Abstract
Estimates of the coalescent effective population size Ne can be poorly correlated with the true population size. The relationship between Ne and the population size is sensitive to the way in which birth and death rates vary over time. The problem of inference is exacerbated when the mechanisms underlying population dynamics are complex and depend on many parameters. In instances where nonparametric estimators of Ne such as the skyline struggle to reproduce the correct demographic history, model-based estimators that can draw on prior information about population size and growth rates may be more efficient. A coalescent model is developed for a large class of populations such that the demographic history is described by a deterministic nonlinear dynamical system of arbitrary dimension. This class of demographic model differs from those typically used in population genetics. Birth and death rates are not fixed, and no assumptions are made regarding the fraction of the population sampled. Furthermore, the population may be structured in such a way that gene copies reproduce both within and across demes. For this large class of models, it is shown how to derive the rate of coalescence, as well as the likelihood of a gene genealogy with heterochronous sampling and labeled taxa, and how to simulate a coalescent tree conditional on a complex demographic history. This theoretical framework encapsulates many of the models used by ecologists and epidemiologists and should facilitate the integration of population genetics with the study of mathematical population dynamics.
INTEREST has grown in methods that integrate increasingly abundant genetic data from viruses with the inference of historic prevalence of infection and epidemiologically relevant parameters (Grenfell et al. 2004; Gordo et al. 2009; Pybus and Rambaut 2009; Van Ballegooijen et al. 2009; Bedford et al. 2010; Biek and Real 2010; Kretzschmar et al. 2010; Talbi et al. 2010; Bataille et al. 2011; Koelle et al. 2011; O’Dea and Wilke 2011; Stadler 2011). Modeling the replication of a pathogen requires consideration of epidemiological dynamics that are not always a concern when investigating the population genetics of other organisms. Changes in population size are often rapid and highly nonlinear. Birth rates and transmission rates change dramatically over time and are not strictly proportional to population size. The time between transmission events (the serial interval) contracts over the course of an epidemic (Kenah et al. 2008), and the variance in the number of transmissions per infected unit may not be consistent with conventional models in population genetics (K. Koelle and D. Rassmussen, unpublished results). Finally, in many epidemic scenarios, the sample fraction is quite large and cannot be neglected. For example, sequencing of human immunodeficiency virus (HIV) for drug resistance testing is now routine in developed countries (Hirsch et al. 2000), and a majority of incident infections are now genotyped.
In the following, we consider a haploid population multiplying according to a continuous time birth–death process with varying rates. The population size Y(t) is a deterministic and differentiable function of time. The birth rate, f(t), describes the rate at which the Y(t) extant gene copies replicate. Cases in which f(t) = cY(t) is strictly proportional to population size, such as during exponential growth, are already well understood. The more general case where f(Y, t) is an arbitrary function of time and the state of the system is not well explored from the perspective of coalescent theory. Throughout this article, t denotes time prospectively, while s denotes units of time into the past from the last taxon sampled.
In an epidemiological context, f(t) is the number of transmissions of a pathogen between hosts per unit time (the incidence of infection), and Y(t) is the number of infected hosts (the prevalence of infection). Inference of epidemiological parameters from a gene genealogy requires that the tree reflects history of transmission events between hosts. The validity of this comparison requires that several conditions are satisfied. I assume that each infected host corresponds to a single lineage in a gene genealogy of virus, which is a fair approximation if superinfection is rare. I further assume that the time of transmission corresponds to the potential time at which two lineages coalesce. This is a fair approximation if the intrahost coalescence time is short relative to the rate of epidemic dispersal. This is equivalent to the condition that each infected host corresponds to a single representative gene copy and the rate of replication of gene copies is equal to the rate of transmission between hosts.
For motivation, consider the simple example of an epidemic where the state of the system is described by the number X of susceptibles and the number Y of infected hosts, and the number of transmissions per unit time is f(t) = βX(t)Y(t). Infected individuals recover and gain permanent immunity at a rate γY. In this case, the population size Y(t) appears as the solution to the ordinary differential equations [the well-studied Kermack–McKendrick system (Kermack and McKendrick 1927)]:
| (1) |
| (2) |
A solution for Y(t) may be obtained by integrating these equations forward in time, and a record Y(s) can be kept of the size of the population s units in the past. Given the solution for Y(t), one might assume that the rate λ2(s) that two lineages coalesce s units in the past is which would be similar to the rate in the Kingman coalescent. This assumption underlies much recent work that attempts to correlate estimates of effective population size with the number of prevalent infected hosts. But, this is incorrect; the rate of coalescence depends not only on just the changing population size, but also on changing birth rates, which in turn affect the variance in the number of offspring per unit time.
In previous work (Volz et al. 2009; Frost and Volz 2010), we showed that the rate of coalescence for two extant lineages is the following function of birth rates and population size:
| (3) |
In the above example (Equation 1), we have A more rigorous derivation of Equation 3 than was given in Volz et al. (2009) is provided in Methods. Classical solutions, such as λ2(s) ∝ 1/Y(s), appear as special cases when births are strictly proportional to population size. The simple formula for λ2 is used as the point of departure for exploring the effects of complex population dynamics on genealogical structure. First, it is straightforward to investigate the implications of varying birth rates f(s). From this perspective, it is easy to see when and why skyline estimates of effective population size will be biased for the true population size, and this is explored in The effective number of infections. I revisit the problem of developing useful heuristics (Nee et al. 1995) to infer the mechanism of population growth from the distribution of node heights in a gene genealogy (The number of lineages through time). This solution, which holds within a single homogeneously mixing deme, can be relaxed to a situation with multiple demes and an arbitrarily complex pattern of birth, migration, and death, as described in Population dynamics and gene genealogies in structured populations. The coalescent in structured populations under birth–death processes does not appear to be a well-explored problem, and some solutions are presented that describe the coalescent in the presence of concurrent processes of birth and migration. Finally, a method for calculating the likelihood of a gene genealogy conditional on a history of f(s) and Y(s) is presented, as well as a simple method to simulate coalescent trees conditional on this history (Population dynamics and gene genealogies in structured populations).
The models developed herein should find similar applications to structured coalescent models used to estimate migration rates between demes (Beerli and Felsenstein 1999, 2001; Bahlo and Griffiths 2000; Bloomquist et al. 2010). Before proceeding, it is worthwhile to remark on how these methods differ from previous approaches. In our case, the coalescent may be used to estimate the parameters of commonly used epidemiological and ecological models of population dynamics, such as birth rates, both within and across demes. The models developed below differ from other structured models in several important respects:
The models allow for potentially large sample fractions.
