Abstract
Uracil-DNA glycosylase (Ung) is a DNA repair enzyme that excises uracil bases from DNA, where they appear through deamination of cytosine or incorporation from a cellular dUTP pool. DNA repair enzymes often use one-dimensional diffusion along DNA to accelerate target search; however, this mechanism remains poorly investigated mechanistically. We used oligonucleotide substrates containing two uracil residues in defined positions to characterize one-dimensional search of DNA by Escherichia coli Ung. Mg2+ ions suppressed the search in double-stranded DNA to a higher extent than K+ likely due to tight binding of Mg2+ to DNA phosphates. Ung was able to efficiently overcome short single-stranded gaps within double-stranded DNA. Varying the distance between the lesions and fitting the data to a theoretical model of DNA random walk, we estimated the characteristic one-dimensional search distance of ~100 nucleotides and translocation rate constant of ~2×106 s−1.
Keywords: DNA repair, uracil-DNA glycosylase, one-dimensional diffusion, random walk, processivity
Introduction
One of the most daunting tasks faced by proteins that recognize specific elements in DNA is one of location of their targets within a physiologically relevant period of time. The best-known examples of such proteins are sequence-specific transcription factors, endonucleases and methyltransferases of restriction systems, and DNA repair enzymes (reviewed in [1; 2]). The last group of proteins includes enzymes that often need to find and recognize damaged nucleobases that are quite similar to the canonical ones. For example, uracil-DNA glycosylase excises uracil from DNA and thus has to detect a difference between an oxo and an amino moiety at C4 and between a hydrogen and a methyl group at C5 of a pyrimidine heterocycle to distinguish Ura from Cyt or Thy, respectively. As crystal structures show, sampling of bases by DNA repair enzymes often involves gross distortion of DNA such as kinking and eversion of the sampled base from the double helix [1; 3; 4].
Experimentally measured rates of association of specific DNA-binding proteins with their target sites often exceed the formal diffusion limit of ~108 M−1s−1 [2; 5]. To explain this apparent paradox, the search for targets was proposed to involve a significant component of one-dimensional (1D) diffusion along DNA rather than exclusively three-dimensional (3D) diffusion typical of proteins binding small soluble ligands [6; 7; 8]. This concept of “facilitated diffusion” has been applied to a number of cases, including restriction endonucleases EcoRI, EcoRV, HindIII, BamHI, and transcription factors LacI, GalR, Cro, glucocorticoid nuclear receptor, and p53 (reviewed in [1; 2]). For DNA repair enzymes, the 1D diffusion mechanism was most thoroughly studied for bacteriophage T4 endonuclease V (reviewed in [9]), which even has been shown to use 1D search in vivo. Other examples of 1D search include enzymes of base excision repair (E. coli and mammalian uracil-DNA glycosylases [10; 11; 12; 13], human alkyladenine-DNA glycosylase [14; 15], 8-oxoguanine-DNA glycosylases from E. coli [16; 17], Bacillus stearothermophilus [18] and human [17; 18], E. coli MutY DNA glycosylase [16], human abasic site endonuclease [19], and several proteins involved in direct reversal, mismatch repair, and recombination repair (reviewed in [1; 2]).
Traditionally, studies of target search by DNA repair enzymes relied on the use of randomly damaged plasmids or concatemer substrates, with 1D search mechanism giving rise to processivity, the ability of the enzyme to cleave several targets in a multiply damaged DNA molecule without releasing it [1]. Newer approaches, both powerful and expensive, include single-molecule fluorescence microscopy [18] and atomic force microscopy [20]. Recently, we [12; 17] and others [13; 14; 15] proposed a simple biochemical assay based on oligonucleotide substrates containing damaged bases in defined positions (Fig. 1A). The conditional probability of cleavage of the second damaged site after the first site is cleaved, Pcc (probability of correlated cleavage), which can easily be extracted from steady-state kinetic data, serves as a quantitative characteristic of processivity. Unlike in the other methods mentioned above, which require long DNA, the oligonucleotide nature of the substrate allows straightforward manipulation of its structure to pinpoint factors important for 1D search.
