Abstract
The human Abelson helper integration site-1 (AHI1) gene is associated with both neurological and haematological disorders; however, it is also located in a chromosomal region linked to metabolic syndrome phenotypes and was identified as a type 2 diabetes susceptibility gene from a genome-wide association study. To further define a possible role in type 2 diabetes development, AHI1 mRNA expression levels were investigated in a range of tissues and found to be highly expressed in skeletal muscle as well as displaying elevated levels in brain regions and gonad tissues. Further analysis in a rodent, polygenic animal model of obesity and type 2 diabetes identified increased Ahi-1 mRNA levels in red gastrocnemius muscle from fasted impaired glucose tolerant and diabetic rodents compared with normal animals (p<0.002). Moreover, elevated gene expression levels were confirmed in skeletal muscle from fasted obese and type 2 diabetic human subjects (p≤0.02). RNAi-mediated suppression of Ahi-1 resulted in increased glucose transport in rat L6 myotubes in both the basal and insulin-stimulated states (p<0.01). Finally, SNP association studies identified two novel AHI1 genetic variants linked with fasting blood glucose levels in Mexican American subjects (p<0.037). These findings indicate a novel role for AHI1 in skeletal muscle and identify additional genetic links with metabolic syndrome phenotypes suggesting an involvement of AHI1 in the maintenance of glucose homeostasis and type 2 diabetes progression.
1. Introduction
The murine Ahi-1 gene locus is a pro-viral integration site associated with the development of viral-induced murine leukaemias and lymphomas (1, 2). In humans, AHI1 is linked to leukaemia development (2) as well as to several neurological disorders including schizophrenia (3, 4) and Joubert Syndrome, an autosomal recessive brain malformation (5, 6). AHI1 is a large protein containing multiple protein-protein interaction domains including Src homology 3 and WD40-repeat domains, and proline-rich regions (7), which are often present in proteins involved in signal transduction and cytoskeletal organisation (8–10), and was recently described to interact with BCR-ABL and JAK2 and modulate their phosphorylation levels in leukemic cells (11). These features suggest that AHI1 has the potential to act as both a scaffold and signalling protein.
In addition to its associations with neurological and haematological disorders, prior studies have linked 6q23, the AHI1 chromosomal region, to type 2 diabetes (12) and to insulin resistance and metabolic syndrome-related phenotypes (13, 14) including fasting blood glucose levels in Mexican Americans (15). Furthermore, the AHI1-LOC441171 gene locus was identified as a type 2 diabetes susceptibility gene region following a genome-wide association study (GWAS) in European populations (16). A GWAS is a powerful approach to identify novel genomic regions associated with complex diseases and at least six new gene regions have been linked to the risk of type 2 diabetes by GWASs in recent times (17). Two single nucleotide polymorphisms (SNPs) from the AHI1-LOC441171 gene region remained the most significantly associated out of ten SNPs further investigated in a replication study (16). However, a follow-up study was not able to replicate the same SNPs from a study in a Danish population (18) making it unclear as to whether a role for the AHI1 gene in the development of type 2 diabetes and metabolic syndrome exists. Here, we sought to identify novel biological or further genetic evidence for Ahi-1/AHI1 in the metabolic syndrome. We measured Ahi1-1/AHI1 mRNA expression in both insulin sensitive and insulin resistant tissues from an animal model of obesity and type 2 diabetes, and verified significant gene expression changes in humans. We examined the effects of reduced Ahi-1 levels on glucose transport in L6 myotubes and also investigated a range of AHI1 SNPs in a sample of Mexican American subjects to determine if any were associated with fasting blood glucose levels.
2. Materials and Methods
2.1 Human and animal research
All human and animal experimental procedures were approved by the Deakin University ethics committee, and for the human study, informed written consent was obtained from each participant. Psammomys obesus animals were housed at Deakin University and maintained at 22±1°C with a 12 h light, 12 h dark cycle. When fed ad libitum a standard laboratory diet from which 63% of energy was derived from carbohydrate, 25% from protein and 12% from fat (Barastoc, Victoria, Australia), a proportion of P. obesus display a range of metabolic responses including obesity, dyslipidemia, insulin resistance and type 2 diabetes (19–21). At 16 weeks of age, animals were classified as either lean, normal glucose tolerant (NGT), obese and impaired glucose tolerant (IGT) or obese, type 2 diabetic (T2D) according to circulating plasma insulin levels, blood glucose levels and body weight as previously described (19, 21). At 18 weeks of age, fed and animals fasted for 24 h were sacrificed by anaesthetic overdose (120 mg/kg of pentobarbitone; Sigma Chemical Co., St Louis, MO, USA). See Table 1 for the body weight, plasma insulin and whole blood glucose levels of the animals used for this study. Human participants consisting of 10 lean (3 males, 7 females), 13 obese (1 male, 12 female) and 9 obese, T2D (6 male, 3 female) were fasted for 12–18 h prior to skeletal muscle biopsies taken from the vastus lateralis. See Table 2 for the subjects’ age, weight, body mass index, fasting blood glucose and fasting plasma insulin phenotypic characteristics. All biopsies and tissues were snap frozen in liquid nitrogen, and stored at −80°C. Whole blood glucose levels were measured using an enzymatic glucose analyzer (model 27; Yellow Springs Instruments, Yellow Springs, OH), and plasma insulin concentrations were determined using an insulin double antibody radioimmunoassay kit (Linco Research, St. Charles, MO, USA).
Table 1.