Birth and migration rates may be arbitrary functions of the state of the system and time.
Gene copies may reproduce both within and across demes. Consequently, two gene copies in different demes may coalesce without being preceded by a migration event.
These models were developed with viral epidemics in mind and may be most appropriate for populations that change and evolve rapidly over short timescales. While no assumptions are made about the sample fraction, this comes with a cost: The population size is an extremely important parameter of the system, and the models presented herein will work best when there is good prior information about this quantity, such as reported incidence of infection from public health authorities.
Methods
In this section, the processes governing reproduction and death are made precise and Equation 3 is rederived from first principles. Multiple stochastic processes may generate the asymptotic dynamics of a deterministic system such as Equation 1, and the details of that process are key to understanding the behavior of the coalescent.
At time t there will be a homogeneous population of Y(t) gene copies. The rate that the entire population reproduces is the function f(t, Y) that is differentiable with respect to time and population size. The rate that a single gene copy reproduces (generates a single new copy) is f(t, Y)/Y(t). Similarly, the death rate is μ(Y, t), although this will not appear in the solutions for the coalescent. Birth and death events are asynchronous. This is similar to a continuous-time birth–death (BD) process, but with varying rates. BD processes have previously been studied from the perspective of the coalescent with constant rates (Stadler 2011). As noted by Harris (2002), the BD process does not seem to be a good model for the spread of an epidemic in a finite population, since when a large proportion of the population has been infected, we cannot suppose that the rate of new infections is independent of past history.
The processes considered here are different from the standard form of BD models, since the birth rate of a single gene copy (f(t, Y)/Y(t)) is both time and state dependent. While at a particular point in time, all gene copies reproduce and die at identical rates, the birth rate, and possibly the death rate, may depend on the state of the system. f(t, Y) may be any differentiable function of Y and t.
Given how the BD process is defined for gene copies, the quantity Y(t) is a density- and time-dependent Markov jump process. At rate f(t, Y), the number of gene copies Y(t) is incremented by one, and Y(t) is decremented at rate μ(t, Y). In the remainder of this section, Y(t) is assumed to be very large at all times, which will enable us to approximate Y(t) with a system of ordinary differential equations (ODEs). For epidemic models in particular, the relationship between Markov-jump processes with varying rates and ODEs was made precise in Kurtz (1981). In the limit of large population size, the ODE approximation becomes exact.
Although this model differs from the model of Moran (births and deaths are not contemporaneous), it leads to similar expressions in terms of the variance in the number of offspring and the probability of a coalescence following a birth event. The connection to Moran is made precise below.
The rate of coalescence
The cumulative number of births prior to time s on the reverse time axis is
| (4) |
Denote the population size when the jth birth happens (retrospectively) as , and is the variance of offspring at the jth birth event. It follows that the cumulative hazard of coalescence is
| (5) |
as with the Cannings-type models, but summing over individual birth events. Following the jth birth there is probability 2/(Y(j) + 1) of either having replicated or being the new copy, so the number of offspring ν(j) is
| (6) |
It follows that is the variance in the number of offspring for a single birth event, which is the same as for a Moran model of constant size . It follows that
| (7) |
At this point, we could rescale time to units of ΔF births, and we would retrieve the Kingman coalescent. Alternatively, I modify the coalescent rate to take the nonconstant birth rates into account. By breaking the interval t into units of equal duration h, we have
| (8) |
where υ(k) is the average of over the kth interval,
| (9) |
and ΔkF = F(kh) – F((k – 1)h). Similarly, I define the cumulative function
| (10) |
so that υ(k) = (ϒ(F(kh)) − ϒ(F((k − 1)h)))/ΔkF. Using Taylor expansion of υ(k), I rewrite Equation 8:
| (11) |
| (12) |
Multiplying and dividing by h and taking the limit as h goes to zero gives
| (13) |
| (14) |
where I have also used the fact that O(ΔkF) = O(h), assuming F is continuous and h << 1. The integrand is the solution for the rate of coalescence.
The above calculation can be repeated if there are A extant lineages, which yields the coalescent rate
| (15) |
This rate is correct even if A is very large, and this model should accomodate large sample fractions. To see this, note that the ratio can be interpreted as the probability of picking two extant lineages if selecting two gene copies from the total population. This probability tends to one as A → Y, in which case the rate of coalescence is simply the rate of births. Furthermore, in the continuous birth–death model, we need not worry about multiple mergers (Fu 2006), as there is zero probability of simultaneous events.
The effective number of infections:
It is common to compare estimates of the effective population size Ne, such as the Bayesian skyline plot (BSP) (Drummond et al. 2005), to the number of infected individuals (Frost and Volz 2010). For the remainder of this article, Ne refers to the coalescent effective size (Sjödin et al. 2005; Wakeley and Sargsyan 2009). I operationalize this definition somewhat differently from in previous work: Where the coalescent rate is known under our model, I define Ne = 1/λ2. I distinguish Ne from the true size Y(t), under the given model. The skyline estimators are unbiased for the harmonic mean of Ne within each internode interval of the genealogy (Pybus et al. 2000).
Comparison of Ne to Y is typically based on a coalescent rate that is valid when birth rates are constant. In this section, I explore the relationship between and λ2 = 2f/Y2. In some situations there is close correspondence between λ2 and , while in other cases the correspondence can be very poor, and this can lead estimates of Ne to be very biased for the true population size. Furthermore, even when there is a linear relationship between λ2 and , it can be a complex task to derive the correct scale of proportionality. The main conclusion of this section is that there is a poor correspondence between λ2 and when the birth rate (i.e., incidence of infection) is not strictly proportional to the population size (i.e., the prevalence of infection).