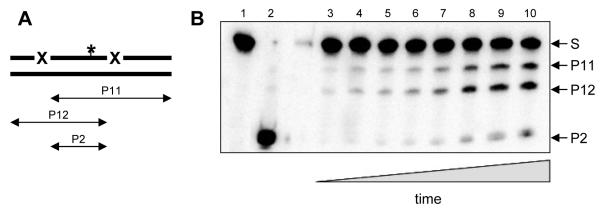
Fig. 1.
Outline of the oligonucleotide-based processivity assay. A, general scheme of the substrate and products formed during the reaction. X, enzyme-specific site (Ura for uracil-DNA glycosylase). P11, P12, products of cleavage at one of the sites; P2, product of cleavage at both sites. The asterisk marks the position of a radioactive label. B, representative gel of a cleavage reaction. S, substrate (U20L-*U20R//comp40), P11, P12, P2, products of different lengths. The reaction times were 0 min (lane 1), 0.5 min (lane 3), 1 min (lane 4), 1.5 min (lane 5), 2 min (lane 6), 3 min (lane 7), 5 min (lane 8), 7 min (lane 9), and 10 min (lanes 2 and 10). The concentration of Ung was 15 pM(lanes 1, 3-10) or 15 nM (lane 2); the concentration of KCl was 50 mM.
In this work, we apply the oligonucleotide-based method to dissect several important features of the mechanism of target search by E. coli uracil-DNA glycosylase, including the effect of divalent cations, DNA structure interruptions, and the distance between the lesions. We also use a theoretical model of random walk developed by Belotserkovskii and Zarling [21] to estimate the rate constants of search by Ung.
Materials and Methods
Preparation of substrates
Oligonucleotides were synthesized on an ASM-800 DNA/RNA Synthesizer (Biosset, Novosibirsk, Russia) from commercially available phosphoramidites (Glen Research, Sterling, VA) according to the manufacturer’s protocols and purified by high-performance liquid chromatography followed by electrophoresis in 8% or 20% polyacrylamide gel containing 8 M urea. Sequences of the oligonucleotides are listed in the Table, and the procedures of construction of substrates are summarized in the Supplementary Chart. To prepare the shoulders for ligation, 1000 pmol of a 20-mer oligonucleotide U20R (Table) was 5′-labeled using γ[32P]ATP (Radioizotop, Moscow, Russia) and 20 units of T4 polynucleotide kinase (SibEnzyme, Novosibirsk, Russia) at 37°C for 40 min according to the manufacturer’s protocol. Another 10 U of polynucleotide kinase were then added and the incubation was continued for 40 min. The reaction was stopped by heating for 5 min at 95°C. In a separate reaction, the same oligonucleotide was phosphorylated using 1 mM non-radioactive ATP in the same way. Both reaction mixtures were mixed and used for subsequent ligation.
Table 1.
Sequences of the oligonucleotides used in this work
| ID | sequence |
|---|---|
| comp40 | 5′-TCTGGAAAGGAGAGAAGTCCGAAGGAAAGGAGAGAAGGGA-3′ |
| comp40R | 5′-TCTGGAAAGGAGAGAAGTCCGAGGTCTGAACGAGAGGAAA-3′ |
| comp41 | 5′-TCTGGAAAGGAGAGAAGTCCGGAAGGAAAGGAGAGAAGGGA-3′ |
| comp41L | 5′-pGATCGCACAAATGAAAGGTCCGAAGGAAAGGAGAGAAGGGA-3′ |
| comp46 | 5′-TTTTCTGGAAAGGAGCGAAGTCCGAAGGAAAGGAGCGAAGGGATTT-3′ |
| comp50L | 5′-pAAATTCACTCATCGCACAAATGAAAGGTCCGAAGGAAAGGAGAGAAGGGA-3′ |
| comp51R | 5′-TCTGGAAAGGAGAGAAGTCCGAGGTCTGAACGAGAGGAAAGCTAAATCCCG-3′ |
| comp61 | 5′-TCTGGAAAGGAGAGAAGTCCGCTCTAACGCAAGTAAAGTCCGAAGGAAAGGAGAGAAGGGA-3′ |
| gap0L | 5′-GAAGGAAAGGAGGGAAGGGA-3′ |
| gap0R | 5′-TCTGGAAAGGAGGGAAGTCC-3′ |
| gap2L | 5′-AAGGAAAGGAGGGAAGGGA-3′ |
| gap2R | 5′-TCTGGAAAGGAGGGAAGTC-3′ |
| gap4L | 5′-AGGAAAGGAGGGAAGGGA-3′ |
| gap4R | 5′-TCTGGAAAGGAGGGAAGT-3′ |
| gap6L | 5′-GGAAAGGAGGGAAGGGA-3′ |
| gap6R | 5′ -TCTGGAAAGGAGGGAAG-3′ |