Phenotypic characteristics of fed and 24 h fasted NGT, IGT and T2D P. obesus
| Characteristic | NGT P. obesus | IGT P. obesus | T2D P. obesus | |
|---|---|---|---|---|
| Weight (g) | Fed (n=6) | 192.6 ± 7.7 | 227.6 ± 7.1* | 238 ± 6.6*# |
| Fasted (n=7) | 194.8 ± 8.2 | 225.9 ± 8.2* | 237 ± 6.6*# | |
|
| ||||
| BG (mM) | Fed (n=6) | 4.3 ± 0.2 | 4.5 ± 0.3 | 13.5 ± 1.2*# |
| Fasted (n=6) | 3.4 ± 0.3 | 3.6 ± 0.3 | 5.1 ± 0.3*# | |
|
| ||||
| PI (μU/L) | Fed (n=6) | 80.7 ± 10.9 | 407.4 ± 85.2* | 481.3 ± 78.2* |
| Fasted (n=7) | 21.3 ± 3.2 | 119.5 ± 28.1* | 173.8 ± 59* | |
Data presented as mean ± SEM.
denotes p<0.05 to NGT and IGT values, respectively.
BG = blood glucose; PI = plasma insulin.
Table 2.
Phenotypic characteristics of fasted lean, obese and T2D human subjects
| Characteristic | Lean (n=10) | Obese (n=13) | T2D (n=9) |
|---|---|---|---|
| Age (years) | 44.1 ± 1.6 | 36.2 ± 2.2* | 51.1 ± 2.3*# |
| BMI (kg/m2) | 22.6 ± 0.8 | 46.4 ± 1.9* | 40.0 ± 1.0*# |
| Weight (kg) | 66.7 ± 4.0 | 123.7 ± 5.0* | 121.8 ± 7.2* |
| FBG (mM) | 5.0 ± 0.3 | 5.1 ± 0.1 | 7.7 ± 0.8*# |
| FPI (μU/L) | 7.9 ± 3.0 | 16.6 ± 1.2* | 27.2 ± 2.8*# |
Data presented as mean ± SEM.
denotes p<0.05 to lean and obese values, respectively.
BMI = body mass index; FBG = fasting blood glucose; FPI = fasting plasma insulin.
2.2 Cell culture and siRNA transfection
Rat L6 myoblasts were a gift from Professor A. Klip (The Hospital for Sick Children, Toronto, Canada) and were maintained in 25 mM glucose Dulbecco’s minimum essential medium (HG DMEM) (Invitrogen, Melbourne, Australia) with 10% FCS (Invitrogen) at 37°C in 5% CO2. When confluent, myoblasts were differentiated in HG DMEM plus 2% FCS (differentiation medium) for three days prior to transfection with 25 nM small interfering RNAs (siRNAs; Silencer™ siRNA Construction Kit, Ambion, Texas, USA) using Lipofectamine 2000 (Invitrogen) in serum-free Opti-MEM (Invitrogen). The Ahi-1-specific siRNA sequence used in this study was 5′-aagaaagaugacagugagcgu and 5′-aaacgcucacugucaucuuuc for the siAhi-1 antisense and sense strands, respectively. To control for siRNA transfection effects, a negative control siRNA (siNeg) was used. The antisense and sense sequences were 5′-aaguaucuagguacacacuca and 5′-aaugaguguguacguagauac, respectively. The cells were harvested for gene expression analysis and for 2-deoxyglucose transport assays 48 h post-transfection.
2.3 2-Deoxyglucose transport
2-Deoxy-D-[1-3H] glucose transport measurements were carried out as previously described (22) with minor modifications. Following a 4 h starvation in serum-free HG DMEM, L6 myotubes were incubated at 37°C in Krebs Ringer phosphate buffer for 30 min with 0 or 100 nM insulin (HumulinR, Eli Lilly, Indianapolis, Indiana, USA). 2-Deoxyglucose (1 mCi/ml; Amersham Biosciences, Buckinghamshire, UK) at a final concentration of 50 μM was added to the cells, and after 10 min, glucose transport was stopped by the addition of ice-cold PBS. Cells were washed 3 times in PBS prior to lysis with 0.01% SDS and counting for 3H incorporation. Glucose transported was calculated as pmols glucose/min.
2.4 Real time PCR
Total RNA was extracted and isolated using TRIzol (Invitrogen) and RNeasy mini kit columns (QIAGEN, Chatsworth, CA, USA), respectively. The RNA quality and quantity was assessed using the RNA 6000 Nano Assay kit and the Agilent 2100 bioanalyzer (Agilent Technologies, Waldbronn, Germany). The first strand cDNA was generated using the Superscript II First Strand Synthesis system (Invitrogen). Primers for real time PCR analyses were as follows: Rat Ahi-1 (5′-agagcttcgtcaactccatctgt and 5′-cgatgacgccgatgca), P. obesus Ahi-1 (5′-actgctcaaagtatcgtactgttcattagt and 5′-cagcggtgcaattctgatgta), human AHI1 (5′-atggatctccggatattagtagcaa and 5′-aaaagtcccacatggagtcaaagta), rat cyclophilin (5′-cccaccgtgttcttcgaca and 5′-ccagtgctcagagcacgaaa), P. obesus cyclophilin (5′-cccaccgtgttcttcgaca and 5′-ccagtgctcagagcacgaaa) and human cyclophilin (5′-catctgcactgccaagactga and 5′-ttcatgccctcctttcactttgc). Real time PCR was performed on an ABI PRISM 7700 sequence detector (Applied Biosystems, Foster City, CA, USA) using SYBR Green PCR master mix (Applied Biosystems) using the following conditions: 95°C for 10 min (1 cycle), 95°C for 30 s, and 60°C for 1 min (40 cycles).