As a motivating example, consider the following model that describes the prevalence of infection over time for an epidemic where infected individuals have nonpermanent immunity and recover to the susceptible state:
| (16) |
X, Y, and Z are, respectively, the numbers of susceptible, infectious, and recovered individuals in the population. Recovered individuals lose immunity at the rate η. The way that incidence is modeled allows us to explore the relationship between birth rates and the rate of coalescence. Incidence of infection is proportional to Y1+α. The standard susceptible-infected-recovered (SIR) model is recovered if α = 0. The purpose of this model is not to provide a realistic description of dynamics for a particular disease, but to illustrate the relationship between the population size Y and the rate of coalescence λ as the scaling factor α is changed. Nevertheless, there is good reason to believe that real systems often deviate from simple mass action dynamics, and this toy model is a special case of those investigated in Liu et al. (1987). From Equation 3,
| (17) |
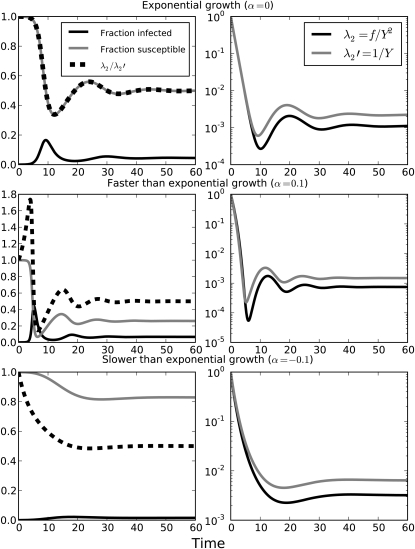
When α = 0, λ2 has a similar functional form to and is different by a factor of βX/N. When α > 0, the growth rate of the epidemic accelerates early on. This type of growth is termed faster than exponential (FTE). And when α < 0, the growth rate decelerates. This type of growth is termed slower than exponential (STE). The population size where α = 0 resembles exponential growth early on, but this growth is transitory.
The relationship between λ2 and is readily understood by examination of the ratio
| (18) |
Early in the epidemic X ≈ N. And if α = 0, the ratio and the correspondence between the coalescence rates is linear. In this situation, may be a good approximation early in the epidemic. However, if α ≠ 0, the ratio depends on Y. If growth is FTE, the ratio is increasing, and if growth is STE, the ratio is decreasing.
This is illustrated in Figure 1, where three solutions of Equation 16 are shown for α = 0, , and . The coalescent rates are normalized so that the maximum is unity. The ratio of the normalized coalescent rates is shown in the left column. When α = 0, this ratio (after normalization) simply corresponds to the susceptible fraction of the population. There is a brief transient where the rates are quite similar, and since most coalescent events will take place during exponential growth, may be a good approximation for the α = 0 case. But in the FTE and STE cases, the correspondence is very fleeting and can be made arbitrarily bad by increasing the exponent α.
Figure 1.
(Left) The fraction of the population susceptible and infected is shown over time for model (16). (Right) The rates of coalescence λ2 = f/Y2 and . In all solutions to Equation 16, N = 104, β = 2, γ = 1, η = . The incidence scaling factor α is varied for each row: α = 0 (top), α = (middle), and α = – (bottom).
The number of lineages through time:
The number of lineages as a function of time (NLFT) is informative of historical population dynamics (Pybus and Rambaut 2009). A variety of approaches have been developed to infer population size from the NLFT. Qualitative conclusions are readily drawn from plots of the NLFT. If the NLFT drops close to the root of the tree, it is considered a signature of exponential growth, whereas if the NLFT decreases close to the time of sampling, the population is usually considered to have a constant or decreasing size (Grenfell et al. 2004). A problem that is apparent from the approach developed in the preceding sections is that the NLFT is sensitive to the history of birth rates f(t), not just the population size Y(t). Consequently, it is possible to contrive situations where the NLFT has a counterintuitive relationship with the true population dynamics. As demonstrated in this section, star-like trees are not necessarily an indication of a rapidly expanding population, and populations that are growing faster than exponentially do not necessarily produce star-like trees.
The NLFT at time s in the past is denoted A(s), and the set of lineages at time s is (s). As discussed in Frost and Volz (2010), a useful deterministic approximation to the NLFT is
| (19) |
This approximation becomes exact in the limit of large sample size n = A(0). For the remainder of this section, I consider the pure birth process with . The coalescent rate λ2 will be the same as for the FTE and STE models early in the epidemic (Equation 17), where X/N ≈ 1. We have
| (20) |
where the latter approximation is valid when the number of lineages is large.
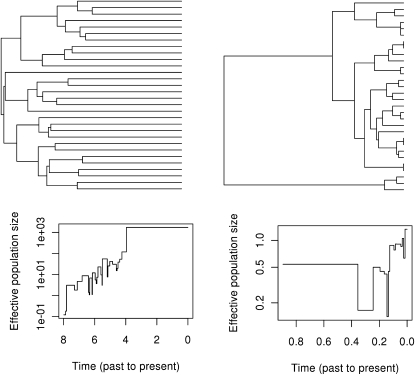
Figure 2 shows simulated genealogies for the pure-birth FTE and exponential growth model: . In the FTE case, α = 1 and in the exponential growth case, α = 0. β = 1 in both cases. Trees were generated using a chain-binomial simulation as described in supporting information, File S1. Samples of 30 taxa were taken during a period of growth when the population reached Y = 2 × 104.
Figure 2.
Simulated genealogies (top) and corresponding skyline estimates of Ne (bottom) for exponential growth (left) and FTE growth (right). Simulations were of a pure-birth process with monotonically increasing population sizes. Samples of 30 taxa were taken during a period of growth (either exponential or FTE) at the point when a population size of Y = 2 × 104 was reached. In the exponential case, the skyline is unbiased for the harmonic mean of Y/2β within each interval. In the FTE case, the skyline underestimates population size.
The coalescence rate in an FTE population is highest close to the present, and most coalescent events will happen close to the present. Somewhat counterintuitively, FTE growth produces trees such that node heights are concentrated toward the tips of the tree and are qualitatively similar to those in trees produced by populations of constant size. This is contrary to the expectation that a star-like tree will be generated by a rapidly growing population. Trees that are more star-like than the exponential case are actually generated by STE.
The FTE and STE models provide a simple illustration of how estimates of effective population size can be biased in models where the birth rate is not strictly proportional to population size. The skyline estimate of effective population size (Pybus et al. 2000) is premised on the duration of internode intervals being proportional to . The estimated effective population size during the interval of duration Δs when there are A lineages is
| (21) |
This estimator is unbiased for the harmonic mean of the population size during each interval, provided the coalescent rate is valid (Pybus et al. 2000). But the skyline will generally underestimate population size in the FTE case because for a given population size; and the skyline will overestimate population size in the STE case because , as illustrated in Figure 2. Equation 17 makes it clear that for this pure birth model, will actually be estimating the harmonic mean of Y1–α/2β within each internode interval. The skyline will have a linear relationship with Y when α = 0; the correct scale is 1/2β (Frost and Volz 2010). When α > 1, and the rate of growth is accelerating rapidly, the skyline will erroneously predict a decreasing population size; however, such large values of the exponent are unlikely to be seen in reality.