| Sp21 | 5′-pGGACTTTACTTGCGTTAGAGC-3′ |
| Sp41 | 5′-pGGACCTTTCATTTGTGCGATCTTTCCTCTCGTTCAGACCTC-3′ |
| Sp61 | 5′-pGGACCTTTCATTTGTGCGATGAGTGAATTTCGGGATTTAGCTTTCCTCTCGTTCAGACCTC-3′ |
| U20L | 5′-TCCCTTCUCTCCTTTCCTTC-3′ |
| U20R | 5′-GGACTTCUCTCCTTTCCAGA-3′ |
| U21L | 5′-TCCCTTCUCTCCTTTCCTTCC-3′ |
p, 5′-terminal phosphate introduced during oligonucleotide synthesis
The 40-mer double-stranded substrate with two Ura residues in one strand separated by 19 normal deoxynucleotides, U20L-*U20R//comp40, was prepared by ligation of 5′-[32P]-U20R with a 20-mer oligonucleotide U20L, both annealed to a complementary 40-mer scaffold comp40 as follows: 1 min at 95°C, 10 min at 57°C, 2 h gradual cooling to 25°C. U20L and C40 oligonucleotides were taken in a 1.5-fold molar excess over U20R. The mixture was supplemented with 200 units of bacteriophage T4 DNA ligase (SibEnzyme, Novosibirsk, Russia), ligase reaction buffer (final, 50 mM Tris-HCl pH 7.8, 10 mM MgCl2; 10 mM dithiothreitol) and ATP (final, 5 mM) and incubated overnight at 4°C. The reaction product was purified by electrophoresis in 20% polyacrylamide gel containing 8 M urea and annealed to a 1.5-fold excess of comp40 complementary scaffold. The completeness of annealing was tested by electrophoresis in a non-denaturing 8% polyacrylamide gel at 4°C.
The 40-mer single-stranded substrate, U20L-*U20R, was prepared similarly to a 40-mer double-stranded substrate but with a 46-mer complementary scaffold comp46 used for annealing instead of 40-mer comp40 to achieve a complete separation of the scaffold strand during gel electrophoresis. The 40-mer substrates contained gaps between uracils was prepared by annealing with complementary oligonucleotides gap0R and gap0L (20-mers, forming a nick), gap2R and gap2L (19-mers, forming a 2-nt gap), gap4R and gap4L (18-mers, forming a 4-nt gap), gap6R and gap6L (17-mers, forming a 6-nt gap).
The 41-mer double-strand substrate with 20 nucleotides between the Ura residues, U21L–*U20R//comp41, was prepared similarly to the 40-mer substrate, but with a 21-mer U21L and 41-mer comp41 oligonucleotides instead of U20L and C40, respectively. The 61-mer double-stranded substrate with 40 nt between two Ura residues, U20L-Sp21-*U20R//comp61, was prepared by annealing 5′-[32P]-U20R with U20L, a 61-mer oligonucleotide scaffold comp61, and a 21-mer Sp21 (forming a spacer between U20R and U20L on the scaffold), which was 5′-phosphorylated with a non-radioactive phosphate group during the synthesis. The 81-mer double-strand substrate with 60 nt between two Ura residues, U20L-Sp41-*U20R//comp41L–comp40R, was prepared by annealing 5′-[32P]-U20R with U20L, a 41-mer 5′-phosphorylated spacer Sp41, and two scaffold oligonucleotides: a 5′-phosphorylated 41-mer comp41L and a 40-mer comp40R. The 101-mer double-strand substrate with 80 nt between two Ura residues, U20L-Sp61-*U20R//comp50L-comp51R, was prepared similarly to the 81-mer substrate, but using a 61-mer 5′-phosphorylated spacer Sp61, and two scaffold oligonucleotides: a 5′-phosphorylated 50-mer comp50L and a 51-mer comp51R. The ligation was performed as described above, and the resulting 61-, 81- and 101-mer double-stranded oligonucleotides were purified by electrophoresis in a non-denaturing 8% polyacrylamide gel at 4°C.
