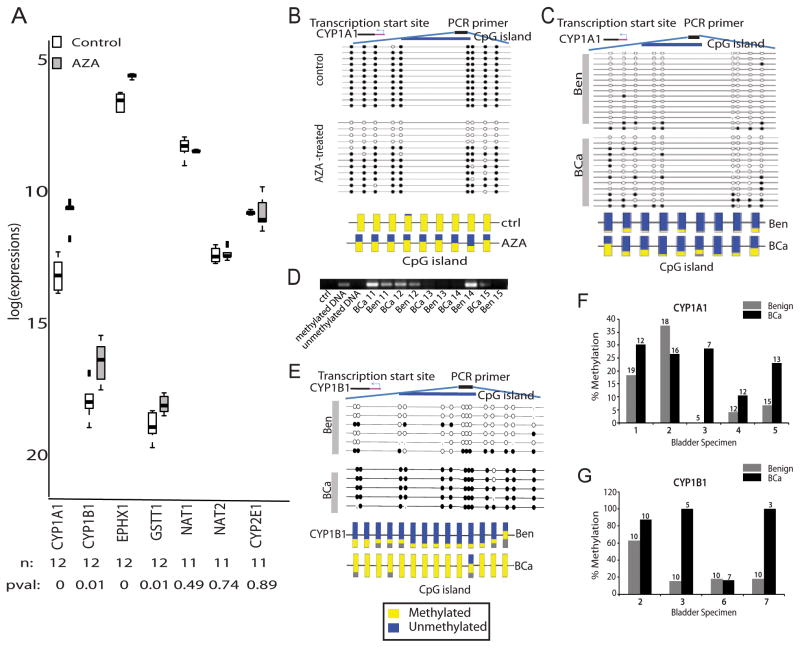
To test this, we first examined the transcript levels for enzymes in phase I/II metabolism in matched BCa and benign adjacent tissues (n = 20) using QPCR (primer sequence in Supplementary Table S10). The transcript levels for cytochrome P450 1A1 (CYP1A1) and cytochrome P450 1B1 (CYP1B1) were significantly reduced in BCa tissues compared to benign adjacent tissues (p < .01, FDR q-value ≤ 5%, Fig. 4B); a finding corroborated by reduced protein expression of CYP1A1 and CYP1B1 in BCa (Fig 4C) and justifying the OCM-based prediction of altered “cytochrome P450-dependent metabolism” in bladder tumors. Additional alterations included reduced transcript levels for cytochrome P450 2E1 (CYP2E1) and glutathione S-transferase T1 (GSTT1, both p < 0.1, FDR q-value ≤ 20%), as well as higher mRNA levels for aromatic hydrocarbon receptor (AHR) and cathechol-O-methyltransferase (COMT, p < 0.001, FDR q-value ≤ 5%) in BCa tissues as compared to benign adjacent tissues (Fig. 4B).
To confirm the role of methylation in regulating the expression of phase I/II metabolic enzymes in BCa tumors, we treated the BCa cell line T-24 with the DNA methyl transferase inhibitor 5-aza-2′-deoxycytidine (AZA, 5 μm for 48h). AZA treatment resulted in a de-repression of CYP1A1, 1B1, and EPHX1 (Fig. 5A) compared to untreated cells (p < .01 and FDR q-value ≤ 5%, for all). Further, bisulfite sequence analysis of these cells confirmed a 50% decrease in methylation of CpGs located ~1,000 bp upstream of the transcription start site on the CYP1A1 promoter (Fig. 5B). Additionally, bisulphite analysis and methylation-specific PCR (MSP) confirmed hypermethylation of CYP1A1 and CYP1B1 promoters in 75% of patient-derived bladder tumors analyzed, compared to matched adjacent benign tissues (Figs. 5C–G). These results for the first time confirm a role for methylation as an alternate mechanism in regulating CYP1A1 and CYP1B1 expression in BCa tumors.
Figure 5. Methylation-induced Silencing of Phase I/II Metabolic Enzymes.
A. Box plots for real time PCR data showing relative transcript levels in T-24 BCa cells with and without treatment with 5 μm 5-aza-2′-deoxycytidine. n = number of samples; pval = p-value. B. Bisulfite sequencing of CYP1A1 in control and AZA-treated T24 BCa cells. Each row indicates the methylation profile for the nine CpG islands in an independent clone (top panel). Multiple clones were analyzed from the treated (n = 11) and untreated groups (n = 12). Filled circles indicate methylated cytosine and open circles denote the unmethylated residue (top panel). The bottom panel shows the average methylation profile for each CpG island. C. Same as B, but for CYP1A1 in BCa and matched benign adjacent tissue. D. Methylation-specific PCR showing higher CpG methylation in CYP1A1 promoter (product size = 127 bp) in matched BCa and benign adjacent tissues (n = 6 pairs). E. Same as B, but for CYP1B1 in BCa and matched benign adjacent tissue. Methylation profile of 14 CpG islands were examined. F. Plot summarizing the bisulfite sequence analysis results for CYP1A1 promoter in BCa and benign adjacent tissues. The median percentage methylation was calculated for each tissue using data obtained from all the sequenced clones. G. same as F, but for CYP1B1.