Simple heuristics have been developed for detecting exponential growth, FTE, and constant size (Nee et al. 1995) on the basis of plots of the NLFT. An alternative heuristic is provided here and compared to existing heuristics. In the particular case of exponential growth (α = 0), the solution of Equation 20 is
| (22) |
We define the inverse function m(A) by solving the preceding equation for s, which yields
| (23) |
The function m(A) has been called the “epidemic transformation” of the NLFT (Nee et al. 1995), and when the plot of m(A(s)) vs. s appears linear, this indicates that the population growth is exponential. Our solution for m(A) has a similar functional form but is slightly different from that provided in Nee et al. (1995), which was based on the distribution of internode intervals:
| (24) |
| (25) |
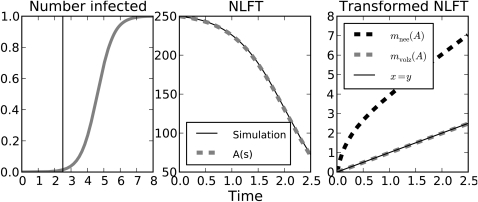
The latter approximation is a good one when Y(0) is very large. These alternative transformations are compared in Figure 3 for a sample taken during the early stages of an susceptible-infected (SI) epidemic for which growth is approximately exponential. When Y(0) and β are known, Equation 23 is exact. However, when testing for exponential growth, Y(0) is often not known. If Y(0) is assumed to be large, Equations 24 and 23 will both appear linear aside from a brief transient for small s. To see this, consider the time derivative of the epidemically transformed NLFT,
| (26) |
and substituting Equations 20 (with α = 0) and 22, this gives
| (27) |
So the growth of mn asymptotically becomes linear with slope β. Therefore, Equations 22 and 23 provide an alternative justification for the approximation (24).
Figure 3.
(Left) The normalized prevalence of infection in an SI epidemic. . The vertical line indicates the point where a sample of n = 250 is collected. (Center) The number of lineages through time. The dashed line shows the solution to Equation 22. (Right) The transformed number of lineages through time.
Population dynamics and gene genealogies in structured populations
The derivations of the preceding section provided the coalescent rate for a population such that each gene copy has equal potential to reproduce. More generally, we consider the case where a gene copy may occupy any of m discrete states that may influence both birth and death rates. These states may represent heterogeneity stemming from a variety of factors, such as spatial and other population structures. In the case of infectious diseases, states may reflect the different properties of hosts in which the pathogen resides. For example, infected hosts of different age, behavior, or clinical stages of infection may provide different potential for a virus to spread to new hosts.
Formally, the models of population dynamics (prospectively) assume that birth rates are deterministic and specified by a time-dependent matrix F(t). There are m states and the indexes k and l always refer to one of these states. In contrast to most island models in population genetics, it is possible for birth events to cross demes. For example, a gene copy in state k may generate a new copy in state l. The rate at which this occurs is the element fkl(t) of the matrix F(t).
To accommodate a larger range of population dynamic models of epidemiological and ecological interest, these models must also include migration of gene copies between states that are independent of reproduction. The matrix G(t) with elements gkl(t) specifies the time-dependent deterministic rate at which a gene copy in state k migrates to state l.
The matrices F and G uniquely specify the vector of population sizes in each state Y(t) along with initial conditions Y(0). Additionally we model exogenous births and deaths in state k with the functions ηk(t) and μk(t), respectively. These birth and death terms are necessary for some models, but as will be shown have no direct effect on the rate of coalescence. The population size Y is the solution of m ordinary differential equations of the form
| (28) |
To motivate this framework, it is shown how to decompose two simple epidemiological models into processes involving births F(t) and migrations G(t): an epidemic such that infected hosts progress through two stages of infection and an epidemic spreading in a host population with age structure.
An epidemic with two stages of infection:
Variations on this model can be useful for epidemics such as HIV, where the transmission probability per contact changes dramatically over the course of infection (Longini et al. 1989). Upon infection, hosts enter the state ℐ1 of average duration 1/γ1 and transmit infection at the rate β1. Infected hosts in ℐ1 progress to state ℐ2 of average duration 1/γ2 with transmission rate β2. Hosts then progress to a recovered state and no longer transmit. There is no birth or natural mortality. The numbers of hosts that are susceptible in state 1 and state 2 are denoted, respectively, S, I1 and I2. The equations that describe this system are as follows:
| (29) |
| (30) |
The birth and migration rates for this model are
| (31) |
An epidemic in an age-structured host population:
For many infectious diseases such as influenza, juvenile hosts have higher contact rates and are more susceptible to infection. In this model we divide the population into juvenile (S1 susceptible and I1 infectious) and adult states (S2 susceptible and I2 infectious), each of which has distinct transmission rates within and between states. For example, the rate that adults transmit to juveniles is β21. Juveniles and adults recover from infection at the same rate γ. The equations that describe this system are as follows:
| (32) |
We assume that the rate of epidemic dispersal is very fast relative to the rate that hosts age, so there is zero migration from the juvenile to the adult states. There is no birth or natural mortality. The birth and migration rates for this model are
| (33) |
The structured coalescent:
In developing the structured coalescent model, the number of taxa sampled at time s in state k is nk(s), and the number of lineages in state k at time s in the past is Ak(s). The indexes i and j always refer to a lineage in the genealogy. A(s) denotes the total NLFT.
The coalescent model developed here does not assume homochronicity of sampling. In general, each taxon may be sampled at a distinct time point. Furthermore, the rate of coalescence depends on the state of the taxa when they are sampled. Therefore it is possible for each branch of the tree to coalesce at a rate distinct from all other branches, and the rate of coalescence between a given pair of branches may be distinct from all other pairs of branches. The goal of this section is to develop a master equation for the rate of coalescence. The solution should be of sufficient generality to capture the rates of coalescence between all pairs of branches as a function of time-dependent births, migrations, and population size.
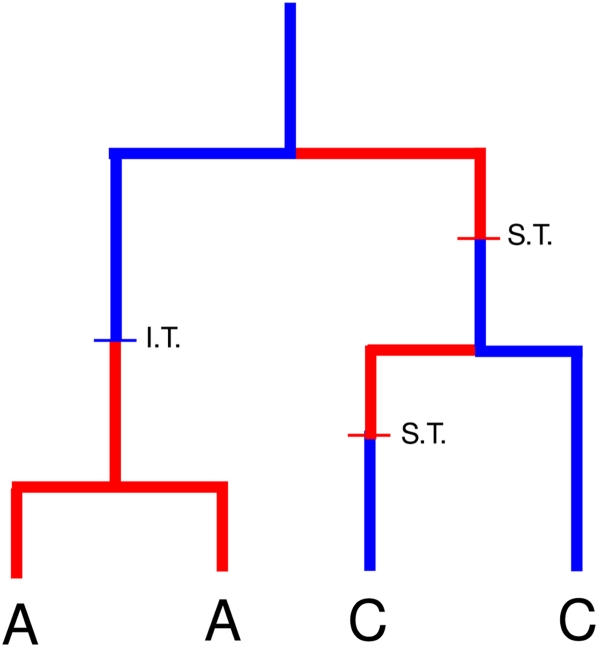
As a motivating example, consider the two-stage epidemic model (Equation 29). Figure 4 shows a gene genealogy that might be generated by this process. The red branches represent infected hosts in the first stage of infection, and blue branches represent hosts in the second stage. Our goal is a mathematical description of the probability that a branch occupies each state at some time s in the past and the rate at which a given pair of branches coalesce. Moving upward in the tree (backward in time), four events can occur in this model:
Two red branches can coalesce, representing transmission by a stage-1 infection.