Correlated cleavage assay
Escherichia coli Ung was purchased from New England Biolabs (Beverly, MA). The reaction mixture (50 μl) contained 50 nM oligonucleotide substrate, 25 mM sodium phosphate buffer (pH 7.5), 1 mM dithiothreitol and 1 mM EDTA. Ung was diluted on ice to 0.4 U/ml in 12.5 mM sodium phosphate buffer (pH 7.5) supplemented with 0.5 mg/ml bovine serum albumin (New England Biolabs), and the reaction was initiated by adding Ung to 0.04 U/ml (15 pM) to the reaction mixture at 37°C. After 0.5, 1, 1.5, 2, 3, 5, 7, and 10 min, 5-μl aliquots were withdrawn and immediately quenched with NaOH (final concentration, 100 mM), heated for 2 min at 95°C, and neutralized with an equimolar amount of HCl. The products were resolved by electrophoresis in polyacrylamide gel (20% for 40- and 41- nt substrates and 8% for longer substrates) containing 8 M urea (Fig. 1B) and quantified by phosphorimaging using a Molecular Imager FX system (Bio-Rad Laboratories, Hercules, CA). Initial rates of accumulation of products of different lengths were determined from the linear parts of the time courses using SigmaPlot v9.0 (Systat Software, Chicago, IL). If necessary, the reaction mixture was supplemented with 10-200 mM KCl, 5-20 mM MgCl2 (both from Sigma-Aldrich, St. Louis, MO), or 0.05-5% polyethelenglycol-8000 (MP Biomedicals, Solon, OH). The probability of correlated cleavage was determined as
where vP11 and vP12 are rates of accumulation of the products of cleavage at one of the Ura sites, and vP2 is the rate of accumulation of the product of cleavage at both Ura sites [12]. At all time points in all experiments, the ratio of P11 and P12 products was 1.00 ± 0.15, confirming equal efficiency of cleavage by Ung at both Ura sites.
Results and Discussion
Effect of magnesium on the processivity of Ung
Unlike monovalent cations such as Na+ or K+, which do not have preferred binding sites in double-stranded DNA and interact with DNA rather non-specifically, Mg2+ ions are mostly coordinated by phosphates and Gua bases in the major groove (either directly or through the first solvation shell) and stabilize DNA duplexes to a much higher degree than monovalent cations [22; 23]. This may cause some DNA-dependent enzymes to be much more sensitive to the concentration of Mg2+ than of monovalent cations even if they, like DNA glycosylases, do not require Mg2+ for their activity [24]. Therefore, it was interesting to compare the effect of Mg2+ and K+ on the processivity of Ung.
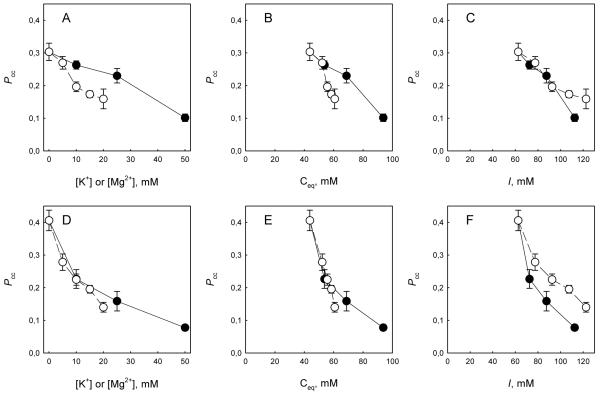
Fig. 2 shows the dependence of Pcc on the concentration of K+ and Mg2+ determined for double-stranded (Fig. 2A) and single-stranded substrates (Fig. 2D). It can be seen that at equal molar concentrations, Mg2+ caused a more pronounced decrease than K+ in the ability of the enzyme to processively cleave double-stranded DNA, while the effect of both ions on Pcc for the single-stranded substrate was similar. To get a deeper insight in the nature of interference of Mg2+ with correlated cleavage, the same Pcc data were re-plotted against the equivalent concentration (Ceq) of all cations present in the reaction mixture. Ceq is by definition a concentration of monovalent cations at which an oligonucleotide duplex has the same melting temperature as in the presence of a given mixture of cations, and accounts for more efficient stabilization of DNA by divalent cations [23]. Several empirical equations for calculating Ceq are available [23]; of those, we selected the function that most accurately approximates the thermodynamic data within the range of salt concentration and oligonucleotide lengths used:
where [K+] and [Mg2+] are concentrations of the added ions and [Na+]buf is the concentration of Na+ in the sodium phosphate buffer (43.75 mM). As expected, no significant difference between K+ and Mg2+ was observed for a single-stranded substrate, which is not stabilized thermodynamically (Fig. 2E); it should be noted that the single-stranded oligonucleotide U20L–U20R does not form stable hairpins or dimers. With a double-stranded substrate, Mg2+ still was more detrimental for correlated cleavage than K+ (Fig. 2B), suggesting that the effect of Mg2+ is not due to local thermodynamic stabilization of DNA by positive counterions. However, when Pcc values were plotted against total ionic strength of the buffer, considering all ionic species, there was either little difference between Mg2+ and K+ for double-stranded DNA (Fig. 2C) or K+ had a stronger effect than Mg2+ for single-stranded DNA (Fig. 2F), ruling out simple electrostatic screening of DNA charge as a factor in suppressing correlated cleavage. We conclude that Mg2+ likely influences processive search by Ung due to its tight coordination to phosphates in double-stranded DNA and competition for these phosphates with the DNA-binding groove of the enzyme. K+ or Na+ are not coordinated and therefore can be much easily displaced from DNA by the moving enzyme.
Fig. 2.
Effect of K+ (●) and Mg2+ (○) on Pcc of Ung. The substrates were double-stranded U1-U2//C2 (A-C) or single-stranded U1-U2 (D-F). A, D, dependence of Pcc on the K+ or Mg2+ concentration. B, E, dependence of Pcc on the equivalent cation concentration. C, F, dependence of Pcc on the ionic strength of the reaction mixture. Mean ± SD of three independent experiments are shown.
Ung can search through short gaps in DNA
Normally, DNA in the cell is associated with many DNA-binding proteins and small molecules that may interfere with processive search. Earlier, we have shown that in E. coli extracts, Ung is capable of correlated cleavage despite the competition with other proteins, albeit Pcc values are lower than in the case of the purified protein [12]. This residual correlated cleavage may be ascribed to the hopping mode of search, as was also shown in a similar type of assay for human alkyladenine-DNA glycosylase bypassing a bound EcoRI molecule [15].
Here we have investigated whether obstacles of another nature, where double-stranded DNA is interrupted by a single-stranded stretch, affect the processive search by Ung. Although no structure of the complex of E. coli Ung with any DNA is available, the human homolog forms contacts with both strands when searching undamaged double-stranded DNA [25], so it is reasonable to suggest that the protein must undergo a conformational change when transferred from double- to single-stranded DNA, or release DNA and re-associate with it in another binding mode. We have constructed double-stranded substrates containing 2-, 4-, or 6-nt long gaps in the non-damaged strand, or containing a nick (essentially, a one-phosphate gap), and compared the correlated cleavage of such substrates by Ung (Fig. 3). The presence of a gap up to 6 nt did not significantly affect the correlated cleavage, suggesting that either the hopping mode of search can successfully act at these distances, or that sliding of Ung along DNA can proceed seamlessly at the single-stranded/double-stranded DNA junctions. For E. coli and human 8-oxoguanine-DNA glycosylases, it has been shown that a nick in the undamaged strand does not decrease Pcc and in some cases even increases it [17]. Addition of polyethylene glycol (PEG8000) to the reaction mixture up to 5% (w/v) had no appreciable effect on Pcc (not shown), suggesting little dependence of Pcc on the association rate and consistent with the continuous sliding model.
Fig. 3.