Discussion
To understand BCa development and progression, and to obtain insights into bioprocesses that could possibly be associated with defective xenobiotic metabolism, we conducted a mass spectrometry-based comprehensive and unbiased metabolomic profiling of tissue specimens from BCa patients and control individuals. Similar to our earlier observations in PCa (14), the BCa-associated metabolome is enriched with amino acids, confirming this metabolic process to be a consistent hallmark of tumor development. Furthermore, we observed that the BCa-associated signature contains aromatic compounds and hydroxylated metabolic derivatives, both of which pinpoint aberrations in xenobiotic metabolism. Although these alterations have been previously reported, their occurrence in BCa has been mainly attributed to defects in the genes regulating detoxification processes (28). However, using a data-driven approach, we confirmed a role for methylation as an additional contributing factor in regulating the expression of xenobiotic metabolizing enzymes. In addition to delineating mechanistic insights into BCa development, our study also defines a compendium of metabolites that reliably distinguishes BCa tissues from benign specimens, as well as in a pilot-setting, delineates muscle-invasive and non-muscle-invasive BCa using patient-derived urine specimens. The PLS-DA-based classification model for detecting BCa in urine had an overall accuracy between 67% and 72% in four independent cohorts (Fig. 3B). This is encouraging considering the benign controls in two of the cohorts were obtained from individuals with antecedent BCa, who were in remission following immuno/chemotherapeutic treatment. These proof-of-concept studies set the stage for evaluating BCa-associated metabolites as non-invasive markers to complement current diagnostic modalities with the goal of optimizing individual patient treatment selection.
In summary, this study describes the global metabolomic map for BCa. Utilizing this approach, we have identified several potential markers for future development and have highlighted the importance of this technique in uncovering epigenetic alterations in genes (i.e., CYP1A1 and CYPB1) involved in pathways implicated in bladder carcinogenesis.
Supplementary Material
Acknowledgments
The authors acknowledge Ravi Kolhe for help with pathology and Sitaram Gayathri for technical assistance. This work is supported in part by the National Cancer Institute grant RO1CA13345 (AS), RO3CA139489-01 (AS) and RCA145444A (AS and GM) and funds to AS from the Alkek Center for Molecular Discovery, Baylor College of Medicine and Georgia Cancer Coalition.
Footnotes
Conflict of Interest: Drs Theodore Sana and Steven Fischer are employees of Agilent Technologies and hold stock options in the company. None of the other authors have any conflict of interest.
References
- 1.ACS. Cancer Facts and Figures 2010. American Cancer Society; Atlanta, GA: 2010. [Google Scholar]
- 2.van der Aa MN, Steyerberg EW, Sen EF, Zwarthoff EC, Kirkels WJ, van der Kwast TH, et al. Patients’ perceived burden of cystoscopic and urinary surveillance of bladder cancer: a randomized comparison. BJU international. 2008;101:1106–10. doi: 10.1111/j.1464-410X.2007.07224.x. [DOI] [PubMed] [Google Scholar]
- 3.Lotan Y, Roehrborn CG. Sensitivity and specificity of commonly available bladder tumor markers versus cytology: results of a comprehensive literature review and meta-analyses. Urology. 2003;61:109–18. doi: 10.1016/s0090-4295(02)02136-2. discussion 18. [DOI] [PubMed] [Google Scholar]
- 4.Pasikanti KK, Esuvanranathan K, Ho PC, Mahendran R, Kamaraj R, Wu Q, et al. Noninvasive Urinary Metabonomic Diagnosis of Human Bladder Cancer. J Proteome Res. 2010 doi: 10.1021/pr901173v. [DOI] [PubMed] [Google Scholar]
- 5.Issaq HJ, Nativ O, Waybright T, Luke B, Veenstra TD, Issaq EJ, et al. Detection of bladder cancer in human urine by metabolomic profiling using high performance liquid chromatography/mass spectrometry. J Urol. 2008;179:2422–6. doi: 10.1016/j.juro.2008.01.084. [DOI] [PubMed] [Google Scholar]
- 6.Shariat SF, Lotan Y, Vickers A, Karakiewicz PI, Schmitz-Drager BJ, Goebell PJ, et al. Statistical consideration for clinical biomarker research in bladder cancer. Urol Oncol. 28:389–400. doi: 10.1016/j.urolonc.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Putluri N, Shojaie A, Vasu VT, Nalluri S, Vareed SK, Putluri V, et al. Metabolomic profiling reveals a role for androgen in activating amino Acid metabolism and methylation in prostate cancer cells. PLoS One. 2011;6:e21417. doi: 10.1371/journal.pone.0021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smyth GK. Limma: linear models for microarray data’ Bioinformatics and Computational Biology Solutions using R and Bioconductor. In: Gentleman RVC, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 9.Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, et al. gplots: Various R programming tools for plotting data. In: Warnes GR, editor. R package version 2.8.0. ed. 2010. [Google Scholar]
- 10.Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–2. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- 11.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach in multiple testing. Journal of Royal Statistical Society. 1995;Series B:57. [Google Scholar]
- 12.Wehrens RM, BH pls: Partial Least Squares Regression (PLSR) and Principal Component Regression (PCR) R package version 21–0. 2007 [cited; Available from: http://mevik.net/work/software/pls.html.