A red and a blue branch can coalesce, representing transmission by a stage-2 infection.
A blue branch can become red, representing stage transition from the early to the late stage.
A red branch can become blue, representing transmission by a stage-2 infected that is not ancestral to the sample; these are subsequently called “invisible transmission” events.
The fourth event is important to include and easy to forget. When a stage-2 infected transmits and initiates a line of descent that is eventually sampled, but has no other extant progeny in the sample, it will not be manifested in a genealogy as a coalescence, but rather as the branch changing state from a stage-1 host to the transmitting stage-2 host. Note that two blue branches never coalesce since there are no birth events between blue lineages. The coalescent model is specified by the rates that these four events happen, and these rates are straightforward to calculate given prior knowledge of F(s) and G(s).
Figure 4.
An example gene genealogy that could be generated by the HIV model (Equation 29). Red branches correspond to stage-1 infected hosts. Blue branches correspond to stage 2.
Suppose that at time s we have two lineages i and j. The probability that lineages i and j are in state 1 are, respectively, pi1 and pj1. And the probabilities of being state 2 are, respectively, pi2 and pj2. Given that a transmission event by a stage-1 infected occurs, what is the probability of observing coalescence between i and j? Such transmissions occur at the rate f11(s). The probability that i or j transmitted is pi1/Y1 or pj1/Y1. The probabilities that i or j became infected from this event are the same: pi1/Y1 and pj1/Y1. The probability that either i or j transmitted and either i or j was infected by this event is
| (34) |
Extrapolating to the entire tree, the probability that two stage-1 lineages coalesce is .
Now suppose a stage-2 unit transmitted. This happens at rate f21(s). The probability of this resulting in a coalescence between i and j is found by considering the probabilities that i or j transmitted or was infected. Note that the lineage that is infected must be in the first state, and the lineage that transmitted is in the second state by assumption:
| (35) |
Extrapolating to the entire tree, the probability that a coalescence occurs between lineages in states 1 and 2 is .
The probability of observing an invisible transmission event (the fourth type of event) is more complex and depends on the probability that the transmitting host is not ancestral to the sample. The probability that i changes state from 1 to 2 is then the probability that i is type 1, times the probability that the transmitting host is not among the j ≠ i lineages:
| (36) |
Extrapolating to the entire tree, the probability that a lineage ancestral to the sample changes state from 1 to 2 is .
Handling migration events (that is, “stage transitions”) is more straightforward. Given that a stage transition occurs, which takes place at the rate g12(s), the probability that lineage i changes state from 2 to 1 is pi2/Y2. The probability that a lineage ancestral to the sample changes state from 2 to 1 is A2/Y2.
Using these derived probabilities and the given rates at which birth and migration events occur, the rate that lineages of type 1 are lost to coalescence is
| (37) |
The first term can be recognized as the rate of coalesce from Equation 1. Furthermore, a convenient deterministic approximation to the NLFT is available when the sample size is large:
| (38) |
| (39) |
The initial conditions of these equations may be based on the number of infected sampled in states 1 and 2. These equations could be used for inference and model fitting. In Volz et al. (2009) a deterministic model similar to this one was fitted to a phylogeny of HIV-1 genetic sequences by comparing the observed distribution of internode intervals to the theoretical expectation with homochronous sampling,
| (40) |
where ti is the time of the ith coalescent event (or “node height”), n is the sample size, and A(t) = A1(t) + A2(t). And this distribution is easily generalized to heterochronous sampling by integrating a different set of equations like (38) between each sample time. While the use of this approximation is computationally efficient, it does not use all information available in the tree. The distribution of node heights contains information about the demographic history, as does the topology of the tree. For example, if we observe that taxa in state 1 coalesce with one another at a different rate than with taxa in state 2, that provides information about the relative transmission rates of β1 and β2. This issue is explored in greater detail in the next section for models with generalized structure.
The coalescent for populations with generalized structure:
The derivations of the preceding section for a structured population with two states can be extended to models with an arbitrary number of states and arbitrary structure. More generally, is the rate of coalescence between lineages i and j. The state of each branch is governed by the variables pik(s), which is the probability that branch i is in state k at time s in the past. The function S(i) ∈ (1, m) returns the state of lineage i, which is usually unknown. Note that the number of lineages in state k is . With the understanding that all state variables and rates are time dependent, the variable s is dropped from future expressions.
Given that a birth event from state k to state l occurs, the probability that lineages i and j coalesce is . The rate that such births occur is fkl, and summing over all combinations of states k and l yields the rate that i and j coalesce:
| (41) |
The rates of coalescence depend on the state vectors , and these change going backward in time as a function of the numbers of migration and invisible birth/transmission events:
Migrations from state k to l at rate gkl cause the state to change l → k with probability pil/Yl.
Migrations from state l to k at rate glk cause the state to change k → l with probability pik/Yk.
Births from state k to l at rate fkl cause the state to change l → k with probability .
Births from state l to k at rate flk cause the state to change k → l with probability .
Putting these terms together yields the master equation for the state of branch i:
| (42) |
Following a coalescent event, the state of the new branch α depends on the state of the daughter lineage i or j when it reproduced. The rate that either i generates j or j generates i while in state k is . Summing over all states l and normalizing by the total coalescent rate between i and j yields
| (43) |
An additional subtlety arises for small populations in which it may not be presumed that the state of a lineage not involved in the coalescent α′ ≠ i, j, α is independent of the state of the new lineage α. For example, if F(tα) is such that the reproducing unit is likely in state k, it is correspondingly less likely that a lineage not involved in the coalescent event is in state k. Adjusting the probabilities pα′k requires a small adjustment that approaches zero in the limit of large population size.