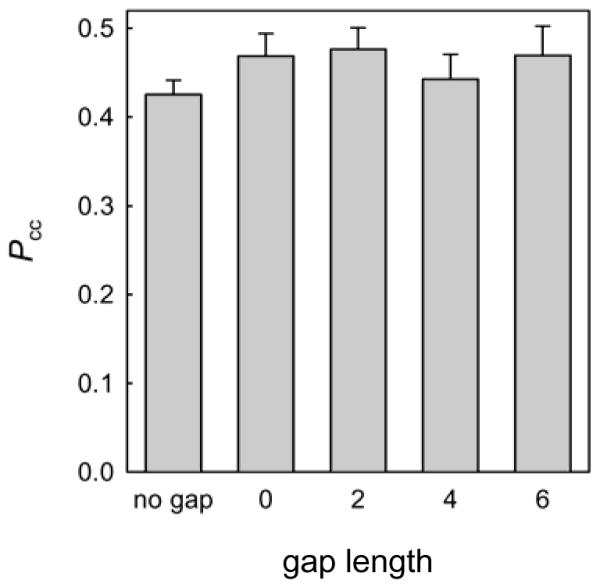
Correlated cleavage of gapped substrates by Ung. The length of the gap in nucleotides is indicated (0 corresponds to lack of a single phosphate). Mean ± SD of three independent experiments are shown.
Random walk description of search by Ung
It has long been realized that the process of searching DNA by strictly one-dimensional diffusional sliding between adjacent positions can be described as a random one-dimensional walk (reviewed in [1]). Among several mathematical models developed to describe random walks of proteins along DNA, the Belotserkovskii-Zarling model [21] provides an analytical dependence of the probability that the protein, starting from a given coordinate (x0) in DNA reaches another given coordinate (x0 ≤ x):
| Eq. 1 |
with a = koff/ktr, b = kloss/ktr, where ktr, koff and kloss are rate constants for translocation by one nucleotide, dissociation from an internal position in DNA, and loss from a DNA end (x = 0) (Supplementary Fig. S1). The parameters a and b, can be used to obtain the characteristic distance of protein translocation before dissociation from DNA (a−1/2) and the probability of loss from the end of DNA (b/(1+b)). This model was recently successfully used to analyze the movement of EcoRI restriction endonuclease along DNA [26].
Applying Eq. 1 to our Pcc data, it is possible to provide an estimate for the parameters of correlated search. To do this, we have constructed double-stranded DNA substrates in which two uracil residues in the same strand were separated by 20, 40, 60, or 80 nucleotides, and determined Pcc values for them (Fig. 4). Over these distances, continuous Eq. 1 describes a discrete random walk with a very good accuracy (see Supplementary Results and Discussion). The particular distances between the targets were selected to keep both Ura bases facing the same side of the DNA helix in order to minimize possible variability if hopping contributes partly to the search process. Not surprisingly, the probability of correlated cleavage decreased over distance, reflecting the loss of some enzyme molecules during the transfer between the first and the second cleavage site. The data were then fit to the Belotserkovskii-Zarling model. Note that W(x0, x) is the probability of the first passage through the target site, whereas Pcc is the probability of cleavage, and thus can be expressed as Pcc = PT×PE, where PT is the probability of transition to the target site, and PE is the probability of uracil excision upon an encounter. Based on stopped-flow data for E. coli Ung, an available estimate of PE is 0.73 [13]. We therefore used PT = Pcc/0.73 as the dependent variable in Eq. 1. Since our detection system requires that the products of the first cleavage would be of different lengths to be resolved by electrophoresis, our substrates contain uracils separated from the nearest end by different spans of DNA: one Ura is in position 8 from the 5′-end, and another, in position 13 from the 3′-end. Since the first cleavage may occur with an equal probability at any of the two sites, we have accounted for the substrate asymmetry by using the average, 1/2×[W(8, N+8) + W(13, N+13)] as the fitting function. The fitting data produced an estimate of a = 10−4 and b = 0.2, corresponding to the characteristic distance covered before dissociation ~100 nt and the probability of dissociation from an end, 0.17.
Fig. 4.