2.5 Immunoblotting analysis
Immunoblotting was performed as essentially described previously (23). Briefly, cells were lysed in ice-cold radio-immunoprecipitation assay (RIPA) buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing protease inhibitors (2 μg/ml aprotinin, 10 μg/ml leupeptin, 1 μg/ml pepstatin A and 1 mM phenylmethylsulfonyl fluoride) and phosphatase inhibitors (10 nM sodium fluoride and 1 mM sodium orthovanadate) before centrifuging at 13,000 g for 20 min at 4°C to remove insoluble material. The protein concentration was determined using Bio-Rad protein assay reagent (Bio-Rad Laboratories, USA) and 20 μg of total protein were separated by SDS-PAGE prior to transferring to polyvinylidene difluoride membranes and immunoblotting with anti-Akt (1:1000; Cell Signaling Technology, MA, USA) or with anti-phospho-Akt Ser473 (1:1000; Cell Signalling) for 1 h at room temperature. Detection of the primary antibodies was performed using an anti-rabbit IgG, horseradish peroxidase (HRP) linked antibody (1:5000; GE Healthcare, Rydalmere, NSW, Australia) and developed in ECL chemiluminescence reagent (GE Healthcare). The ChemiGeniusII gel documentation system and Gene Snap software (Syngene, Cambridge UK) were used to visualise protein bands and densitometry was performed with Gene Tools software (Syngene).
2.6 Statistical analysis used for in vitro data
Gene expression and phenotypic data are expressed as the mean ± SEM and were analysed using SPSS 14.0 software (Fullerton, CA, USA). All data were subjected to Kolmogorov-Smirnov test for normality, and statistical differences were assessed using unpaired Student’s t-test for two-group comparisons. For the statistical analysis of more than two groups, data were subjected to a one-way ANOVA with post hoc LSD tests when variances were equal. Statistical significance was defined as p<0.05.
2.7 Human association study
The Institutional Review Board of the University of Texas Health Science Centre at San Antonio approved all protocols and informed consent was obtained from all subjects. The Illumina Sentrix HumanHap550 Genotyping BeadChip was used to genotype 565 Mexican American subjects from the San Antonio Family Heart Study (SAFHS) (24). Of the 565 individuals, 199 were male and 366 were female from 18 to 81 years. A total of 75 (13.3%) of these individuals had type 2 diabetes and were excluded from the analyses of fasting glucose. Association analysis was performed using the measured genotype approach (25) allowing for non-independence among family members. In order to account for multiple tests, we calculated the effective number of SNPs (tests) using a previously described method (26) based on the pattern of linkage disequilibrium among SNPs. The mean normalised fasting plasma glucose levels to determine the direction of effect of the SNP genotypes were generated by inverse normalisation (inormal) using the computer package SOLAR (http://solar.sfbrgenetics.org/).
2.8 Statistical genetic methods used in human cohort
Prior to genetic analysis, fasting plasma insulin and glucose concentrations were adjusted for a number of covariates (sex and age and their interactions, smoking behaviour, and menopausal status) using standard regression methods. Residuals from this regression analysis were then directly normalized using an inverse Gaussian transformation. Associations between these normalised traits and the chosen SNPs were then analysed by Bayesian quantitative trait nucleotide analysis using SOLAR (27). In this analysis, all parameter estimations were performed by maximum likelihood under the assumption of a multivariate normality. A formal test of association was obtained by calculating a likelihood ratio test statistic comparing a model in which various regression coefficients for SNPs are held equal against a model in which the parameters are allowed to vary. To account for hidden population stratification the method of Abecasis et al (2000) (28) was used. This method is incorporated into the SOLAR program analysis, which automatically performs a series of association tests for a given set of SNPs.
3. Results
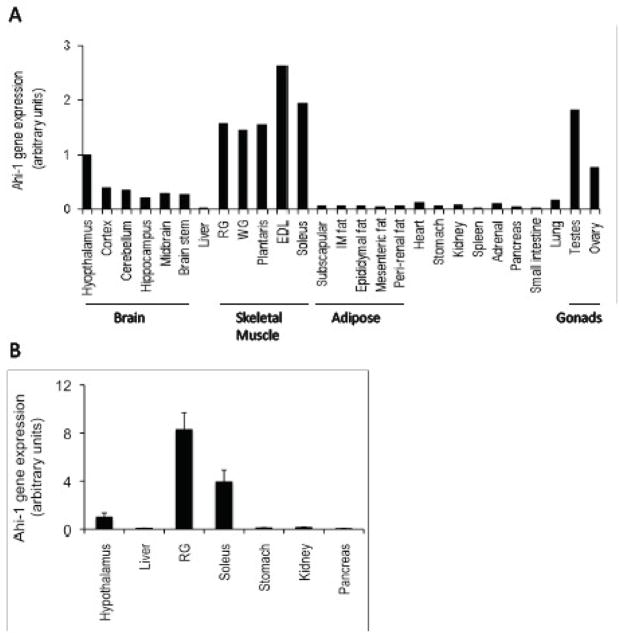
Previous analyses of Ahi-1 tissue-specific expression revealed predominant mRNA levels in the brain and testes of mice (7, 29) but Ahi-1 expression in other insulin-sensitive tissues such as adipose tissue and skeletal muscle depots has not been described to our knowledge. We performed a preliminary analysis of Ahi-1 mRNA expression in an extended range of tissues from one lean, normal glucose tolerant (NGT) Psammomys obesus, a polygenic animal model of obesity and T2D that display a range of metabolic phenotypes (19, 21). This bank of tissues has been utilised in previous studies (30, 31) and the purpose of this initial screen was to identify which tissues or tissue regions display predominant Ahi-1 gene expression. As expected, Ahi-1 was expressed throughout the brain regions and in gonad tissues (ovary and testes) but Ahi-1 mRNA levels were also evident in all skeletal muscle groups measured (Figure 1a). Lower Ahi-1 gene expression was observed in lung, heart and adrenal gland and in various other tissues. To confirm its predominant expression in P. obesus skeletal muscle, Ahi-1 mRNA was next analysed in multiple animals and found to have 4- to 8-fold higher expression in the soleus and red gastrocnemius skeletal muscles, respectively, compared with the hypothalamus, which in turn, displayed at least a 6-fold higher level of Ahi-1 gene expression than in liver, mesenteric fat, kidney, stomach and pancreatic tissues (Figure 1b).