- 13.Wing J, Weston S, Wlliams A, Keefer C, Engelhardt A. caret: Classification and Regression Training. In: Kuhn M, editor. R package. 2010. version 4.67. ed. [Google Scholar]
- 14.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–4. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–8. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- 17.Khan AP, Poisson LM, Bhat VB, Fermin D, Zhao R, Kalyana-Sundaram S, et al. Quantitative proteomic profiling of prostate cancer reveals a role for miR-128 in prostate cancer. Mol Cell Proteomics. 9:298–312. doi: 10.1074/mcp.M900159-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sepai O, Sabbioni G. Biomonitoring workers exposed to arylamines: application to hazard assessment. Adv Exp Med Biol. 1996;387:451–5. doi: 10.1007/978-1-4757-9480-9_55. [DOI] [PubMed] [Google Scholar]
- 19.Carreon T, Hein MJ, Viet SM, Hanley KW, Ruder AM, Ward EM. Increased bladder cancer risk among workers exposed to o-toluidine and aniline: a reanalysis. Occup Environ Med. 2010;67:348–50. doi: 10.1136/oem.2009.051136. [DOI] [PubMed] [Google Scholar]
- 20.Jones TD, Wang M, Eble JN, MacLennan GT, Lopez-Beltran A, Zhang S, et al. Molecular evidence supporting field effect in urothelial carcinogenesis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:6512–9. doi: 10.1158/1078-0432.CCR-05-0891. [DOI] [PubMed] [Google Scholar]
- 21.Rhodes DR, Kalyana-Sundaram S, Tomlins SA, Mahavisno V, Kasper N, Varambally R, et al. Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia. 2007;9:443–54. doi: 10.1593/neo.07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aleman A, Adrien L, Lopez-Serra L, Cordon-Cardo C, Esteller M, Belbin TJ, et al. Identification of DNA hypermethylation of SOX9 in association with bladder cancer progression using CpG microarrays. Br J Cancer. 2008;98:466–73. doi: 10.1038/sj.bjc.6604143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vallot C, Stransky N, Bernard-Pierrot I, Herault A, Zucman-Rossi J, Chapeaublanc E, et al. A novel epigenetic phenotype associated with the most aggressive pathway of bladder tumor progression. J Natl Cancer Inst. 103:47–60. doi: 10.1093/jnci/djq470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baris D, Karagas MR, Verrill C, Johnson A, Andrew AS, Marsit CJ, et al. A case-control study of smoking and bladder cancer risk: emergent patterns over time. J Natl Cancer Inst. 2009;101:1553–61. doi: 10.1093/jnci/djp361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altayli E, Gunes S, Yilmaz AF, Goktas S, Bek Y. CYP1A2, CYP2D6, GSTM1, GSTP1, and GSTT1 gene polymorphisms in patients with bladder cancer in a Turkish population. Int Urol Nephrol. 2009;41:259–66. doi: 10.1007/s11255-008-9444-6. [DOI] [PubMed] [Google Scholar]
- 26.Lower GM, Jr, Nilsson T, Nelson CE, Wolf H, Gamsky TE, Bryan GT. Nacetyltransferase phenotype and risk in urinary bladder cancer: approaches in molecular epidemiology. Preliminary results in Sweden and Denmark. Environmental Health Perspectives. 1979:71–79. doi: 10.1289/ehp.792971. [DOI] [PMC free article] [PubMed] [Google Scholar]; Int J Epidemiol. 2007;36:11–8. [Google Scholar]
- 27.Fanlo A, Sinues B, Mayayo E, Bernal L, Soriano A, Martinez-Jarreta B, et al. Urinary mutagenicity, CYP1A2 and NAT2 activity in textile industry workers. J Occup Health. 2004;46:440–7. doi: 10.1539/joh.46.440. [DOI] [PubMed] [Google Scholar]
- 28.Grando JP, Kuasne H, Losi-Guembarovski R, Sant’ana Rodrigues I, Matsuda HM, Fuganti PE, et al. Association between polymorphisms in the biometabolism genes CYP1A1, GSTM1, GSTT1 and GSTP1 in bladder cancer. Clin Exp Med. 2009;9:21–8. doi: 10.1007/s10238-008-0015-z. [DOI] [PubMed] [Google Scholar]
- 29.Roos PH, Bolt HM. Cytochrome P450 interactions in human cancers: new aspects considering CYP1B1. Expert opinion on drug metabolism & toxicology. 2005;1:187–202. doi: 10.1517/17425255.1.2.187. [DOI] [PubMed] [Google Scholar]
- 30.Thier R, Bruning T, Roos PH, Bolt HM. Cytochrome P450 1B1, a new keystone in gene-environment interactions related to human head and neck cancer? Archives of toxicology. 2002;76:249–56. doi: 10.1007/s00204-002-0349-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.