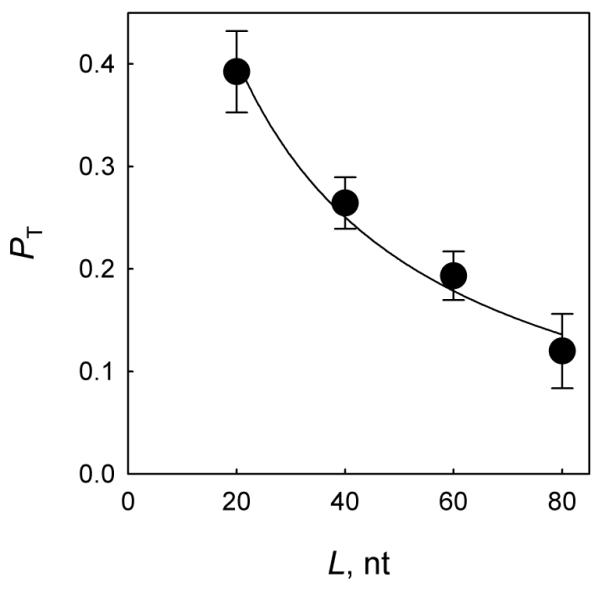
Dependence of the translocation component PT = Pcc/PE on the distance separating the cleavage sites. Mean ± SD of three independent experiments are shown. The curve is the fit of the experimental data to the function P = 0.5×(W(8, N + 8) + W(13, N + 13)) (see main text).
To estimate the rate constants ktr and kloss, we have assumed koff = 200 s−1, a value obtained in [13] using stopped-flow methods under the reaction conditions similar to ours. Using this value of koff, the calculated values of ktr and kloss are 2×106 s−1 and 4×105 s−1, respectively. The ktr value obtained in this way is in the range of ktr extracted from single-molecule experiments with two other DNA glycosylases, Bacillus stearothermophilus Fpg (0.7×106 s−1) and human OGG1 (10×106 s−1) searching double-stranded DNA [18].
In conclusion, we have used the oligonucleotide-based correlated cleavage assay to address the effect of divalent cations and gaps in double-stranded DNA on the process of search of DNA lesions by uracil-DNA glycosylase, and to estimate rate constants of the search. Our results are consistent with the idea that Ung searches for its targets by combining 3D and 1D diffusion to enhance the search efficiency and overcome obstacles that may appear in DNA in living cells.
Supplementary Material
*Highlights.
Uracil-DNA glycosylase (Ung) excises uracil bases from DNA
Ung searches for these lesions by combining diffusion in 3D and random sliding in 1D
Mg2+ ions suppress the 1D search by Ung in double-stranded DNA
Ung can efficiently search through short single-stranded gaps within double-stranded DNA
The characteristic Ung 1D search distance is ~100 nucleotides, and the translocation rate constant is ~2×106 s−1
Acknowledgments
This research was supported by Russian Foundation for Basic Research (10-04-91058-PICS_a, 11-04-00807-a), Russian Ministry of Education and Science (NSh-3185.2010.4, 14.740.12.0819), Molecular Biology Program of the Presidium of the Russian Academy of Sciences (6.14), and National Cancer Institute (CA017395-33).
Abbreviations
- 1D
one-dimensional
- 3D
three-dimensional
- EDTA
ethylenediamine tetraacetate
- nt
nucleotide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Zharkov DO, Grollman AP. The DNA trackwalkers: Principles of lesion search and recognition by DNA glycosylases. Mutat. Res. 2005;577:24–54. doi: 10.1016/j.mrfmmm.2005.03.011. [DOI] [PubMed] [Google Scholar]
- [2].Tafvizi A, Mirny LA, van Oijen AM. Dancing on DNA: Kinetic aspects of search processes on DNA. Chemphyschem. 2011;12:1481–1489. doi: 10.1002/cphc.201100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: New twists and grooves. Mutat. Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- [4].Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: Implications for DNA damage recognition, removal, and repair. DNA Repair. 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- [5].Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J. Mol. Biol. 1970;53:401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 6].Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- [7].Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 2 The Escherichia coli repressor-operator interaction: Equilibrium measurements. Biochemistry. 1981;20:6948–6960. doi: 10.1021/bi00527a029. [DOI] [PubMed] [Google Scholar]
- [8].Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor-operator interaction: Kinetic measurements and conclusions. Biochemistry. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- [9].Lloyd RS. Investigations of pyrimidine dimer glycosylases — a paradigm for DNA base excision repair enzymology. Mutat. Res. 2005;577:77–91. doi: 10.1016/j.mrfmmm.2005.04.003. [DOI] [PubMed] [Google Scholar]