Fig. 1.
Distribution of Ahi-1 mRNA in tissues from an 18 week old, male and female (ovary only) lean NGT P. obesus in the fed state. (A) n=1 and (B) n=6–7. Values are normalised to hypothalamus gene expression in both figures and expressed as fold-change to the hypothalamus values. Abbreviations: RG = red gastrocnemius; WG = white gastrocnemius; EDL = extensor digitorum longus; IM fat = intramuscular fat.
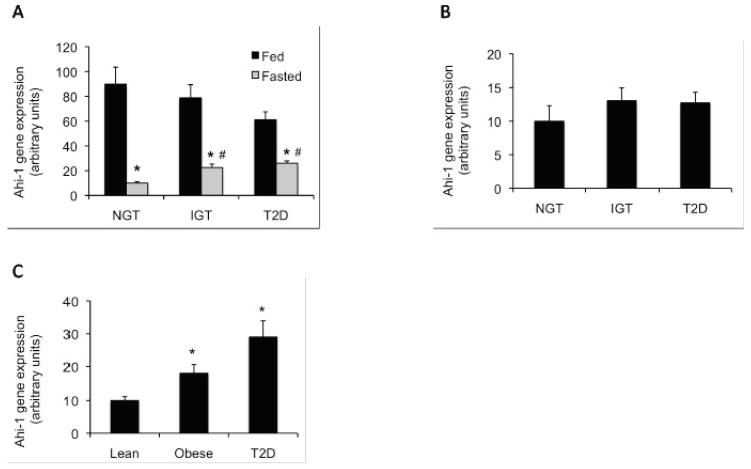
Ahi-1 gene expression in red gastrocnemius muscle from both fed and 24 h fasted NGT, impaired glucose tolerant (IGT) and type 2 diabetic (T2D) P. obesus animals was next examined (see Table 1 for phenotypic charactersistics). Ahi-1 mRNA levels between the fed groups was not found to be significantly different although the T2D animals tended to display decreased expression compared with the NGT groups (p=0.073, Figure 2a). Following 24 h fasting, Ahi-1 gene expression was reduced significantly in all groups (p<0.001); however, Ahi-1 gene expression in both the IGT and T2D animal groups remained significantly greater compared with the fasted NGT animals with nearly a 3-fold difference observed (p<0.002, Figure 2a). This result appeared specific for at least this skeletal muscle group as no significant difference in Ahi-1 gene expression was found in mesenteric fat (Figure 2b) or liver (data not shown) tissues from the same fasted P. obesus groups. To confirm the finding in the P. obesus, AHI1 gene expression in vastus lateralis skeletal muscle from lean, obese and T2D human subjects fasted for 12–18 h was also quantitated. AHI1 was found to be up-regulated significantly by 3-fold in both the obese (p=0.02) and T2D (p<0.001) groups compared with lean individuals (Figure 2c). See Table 2 for the subjects’ relevant phenotypic characteristics.
Fig. 2.
Analysis of Ahi-1/AHI1 gene expression in P. obesus and human skeletal muscle tissues. (A) Fold-change in Ahi-1 mRNA levels in red gastrocnemius from fed (black bars) and fasted (gray bars) NGT, IGT and T2D P. obesus animals (n=6–7 animals per group). *p<0.001 from fed NGT and #p<0.002 from fasted NGT. (B) Fold-change in Ahi-1 mRNA levels in mesenteric fat from NGT, IGT and T2D fasted animals (n=6–7 animals per group) (C) Fold-change in human AHI1 mRNA levels in vastus lateralis muscle from lean, obese and T2D fasted subjects. (n=9–13 individuals per group). *p<0.02 from lean subjects. Values are normalised to cyclophilin gene expression in both figures and expressed as fold-change to NGT or lean values.