The size of this adjustment can be found by application of Bayes’ rule. Denote by p′α′k the probability that α′ is in state k conditional on a particular coalescent event occurring between lineages i and j. First suppose that the reproducing unit was in state k so that S(α) = k. We wish to calculate the probability that α′ is in state k conditional on not reproducing at time tα. This is Pr{α′ not transmitting at tα | S(α′) = k, S(α) = k} times the prior pα′k divided by the probability of not reproducing, Pr{α′ not transmitting at tα | S(α) = k}. We have
| (44) |
| (45) |
It follows that
| (46) |
| (47) |
Now suppose the reproducing unit was in state l ≠ k and S(α) = l.
| (48) |
It follows that
| (49) |
Now integrating over the state of the reproducing unit α with probability mass pαk, we have
| (50) |
Having defined the general framework for modeling the rate of coalescence and the dynamics of branch states, two problems are now open to investigation: simulating coalescent trees and calculating the likelihood of a gene genealogy.
Simulating coalescent trees conditional on complex demographic history:
The total rate that coalescent events occur is denoted (note that and is counted twice in this summation). Similarly, I define to be the rate at which lineage i changes state, and the cumulative rate . The rate that lineage i changes state from k to l is denoted . From the discussion of the preceding section, we have
| (51) |
In the simulations, we do not use a probabilistic description pik for the state of a lineage (although this could be done). Rather, we initialize the state of a lineage at the time of sampling and update the state in a discrete manner, so pik = 0 or 1 at all times.
To simulate the coalescent, we begin at the time s = 0 when the first taxon (retrospectively) is sampled.
At time s, the putative time until the next event se, which may be a coalescent or a change of state, is drawn from a distribution with cumulative distribution function
| (52) |
Denote by a Δs a number generated from this distribution. There are two possibilities:
If Δs is greater than the next sample time, then assign s smaller than the next sample time and add the new lineages to the tree.
Otherwise, assign s ← s + Δs. Then modify the tree with a coalescent or state change.
Select which event occurs with probability proportional to the rates
If a coalescent happens, assign the state to the new lineage selected using Equation 43. Given that i and j coalesce, the state of the new lineage α is selected to be in state k with probability proportional to
After a new lineage is sampled, or a coalescent event occurs, or a state change occurs, a new number is generated from the distribution (52) and the process is repeated.
The likelihood of a gene genealogy conditional on complex demographic history:
The set of coalescent events of a given gene genealogy consists of tuples (i, j, sα): the two lineages that coalesce and the time sα in the past when the event occurred. The probability of observing depends on the rate that each pair of branches coalesce and on the probability that lineages have not coalesced prior to sα. I denote this latter quantity θ(s).
An unbiased calculation of the likelihood of a genealogy given a demographic history can be obtained using the method in Beerli and Felsenstein (1999). Consider an internode interval between times s0 and s. The probability that a coalescent has not occurred by time s is
| (53) |
where is the total rate at which the extant lineages coalesce; note that and is counted twice in the summand. The duration of an internode interval has density . When a coalescent occurs, the probability that it is with lineages i and j is . The probability of i and j coalescing after interval s is
| (54) |
The likelihood is then the product of the probabilities qij(s) after each internode interval:
| (55) |
Regarding the computational efficiency of calculating the likelihood, it requires the solution of m ODEs to describe pik for each of 2(n – 1) branches of the tree; it also requires the solution of 2(n – 1) ODEs for the survivor functions θi of each branch. It is possible to improve on this and reduce the calculation to a system of m2 + m equations in each internode interval that does not depend on the sample size.
The first simplification is to solve for the dynamics of the total number of ancestors of each type, Ak, rather than the state of each lineage. The following equation describes the dynamics of Ak within each internode interval conditional on no coalescent events occurring within that interval:
| (56) |
The second simplification consists of solving for m representative state vectors in each internode interval. At the beginning of the interval, each vector is initialized with unit mass on the κth element for κ = 1, ⋅ ⋅ ⋅, m. The dynamics of the kth element of each state vector are found as above:
| (57) |
Given the solutions for Ak and , the state of each lineage can be calculated at the end of the internode interval that begins at time s0 and ends at time s1:
| (58) |
This can also be written as the dot product
| (59) |
where Q is an m × m matrix with the κth column equal to .
Results and Discussion
Computational experiments have been carried out to corroborate the derivations of the preceding sections and to provide examples of how the structured coalescent model may be used to explore system behavior and for inference. Corroboration of the coalescent model is obtained by event-driven simulation of large populations while maintaining a record of who begot whom. The simulation methods are described in File S1.
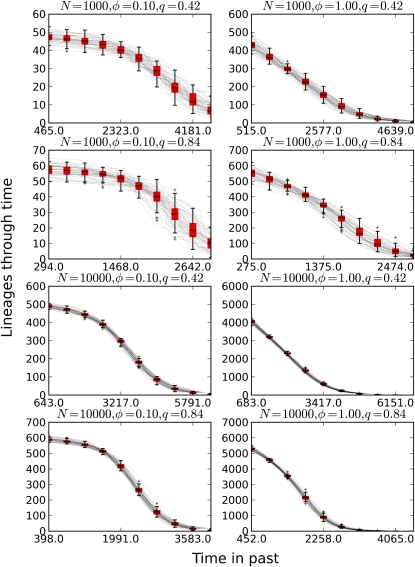
Figure 5 shows the NLFT for coalescent trees generated as described in Simulating coalescent trees conditional on complex demographic history and forward-time simulations. Replication and migration were governed by the HIV model as described in Population dynamics and gene genealogies in structured populations. Fifty forward-time simulations were conducted and were based on N = 104 reproducing units. The coalescent simulator used birth and migration rates generated by the deterministic HIV model (Equation 29). Sampling was conducted when a specified cumulative number of transmissions had taken place. The coalescent trees were compared for a range of sample fractions and transmission rates and were found to faithfully reproduce the NLFT of the forward-time simulations in all cases using a fraction of the computational resources. Of interest, the NLFT is highly sensitive to not only the population size (or prevalence of infection), but also the relative transmission rates in the first and second stages of infection. All epidemic scenarios in Figure 5 have similar population dynamics and identical , but vary by the fraction of transmissions attributable to the first stage.
Figure 5.
A comparison of the NLFT generated by the coalescent simulations and forward-time event-driven simulations (box plot). Fifty simulations were conducted using both methods. Dynamics were governed by the HIV model in Population dynamics and gene genealogies in structured populations. γ1 = 1/365, γ2 = 1/3650. Homochronous samples were collected at the time such that N cumulative transmissions had occurred. System behavior is largely controlled by the ratio q = (β1γ2)/(β2γ1) of the stage-1 reproduction number to the stage-2 reproduction number, and two values of this ratio are compared in the simulations. Two population sizes (N = 103 and 104) are compared. Two sample sizes are compared: a small sample of 10% and a census of 100% of extant infections.