- [10].Higley M, Lloyd RS. Processivity of uracil DNA glycosylase. Mutat. Res. 1993;294:109–116. doi: 10.1016/0921-8777(93)90019-d. [DOI] [PubMed] [Google Scholar]
- [11].Bennett SE, Sanderson RJ, Mosbaugh DW. Processivity of Escherichia coli and rat liver mitochondrial uracil-DNA glycosylase is affected by NaCl concentration. Biochemistry. 1995;34:6109–6119. doi: 10.1021/bi00018a014. [DOI] [PubMed] [Google Scholar]
- [12].Sidorenko VS, Mechetin GV, Nevinsky GA, Zharkov DO. Correlated cleavage of single- and double-stranded substrates by uracil-DNA glycosylase. FEBS Lett. 2008;582:410–414. doi: 10.1016/j.febslet.2008.01.002. [DOI] [PubMed] [Google Scholar]
- [13].Porecha RH, Stivers JT. Uracil DNA glycosylase uses DNA hopping and short-range sliding to trap extrahelical uracils. Proc. Natl Acad. Sci. U.S.A. 2008;105:10791–10796. doi: 10.1073/pnas.0801612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hedglin M, O'Brien PJ. Human alkyladenine DNA glycosylase employs a processive search for DNA damage. Biochemistry. 2008;47:11434–11445. doi: 10.1021/bi801046y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hedglin M, O'Brien PJ. Hopping enables a DNA repair glycosylase to search both strands and bypass a bound protein. ACS Chem. Biol. 2010;5:427–436. doi: 10.1021/cb1000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Francis AW, David SS. Escherichia coli MutY and Fpg utilize a processive mechanism for target location. Biochemistry. 2003;42:801–810. doi: 10.1021/bi026375+. [DOI] [PubMed] [Google Scholar]
- [17].Sidorenko VS, Zharkov DO. Correlated cleavage of damaged DNA by bacterial and human 8-oxoguanine-DNA glycosylases. Biochemistry. 2008;47:8970–8976. doi: 10.1021/bi800569e. [DOI] [PubMed] [Google Scholar]
- [18].Blainey PC, van Oijen AM, Banerjee A, et al. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl Acad. Sci. U.S.A. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Carey DC, Strauss PR. Human apurinic/apyrimidinic endonuclease is processive. Biochemistry. 1999;38:16553–16560. doi: 10.1021/bi9907429. [DOI] [PubMed] [Google Scholar]
- [20].van Noort SJT, van der Werf KO, Eker APM, et al. Direct visualization of dynamic protein-DNA interactions with a dedicated atomic force microscope. Biophys. J. 1998;74:2840–2849. doi: 10.1016/S0006-3495(98)77991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Belotserkovskii BP, Zarling DA. Analysis of a one-dimensional random walk with irreversible losses at each step: Applications for protein movement on DNA. J. Theor. Biol. 2004;226:195–203. doi: 10.1016/j.jtbi.2003.08.013. [DOI] [PubMed] [Google Scholar]
- [22].Subirana JA, Soler-López M. Cations as hydrogen bond donors: A view of electrostatic interactions in DNA. Annu. Rev. Biophys. Biomol. Struct. 2003;32:27–45. doi: 10.1146/annurev.biophys.32.110601.141726. [DOI] [PubMed] [Google Scholar]
- [23].Owczarzy R, Moreira BG, You Y, et al. Predicting stability of DNA duplexes in solutions containing magnesium and monovalent cations. Biochemistry. 2008;47:5336–5353. doi: 10.1021/bi702363u. [DOI] [PubMed] [Google Scholar]
- [24].Sidorenko VS, Mechetin GV, Nevinsky GA, Zharkov DO. Ionic strength and magnesium affect the specificity of Escherichia coli and human 8-oxoguanine-DNA glycosylases. FEBS J. 2008;275:3747–3760. doi: 10.1111/j.1742-4658.2008.06521.x. [DOI] [PubMed] [Google Scholar]
- [25].Parker JB, Bianchet MA, Krosky DJ, et al. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature. 2007;449:433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rau DC, Sidorova NY. Diffusion of the restriction nuclease EcoRI along DNA. J. Mol. Biol. 2010;395:408–416. doi: 10.1016/j.jmb.2009.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.