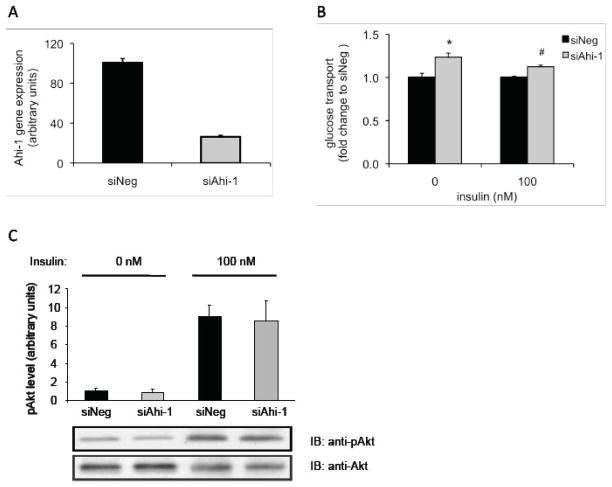
The role of Ahi-1 in myotube insulin sensitivity and/or glucose metabolism was next investigated following the suppression of endogenous Ahi-1 transcripts using Ahi-1-specific siRNA (siAhi-1) in rat L6 myotubes. Ahi-1 gene expression levels were reduced by 75 % when compared with the siNeg-transfected myotubes following transfection of siAhi-1 (Figure 3a). Following the measurement of 2-deoxyglucose transport in the transfected myotubes, reduced levels of Ahi-1 resulted in glucose transport increasing by 23 % and by 9 % in the basal and insulin-stimulated siAhi-1-transfected myotubes, respectively, compared with their siNeg control counterparts (p<0.01; Figure 3b). Insulin stimulation typically caused a 1.4- to 1.8-fold increase in glucose transport in relation to unstimulated myotubes (data not shown). Total protein and phosphorylated Akt levels in either the basal or insulin-stimulated state were not affected by reduced Ahi-1 levels (Figure 3c).
Fig. 3.
Glucose transport and phospho-Akt levels in Ahi-1-deficient L6 myotubes. (A) Ahi-1 gene expression following transfection with the negative control siRNA (siNeg) and Ahi-1-specific (siAhi-1) siRNAs. Values are normalised to cyclophilin gene expression and expressed as a percentage to siNeg. (B) Glucose transport assay in siNeg- (black bars) and siAhi-1- (gray bars) transfected myotubes in both the basal (0 nM insulin) and insulin-stimulated (100 nM insulin) states. Data are expressed as fold-change to siNeg controls (n=5–6 wells per treatment condition) and three independent experiments were performed. *p<0.01 from basal siNeg and #p<0.001 from insulin-stimulated siNeg. (C) Immunoblotting analysis (IB) of Akt and phospho-Akt (ser473) levels in L6 myotubes transfected with siNeg (black bars) or siAhi-1 (grey bars) with or without 15 min insulin treatment. Top panel represents results of the mean ± SEM from four independent experiments. Values are normalised to total Akt protein levels and are expressed relative to siNeg at 0 nM.
To further investigate the possibility that genetic variation in AHI1 may contribute to the development of T2D, 18 non-redundant AHI1 SNPs were genotyped in a sample of Mexican American subjects. An association between the AHI1 SNP, rs6904731, and fasting blood glucose levels in these subjects was observed (p=0.0024), which remained significant after adjusting for multiple testing (target α p<0.0047). This uncommon SNP (minor allele frequency=0.022) is located in intron 25 (Table 3) and accounts for 2.9% of the phenotypic variation in fasting blood glucose levels in these subjects. A nominally significant association with fasting blood glucose levels (p=0.036) was additionally identified for another SNP, rs2614265, located in intron 22 (Table 3). The direction of effect of rs6904731 and rs2614265 with mean normalised fasting plasma glucose levels for each genotype is outlined in Table 4.
Table 3.
Association between AHI1 genetic variants and fasting glucose levels in Mexican Americans subjects.
| SNP | Nucleotide | Exon/Intron | MAF | p-value |
|---|---|---|---|---|
| Variation | ||||
| rs17064526 | A/C | Intron 10 | 0.0207 | 0.6284 |
| rs1535436 | A/G | Intron 10 | 0.0251 | 0.2208 |
| rs717120 | C/T | Intron 12 | 0.0699 | 0.7742 |
| rs1535435 | A/G | Intron 14 | 0.0856 | 0.7331 |
| rs737561 | C/T | Intron 14 | 0.0704 | 0.7251 |
| rs11154801 | A/C | Intron 19 | 0.2846 | 0.1528 |
| rs17778438 | A/G | Intron 20 | 0.0482 | 0.9823 |
| rs13208164 | A/G | Intron 21 | 0.0982 | 0.5778 |
| rs2614265 | A/G | Intron 22 | 0.3342 | 0.0360* |
| rs17064440 | A/G | Intron 23 | 0.0077 | 0.599 |
| rs7772681 | C/T | Intron 23 | 0.3781 | 0.4119 |
| rs2064430 | C/T | Intron 24 | 0.3474 | 0.2699 |
| rs6904731 | G/A | Intron 25 | 0.0217 | 0.0024** |
| rs6931735 | A/G | Intron 25 | 0.3498 | 0.2021 |
| rs2207000 | C/T | Intron 25 | 0.1298 | 0.2652 |
| rs9285480 | C/T | Intron 26 | 0.0598 | 0.8639 |
| rs7766656 | A/G | Intron 26 | 0.0918 | 0.607 |
| rs1052502 | C/T | 3′UTR | 0.0931 | 0.619 |
p< target alpha level 0.00465 for an experiment wide significance after adjusting for multiple testing.
Nominal association p<0.05.
MAF = minor allele frequency.
Table 4.