An additional set of experiments was carried out to corroborate the likelihood function (55). To demonstrate the generality of this solution, a set of complex structured models was sought, and a method was employed to generate a model with an arbitrary number of states and random patterns of births and migrations. Births take place according to the logistic function. Individuals in state k beget units in state l at rate . Migrations take place by the law of independent action. Units in state k move to l at rate γklY. The matrices that specify βkl and γkl are generated by randomly choosing a tunable number d of m2 elements of both matrices to have a normally distributed value. All other rates are zero. There are also small exogenous birth and death terms [ηk(t) and μk(t) from Equation 28] to prevent any population from going to zero.
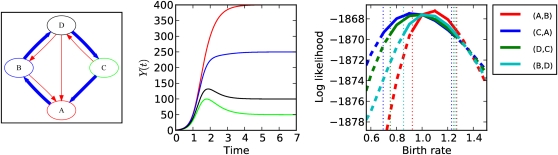
Figure 6 shows the results from a model with m = 5 states, seven migration rates, and four birth rates, with N = 5 × 103 and a sample fraction ϕ = 30%. The potential to estimate each of the four birth rates is evaluated using a likelihood profile. To generate a genealogy, a coalescent tree was simulated using the methods of the preceding section and using birth and migration rates from a deterministic ODE model. Each birth rate was perturbed by ±50%, holding all other parameters constant, and the likelihood of the coalescent tree was calculated. In all cases, the maximum likelihood occurs at or near the true birth rate. From this profile, a 95% confidence interval was calculated on the basis of a difference in likelihood of 1.92 log units. In this experiment, the interval covers the true value in all cases. A multitude of similar experiments with different compartmental models and sample fractions are presented in File S1.
Figure 6.
(Left) A directed graph representing the model structure with m = 5 states, four birth terms, and seven migration terms. Blue arrows represent logistic birth terms. Red arrows represent migration between states. (Center) The population size Yk over time for each of five states. (Right) Likelihood profiles for each of four birth rates and 95% confidence intervals. To generate the profiles, the birth rates were perturbed from the true value by the factor of parameter expansion.
An additional set of experiments was conducted using 35 genealogies simulated using the process described in File S1 and the same model as above with five states and four birth rates. Likelihood profiles and confidence bounds were calculated for each genealogy and each of four birth rates. The carrying capacity of the population was set at N = 5000 and a single homochronous sample was taken after 5N birth events had occurred. The simulations were initialized far out of equilibrium with a single individual in each of m states. The sample fraction was 20% of extant individuals (780 total) and sampling was random without replacement. Across the 35 replicates, I find that the maximum likelihood is unbiased (mean error <1.3%) and the confidence intervals based on likelihood profiles had good coverage of the true value (74.3%).
These computational results suggest that it may be feasible to use multiple sequence alignments as an additional source of data when estimating the parameters of complex demographic models. But these experiments have explored only error generated by the birth–death process itself and not error due to the measurement process or in the inference of the tree topology and branch lengths. The branch lengths and topology of a gene genealogy are never known with certainty, and these quantities must be estimated at the same time as the parameters of a demographic model. This article has addressed only the problem of calculating the likelihood of a gene genealogy, but estimation from genetic data will require incorporation of this likelihood into algorithms for sampling genealogies. Recently developed tools have made it increasingly convenient to conduct such inference for simple demographic models within a Bayesian MCMC framework (Drummond et al. 2002; Beerli 2006; Drummond and Rambaut 2007; Kuhner and Smith 2007; Beerli and Palczewski 2010; Hey 2010). For complex demographic models, which may include many states and more than a dozen parameters, much more care is required for the design of proposal distributions for efficient MCMC and for incorporating detailed prior information about population size and rates.
When fitting complex models with many free parameters, the likelihood calculation proposed here may be most useful when there is abundant prior information about population size and birth rates as well as a large sample of genetic data. The HIV epidemic provides a good example where these methods may find immediate application. In developed countries, genotyping HIV is now standard for drug-resistance testing, and these data are sufficient for phylogenetic analyses despite composing only ∼10% of the genome (Lewis et al. 2008). The study of HIV epidemiological dynamics may also benefit from generally good case reporting, prior estimates of incidence and prevalence (Hall et al. 2008), and scores of studies that have estimated parameters describing the natural history of infection (Longini et al. 1989). Where population size over time is already known with high precision, models using genetic data may be used to estimate parameters that are hard to discern from standard data sources. For example, there is much debate (Yerly et al. 2001; Pao et al. 2005; Lewis et al. 2008; Cohen et al. 2011) about the relative contribution of transmissions during the early/acute stage of HIV infection to total incidence. In this case, a complex model of HIV transmission that features transmission during multiple stages of infection could be fitted to genetic data while making use of highly precise prior information about historic incidence and prevalence.
While deficiencies of the skyline estimate have been discussed regarding estimation of epidemic prevalence, it is important to remember that model misspecification can still be a large source of bias. The SIR models discussed above are also simplifications of reality and are subject to inductive bias. It is hoped that these models may be sufficiently complex and incorporate enough prior information to obtain nearly unbiased estimates of the unkown population size. Ultimately, this work may contribute to the integration of phylogenetic inference over short timescales with the vast and growing literature on mathematical modeling of ecological and epidemiological population dynamics (Bailey 1975; Anderson and May 1991). These models could likely be incorporated into recent statistical approaches that include stochastic effects (Rasmussen et al. 2011). Such an integration would be much harder to achieve with conventional estimates of effective population size, which can correlate poorly with the true population size when birth rates vary through time.
Supplementary Material
Acknowledgments
Participants of the National Institutes of Health (NIH) Research and Policy for Infectious Disease Dynamics workshop on Phylodynamics (May 2011, Durham, NC) provided valuable feedback. The author thanks Katia Koelle, David Rasmussen, Ed Ionides, James Koopman, and Peter Beerli for valuable comments. The author acknowledges support from NIH grants 1-K01-AI-091440-01 and R01-AI078752.
Literature Cited
- Anderson R. M., May R. M., 1991. Infectious Diseases of Humans: Dynamics and Control. Oxford University Press, New York [Google Scholar]
- Bahlo M., Griffiths R., 2000. Inference from gene trees in a subdivided population. Theor. Popul. Biol. 57: 79–95 [DOI] [PubMed] [Google Scholar]
- Bailey N. T. J., 1975. The Mathematical Theory of Infectious Diseases and Its Applications. Griffin, London [Google Scholar]
- Bataille A., van der Meer F., Stegeman A., Koch G., 2011. Evolutionary analysis of inter-farm transmission dynamics in a highly pathogenic avian influenza epidemic. PLoS Pathog. 7: e1002094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford T., Cobey S., Beerli P., Pascual M., Ferguson N., 2010. Global migration dynamics underlie evolution and persistence of human influenza a (h3n2). PLoS Pathog. 6: 1220–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P., 2006. Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 22: 341. [DOI] [PubMed] [Google Scholar]
- Beerli P., Felsenstein J., 1999. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics 152: 763–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P., Felsenstein J., 2001. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc. Natl. Acad. Sci. USA 98: 4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P., Palczewski M., 2010. Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics 185: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek R., Real L., 2010. The landscape genetics of infectious disease emergence and spread. Mol. Ecol. 19: 3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomquist E. W., Lemey P., Suchard M. A., 2010. Three roads diverged? Routes to phylogeographic inference. Trends Ecol. Evol. 25: 626–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M., Shaw G., McMichael A., Haynes B., 2011. Acute hiv-1 infection. N. Engl. J. Med. 364: 1943–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A., Rambaut A., 2007. Beast: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A., Nicholls G., Rodrigo A., Solomon W., 2002. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 161: 1307–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A., Rambaut A., Shapiro B., Pybus O., 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22: 1185. [DOI] [PubMed] [Google Scholar]
- Frost S., Volz E., 2010. Viral phylodynamics and the search for an ‘effective number of infections’. Philos. Trans. R. Soc. B Biol. Sci. 365: 1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., 2006. Exact coalescent for the Wright–Fisher model. Theor. Popul. Biol. 69: 385–394 [DOI] [PubMed] [Google Scholar]
- Gordo I., Gomes M., Reis D., Campos P., 2009. Genetic diversity in the sir model of pathogen evolution. PLoS ONE 4: e4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenfell B. T., Pybus O. G., Gog J. R., Wood J. L. N., Daly J. M., et al. , 2004. Unifying the epidemiological and evolutionary eynamics of pathogens. Science 303: 327. [DOI] [PubMed] [Google Scholar]
- Hall H., Song R., Rhodes P., Prejean J., An Q., et al. 2008. Estimation of HIV incidence in the United States. JAMA 300: 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T., 2002. The Theory of Branching Processes. Dover, New York [Google Scholar]
- Hey J., 2010. Isolation with migration models for more than two populations. Mol. Biol. Evol. 27: 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, M., F. Brun-Vezinet, R. D’aquila, S. Hammer, V. Johnson et al., 2000 Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an international AIDS society-USA panel. JAMA 283: 2417–2426. [DOI] [PubMed]
- Kenah E., Lipsitch M., Robins J., 2008. Generation interval contraction and epidemic data analysis. Math. Biosci. 213: 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermack, W. O., and A. G. McKendrick, 1927 A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. Ser. A 115: 700–721.
- Koelle, K., O. Ratmann, D. A. Rasmussen, V. Pasour, and J. Mattingly, 2011 A dimensionless number for understanding the evolutionary dynamics of antigenically variable RNA viruses. Proc. R. Soc. Ser. B. 278: 3723. [DOI] [PMC free article] [PubMed]
- Kretzschmar, M., M. G. Gomes, R. A. Coutinho, and J. S. Koopman, 2010 Unlocking pathogen genotyping information for public health by mathematical modeling. Trends Microbiol. 18: 406–412. [DOI] [PubMed]
- Kuhner M., Smith L., 2007. Comparing likelihood and Bayesian coalescent estimation of population parameters. Genetics 175: 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz, T., 1981 Approximation of Population Processes, Regional Conference Series in Applied Mathematics (Nos. 36–40). Society for Industrial Mathematics, Philadelphia.
- Lewis F., Hughes G. J., Rambaut A., Pozniak A., Leigh Brown A. J., 2008. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 5: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Hethcote H., Levin S., 1987. Dynamical behavior of epidemiological models with nonlinear incidence rates. J. Math. Biol. 25: 359–380 [DOI] [PubMed] [Google Scholar]
- Longini I., Jr, Clark W., Byers R., Ward J., Darrow W., et al. , 1989. Statistical analysis of the stages of HIV infection using a Markov model. Stat. Med. 8: 831–843 [DOI] [PubMed] [Google Scholar]
- Nee S., Holmes E., Rambaut A., Harvey P., 1995. Inferring population history from molecular phylogenies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 349: 25–31 [DOI] [PubMed] [Google Scholar]
- O’Dea E., Wilke C., 2011. Contact heterogeneity and phylodynamics: How contact networks shape parasite evolutionary trees. Interdiscip. Perspect. Infect. Dis. 2011: 238743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao D., Fisher M., Hué S., Dean G., Murphy G., et al. , 2005. Transmission of HIV-1 during primary infection: relationship to sexual risk and sexually transmitted infections. AIDS 19: 85. [DOI] [PubMed] [Google Scholar]
- Pybus O., Rambaut A., 2009. Evolutionary analysis of the dynamics of viral infectious disease. Nat. Rev. Genet. 10: 540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus O. G., Rambaut A., Harvey P. H., 2000. An integrated framework for the inference of viral population history from reconstructed genealogies. Genetics 155: 1429–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen D., Ratmann O., Koelle K., 2011. Inference for nonlinear epidemiological models using genealogies and time series. PLoS Comput. Biol. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjödin P., Kaj I., Krone S., Lascoux M., Nordborg M., 2005. On the meaning and existence of an effective population size. Genetics 169: 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler T., 2011. Inferring epidemiological parameters based on allele frequencies. Genetics 188: 663–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbi C., Lemey P., Suchard M., Abdelatif E., Elharrak M., et al. , 2010. Phylodynamics and human-mediated dispersal of a zoonotic virus. PLoS Pathog. 6: e1001166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ballegooijen W., Van Houdt R., Bruisten S., Boot H., Coutinho R., et al. , 2009. Molecular sequence data of hepatitis B virus and genetic diversity after vaccination. Am. J. Epidemiol. 170: 1455. [DOI] [PubMed] [Google Scholar]
- Volz E., Kosakovsky Pond S., Ward M., Leigh Brown A., Frost S., 2009. Phylodynamics of infectious disease epidemics. Genetics 183: 1421–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J., Sargsyan O., 2009. Extensions of the coalescent effective population size. Genetics 181: 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerly S., Vora S., Rizzardi P., Chave J. P., Vernazza P. L., et al. , 2001. Acute HIV infection: impact on the spread of HIV and transmission of drug resistance. AIDS 15: 2287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.