Mean normalised fasting plasma glucose values for each genotype
| SNP | p-value | AA | AG | GG |
|---|---|---|---|---|
| rs2614265 | 0.036 | 0.325 ± 0.096 | 0.458 ± 0.089 | 0.590 ± 0.121 |
| rs6904731 | 0.0024 | 0.446 ± 0.086 | −0.112 ± 0.192 | −0.670 ± 0.365 |
4. Discussion
Here, we provide novel biological evidence for a role for Ahi-1/AHI1 in skeletal muscle. Elevated Ahi-1/AHI1 gene expression was measured in skeletal muscle from both insulin resistant/obese and type 2 diabetic P. obesus and human subjects. Moreover, we show that suppression of Ahi-1 enhanced glucose transport in basal and insulin-stimulated L6 myotubes. We also identified two new AHI1genetic variants associated with fasting blood glucose levels in a sample of Mexican American subjects.
Elevated expression of Ahi-1 mRNA in skeletal muscle compared with other P. obesus tissues suggests a biological role in this tissue. To our knowledge, the investigation of Ahi-1/AHI1 mRNA or protein expression levels in skeletal muscle from any species has not been reported. Species differences in some spatial and temporal expression of Ahi-1/AHI1 levels from mouse and human brain regions have been noted (29), but as moderate Ahi-1 gene expression in both brain and gonads tissues was measured in P. obesus, which has also been described in other species (7, 32, 33), Ahi-1 gene expression in skeletal muscle is potentially conserved between orthologs. The increased level of skeletal muscle Ahi-1 gene expression in insulin resistant/obese and T2D animals and humans compared with the lean and healthy groups in the fasted state also reveals that the regulation of Ahi-1/AHI1 transcription is altered in response to fasting in these groups displaying dysregulated metabolic conditions. Furthermore, as fasting causes higher rates of fatty acid oxidation, elevated free fatty acids and reduced glucose disposal in skeletal muscle (reviewed in (34)), these effects in conjunction with metabolic syndrome conditions appear to increase Ahi-1/AHI1 expression, which in turn, could indicate that Ahi-1/AHI1 negatively regulates insulin sensitivity and/or glucose metabolism.
The outcome of the glucose transport studies performed following suppression of endogenous Ahi-1 mRNA in L6 myotubes appear to support the premise of Ahi-1 functioning as a negative regulator of glucose uptake. How Ahi-1 may function to do this is yet not clear. We were unable to detect any change in the total level of Akt protein or in its phosphorylated state and so Ahi-1 may be affecting Akt-independent glucose transport pathways such as through Rac1 (35). There are multiple glucose transporters (GLUT1, GLUT3 and GLUT4) present in L6 muscle cells (36) and whether Ahi-1 regulates their expression and/or activities, directly or indirectly, remains to be determined. However, it is of interest to note the recent publication describing a specific mode of action for Ahi-1 in the regulation of ciliogenesis and vesicle trafficking via the GTPase, Rab8A (37). Given that Rab8A appears to be one of the key modulators of GLUT4 vesicle recruitment and fusion in L6 myotubes (38, 39), it will be of interest to determine if Ahi-1 is involved in Rab8A/GLUT4 activity, and hence, glucose transport in skeletal muscle.
The identification of novel AHI1 intronic SNPs, rs6904731 and rs2614265, associated with fasting blood levels in Mexican Americans in this study contributes further to the findings of past GWAS and linkage studies linking AHI1 to a range of metabolic syndrome phenotypes (12–16), and again suggests that AHI1 is a gene associated with increased susceptibility to dysregulated glucose metabolism and type 2 diabetes. One of the 18 SNPs we tested in this study, rs1535435, was previously found associated with type 2 diabetes (16), but similarly to a follow-up study investigating the association of this SNP with a range of type 2 diabetes-related metabolic traits (18), we were not able to find this specific SNP significantly associated with fasting blood glucose levels in our population of Mexican Americans. The reasons for lack of replication may be several-fold including variation in patterns of linkage disequilibrium and the genetic basis of their quantitative trait variation, which may be due to ethnicity differences, sample size differences between the cohorts resulting in varying statistical power, and different design strategies. As all these SNPs are intronic, they may also be exerting weak effects on metabolic traits that are not easily replicated.
Our study describes novel biological evidence of a role for AHI1 in skeletal muscle, a key metabolic tissue that is responsible for the majority of peripheral glucose uptake (40). In addition, it identifies further genetic links associated with metabolic syndrome phenotypes. Together, these data provide a link for AHI1 to the maintenance of glucose homeostasis and type 2 diabetes progression.
Acknowledgments
We thank Petra Gran, Marissa Trenerry and Rani Watts for their efforts in preparing and analysing the clinical samples, and we are grateful to the participants in the San Antonio Family Heart Study. This study was funded by Verva Pharmaceuticals and by the Molecular Medicine and Nutrition Cluster, Deakin University. The SOLAR statistical genetics computer package is supported by a grant from the US National Institute of Mental Health (MH059490).
Footnotes
There are no known conflicts of financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanlon L, Barr NI, Blyth K, Stewart M, Haviernik P, Wolff L, et al. Long-range effects of retroviral insertion on c-myb: overexpression may be obscured by silencing during tumor growth in vitro. J Virol. 2003;77(2):1059–68. doi: 10.1128/JVI.77.2.1059-1068.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang X, Zhao Y, Chan WY, Vercauteren S, Pang E, Kennedy S, et al. Deregulated expression in Ph+ human leukemias of AHI-1, a gene activated by insertional mutagenesis in mouse models of leukemia. Blood. 2004;103(10):3897–904. doi: 10.1182/blood-2003-11-4026. [DOI] [PubMed] [Google Scholar]
- 3.Amann-Zalcenstein D, Avidan N, Kanyas K, Ebstein RP, Kohn Y, Hamdan A, et al. AHI1, a pivotal neurodevelopmental gene, and C6orf217 are associated with susceptibility to schizophrenia. Eur J Hum Genet. 2006;14(10):1111–9. doi: 10.1038/sj.ejhg.5201675. [DOI] [PubMed] [Google Scholar]
- 4.Ingason A, Sigmundsson T, Steinberg S, Sigurdsson E, Haraldsson M, Magnusdottir BB, et al. Support for involvement of the AHI1 locus in schizophrenia. Eur J Hum Genet. 2007;15(9):988–91. doi: 10.1038/sj.ejhg.5201848. [DOI] [PubMed] [Google Scholar]
- 5.Parisi MA, Doherty D, Chance PF, Glass IA. Joubert syndrome (and related disorders) Eur J Hum Genet. 2007;15(5):511–21. doi: 10.1038/sj.ejhg.5201648. [DOI] [PubMed] [Google Scholar]
- 6.Valente EM, Brancati F, Silhavy JL, Castori M, Marsh SE, Barrano G, et al. AHI1 gene mutations cause specific forms of Joubert syndrome-related disorders. Ann Neurol. 2006;59(3):527–34. doi: 10.1002/ana.20749. [DOI] [PubMed] [Google Scholar]
- 7.Jiang X, Hanna Z, Kaouass M, Girard L, Jolicoeur P. Ahi-1, a novel gene encoding a modular protein with WD40-repeat and SH3 domains, is targeted by the Ahi-1 and Mis-2 provirus integrations. J Virol. 2002;76(18):9046–59. doi: 10.1128/JVI.76.18.9046-9059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. Faseb J. 2000;14(2):231–41. [PubMed] [Google Scholar]
- 9.Li D, Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol Life Sci. 2001;58(14):2085–97. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pawson T, Olivier P, Rozakis-Adcock M, McGlade J, Henkemeyer M. Proteins with SH2 and SH3 domains couple receptor tyrosine kinases to intracellular signalling pathways. Philos Trans R Soc Lond B Biol Sci. 1993;340(1293):279–85. doi: 10.1098/rstb.1993.0069. [DOI] [PubMed] [Google Scholar]
- 11.Zhou LL, Zhao Y, Ringrose A, DeGeer D, Kennah E, Lin AE, et al. AHI-1 interacts with BCR-ABL and modulates BCR-ABL transforming activity and imatinib response of CML stem/progenitor cells. J Exp Med. 2008;205(11):2657–71. doi: 10.1084/jem.20072316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang K, Wang Y, Zheng T, Jia W, Li J, Chen L, et al. Genome-wide search for type 2 diabetes/impaired glucose homeostasis susceptibility genes in the Chinese: significant linkage to chromosome 6q21–q23 and chromosome 1q21–q24. Diabetes. 2004;53(1):228–34. doi: 10.2337/diabetes.53.1.228. [DOI] [PubMed] [Google Scholar]
- 13.Arya R, Blangero J, Williams K, Almasy L, Dyer TD, Leach RJ, et al. Factors of insulin resistance syndrome--related phenotypes are linked to genetic locations on chromosomes 6 and 7 in nondiabetic mexican-americans. Diabetes. 2002;51(3):841–7. doi: 10.2337/diabetes.51.3.841. [DOI] [PubMed] [Google Scholar]
- 14.Duggirala R, Blangero J, Almasy L, Arya R, Dyer TD, Williams KL, et al. A major locus for fasting insulin concentrations and insulin resistance on chromosome 6q with strong pleiotropic effects on obesity-related phenotypes in nondiabetic Mexican Americans. Am J Hum Genet. 2001;68(5):1149–64. doi: 10.1086/320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stern MP, Duggirala R, Mitchell BD, Reinhart LJ, Shivakumar S, Shipman PA, et al. Evidence for linkage of regions on chromosomes 6 and 11 to plasma glucose concentrations in Mexican Americans. Genome Res. 1996;6(8):724–34. doi: 10.1101/gr.6.8.724. [DOI] [PubMed] [Google Scholar]
- 16.Salonen JT, Uimari P, Aalto JM, Pirskanen M, Kaikkonen J, Todorova B, et al. Type 2 diabetes whole-genome association study in four populations: the DiaGen consortium. Am J Hum Genet. 2007;81(2):338–45. doi: 10.1086/520599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007;8(9):657–62. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- 18.Holmkvist J, Anthonsen S, Wegner L, Andersen G, Jorgensen T, Borch-Johnsen K, et al. Polymorphisms in AHI1 are not associated with type 2 diabetes or related phenotypes in Danes: non-replication of a genome-wide association result. Diabetologia. 2008;51(4):609–14. doi: 10.1007/s00125-008-0925-z. [DOI] [PubMed] [Google Scholar]
- 19.Barnett M, Collier GR, Collier FM, Zimmet P, O’Dea K. A cross-sectional and short-term longitudinal characterisation of NIDDM in Psammomys obesus. Diabetologia. 1994;37(7):671–6. doi: 10.1007/BF00417690. [DOI] [PubMed] [Google Scholar]
- 20.Walder K, Oakes N, Fahey RP, Cooney G, Zimmet PZ, Collier GR. Profile of dyslipidemia in Psammomys obesus, an animal model of the metabolic syndrome. Endocr Regul. 2002;36(1):1–8. [PubMed] [Google Scholar]
- 21.Walder KR, Fahey RP, Morton GJ, Zimmet PZ, Collier GR. Characterization of obesity phenotypes in Psammomys obesus (Israeli sand rats) Int J Exp Diabetes Res. 2000;1(3):177–84. doi: 10.1155/EDR.2000.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olefsky JM. Mechanisms of the ability of insulin to activate the glucose-transport system in rat adipocytes. Biochem J. 1978;172(1):137–45. doi: 10.1042/bj1720137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foletta VC, Prior MJ, Stupka N, Carey K, Segal DH, Jones S, et al. NDRG2, a novel regulator of myoblast proliferation, is regulated by anabolic and catabolic factors. J Physiol. 2009;587(Pt 7):1619–34. doi: 10.1113/jphysiol.2008.167882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell BD, Kammerer CM, Blangero J, Mahaney MC, Rainwater DL, Dyke B, et al. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation. 1996;94(9):2159–70. doi: 10.1161/01.cir.94.9.2159. [DOI] [PubMed] [Google Scholar]
- 25.Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet. 1986;50(Pt 2):181–94. doi: 10.1111/j.1469-1809.1986.tb01037.x. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005 Sep;95(3):221–7. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 27.Blangero J, Goring HH, Kent JW, Jr, Williams JT, Peterson CP, Almasy L, et al. Quantitative trait nucleotide analysis using Bayesian model selection. Hum Biol. 2005;77(5):541–59. doi: 10.1353/hub.2006.0003. [DOI] [PubMed] [Google Scholar]
- 28.Abecasis GR, Cookson WO, Cardon LR. Pedigree tests of transmission disequilibrium. Eur J Hum Genet. 2000;8(7):545–51. doi: 10.1038/sj.ejhg.5200494. [DOI] [PubMed] [Google Scholar]
- 29.Doering JE, Kane K, Hsiao YC, Yao C, Shi B, Slowik AD, et al. Species differences in the expression of Ahi1, a protein implicated in the neurodevelopmental disorder Joubert syndrome, with preferential accumulation to stigmoid bodies. J Comp Neurol. 2008;511(2):238–56. doi: 10.1002/cne.21824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bozaoglu K, Bolton K, McMillan J, Zimmet P, Jowett J, Collier G, et al. Chemerin is a novel adipokine associated with obesity and metabolic syndrome. Endocrinology. 2007;148(10):4687–94. doi: 10.1210/en.2007-0175. [DOI] [PubMed] [Google Scholar]
- 31.Trevaskis J, Walder K, Foletta V, Kerr-Bayles L, McMillan J, Cooper A, et al. Src homology 3-domain growth factor receptor-bound 2-like (endophilin) interacting protein 1, a novel neuronal protein that regulates energy balance. Endocrinology. 2005;146(9):3757–64. doi: 10.1210/en.2005-0282. [DOI] [PubMed] [Google Scholar]
- 32.Ferland RJ, Eyaid W, Collura RV, Tully LD, Hill RS, Al-Nouri D, et al. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat Genet. 2004;36(9):1008–13. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- 33.Zhou W, Song P. Molecular cloning of a novel gene ZAhi-1 and its expression analysis during zebrafish gametogenesis. Mol Biol Rep. 2006;33(2):111–6. doi: 10.1007/s11033-006-0005-8. [DOI] [PubMed] [Google Scholar]
- 34.Roden M. How free fatty acids inhibit glucose utilization in human skeletal muscle. News Physiol Sci. 2004;19:92–6. doi: 10.1152/nips.01459.2003. [DOI] [PubMed] [Google Scholar]
- 35.JeBailey L, Wanono O, Niu W, Roessler J, Rudich A, Klip A. Ceramide- and oxidant-induced insulin resistance involve loss of insulin-dependent Rac-activation and actin remodeling in muscle cells. Diabetes. 2007;56(2):394–403. doi: 10.2337/db06-0823. [DOI] [PubMed] [Google Scholar]
- 36.Tsakiridis T, McDowell HE, Walker T, Downes CP, Hundal HS, Vranic M, et al. Multiple roles of phosphatidylinositol 3-kinase in regulation of glucose transport, amino acid transport, and glucose transporters in L6 skeletal muscle cells. Endocrinology. 1995;136(10):4315–22. doi: 10.1210/endo.136.10.7664650. [DOI] [PubMed] [Google Scholar]
- 37.Hsiao YC, Tong ZJ, Westfall JE, Ault JG, Page-McCaw PS, Ferland RJ. Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a localization, ciliogenesis and vesicle trafficking. Hum Mol Genet. 2009;18(20):3926–41. doi: 10.1093/hmg/ddp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishikura S, Klip A. Muscle cells engage Rab8A and myosin Vb in insulin-dependent GLUT4 translocation. Am J Physiol Cell Physiol. 2008;295(4):C1016–25. doi: 10.1152/ajpcell.00277.2008. [DOI] [PubMed] [Google Scholar]
- 39.Randhawa VK, Ishikura S, Talior-Volodarsky I, Cheng AW, Patel N, Hartwig JH, et al. GLUT4 vesicle recruitment and fusion are differentially regulated by Rac, AS160, and Rab8A in muscle cells. J Biol Chem. 2008;283(40):27208–19. doi: 10.1074/jbc.M804282200. [DOI] [PubMed] [Google Scholar]
- 40.DeFronzo RA, Gunnarsson R, Bjorkman O, Olsson M, Wahren J. Effects of insulin on peripheral and splanchnic glucose metabolism in noninsulin-dependent (type II) diabetes mellitus. J Clin Invest. 1985;76(1):149–55. doi: 10.1172/JCI111938. [DOI] [PMC free article] [PubMed] [Google Scholar]