Abstract
Histone post-translational modifications (PTMs) comprise one of the most intricate nuclear signaling networks that govern gene expression in a long-term and dynamic fashion. These PTMs are considered to be ‘epigenetic’ or heritable from one cell generation to the next and help establish genomic expression patterns. While much of the analyses of histones have historically been performed using site-specific antibodies, these methods are replete with technical obstacles (i.e., cross-reactivity and epitope occlusion). Mass spectrometry-based proteomics has begun to play a significant role in the interrogation of histone PTMs, revealing many new aspects of these modifications that cannot be easily determined with standard biological approaches. Here, we review the accomplishments of mass spectrometry in the histone field, and outline the future roadblocks that must be overcome for mass spectrometry-based proteomics to become the method of choice for chromatin biologists.
Keywords: chromatin, combinatorial, discovery, epigenetic, histone, mass spectrometry, post-translational modification, proteomics, quantification
The study of chromatin biology has evolved to reveal another layer of control over the cellular processes of transcription, cellular division, differentiation and cellular repair. Once deemed a solely structural element of chromatin, the nucleosome has emerged as a crucial controller of cellular fate. In eukaryotes, this basic repeating unit of chromatin is comprised of approximately 147 bp of DNA wrapped around an octamer of the highly conserved core histone proteins (H2A, H2B, H3 and H4) [1]. The amino-terminal domains of these proteins project outward from the core particle and are accessible to proteases. Interestingly, these tail domains act as the sites of a myriad of covalent post-translational modifications (PTMs), many of which have been linked directly to the transcriptional output of the genome. Thus far, these PTMS include modifications on lysine residues such as: acetylation, ubiquitination and mono-, di- and tri-methylation; mono- and asymmetric or symmetric di-methylation on arginine residues; and phosphorylation on serine and threonine residues. Specific modified residues, such H3 lysine 9 acetylation (H3K9ac), H3 serine 10 phosphorylation (H3S10ph) and H3 lysine 27 methylation (H3K27me), serve as target epitopes for chromatin remodeling machinery, as well as gene repression and/or activation complexes [1]. However, these marks are usually found in series or in combinations. Out of these observations was born the ‘histone code hypothesis’, which states that “distinct histone modifications, on one or more N-terminal tails, act sequentially or in combination to form a ‘histone code’ that is read by other proteins (containing specialized binding domains) to bring about distinct downstream events” [2,3]. Histone PTMs, along with DNA methylation and small noncoding RNA, collectively make up mechanisms referred to as ‘epigenetic’ controls, which are believed to affect gene expression patterns and phenotype in a heritable manner.
In recent decades, epigenetics has come to the forefront of clinical investigation as researchers are faced with the obstacle of identifying nongenetic components that display the same phenotypical disease state as several previously characterized genetic conditions. Technological advances in the field of genetics and medicine have unveiled new genetic bases for hundreds of human diseases. For a number of these disorders, such as Prader–Willi and Angelman sister syndromes, Beckwith–Wiedemann Syndrome and Rett Syndrome, the phenotypic variability from patient to patient is drastic even though the underlying genetic mutations are well defined [4–12]. Deregulation of controlled cellular pathways is a hallmark of tumorigenesis and epigenetic signatures, such as altered distribution of 5-methylcytosine DNA modifications and hypermethylated gene promoters, have become distinct markers of cancer. Changes in histone modification patterns have also gained attention for possible roles in various types of cancer. For example, misdirected targeting of histone acetyltransferases and histone deacetylases have been found in several types of leukemias [13,14]. Loss of histone H4K20me3 and H4K16ac have been determined to be hallmarks of all common cancers (both in cancer cell lines and primary tumors) [15], and these modification losses were found to appear early, accumulating during tumorigenesis. Changes in global levels of individual histone H4 and H3 acetylation and methylation have also been associated with prostate cancer and these changes were indicative of clinical results [16]. In addition, increased histone kinase activity has been shown to be associated with colorectal cancer, and the list of malignancies correlated with significant changes in histone PTM patterns during specific disease states continues to grow, as recently shown in leukemogenesis [17]. Last, small molecule inhibitors of histone deacetylases are currently in various phases of clinical trials for treating several forms of cancer [18]. The continued comprehensive hybridization of epigenetic and genetic approaches to understanding cancer formation and development could lead to personalized treatment regimens for chemoresistant cancer patients as opposed to blanket treatments.
Nevertheless, while data continue to accumulate regarding disruption of histone modification patterns and links to human disease, the precise epigenetic mechanisms possibly underlying these diseases are not yet fully understood. These observations have made a strong case for the initiation of an international Human Epigenome Project on the scale of the Human Genome Project, and organization of this possible effort by the Alliance for the Human Epigenome and Disease and the Epigenome Network of Excellence has begun [19]. The Alliance for the Human Epigenome and Disease, backed by the American Association for Cancer Research, has very recently published a rough formal plan to begin indexing specific histone PTMs in a defined subset of selected ‘reference epigenomes’ that could provide a reference standard to which disease states could be compared [19]. The NIH has also committed over US$100 million over the next several years to fund and accelerate epigenetics/epigenomics research. Understanding the epigenetic changes in normal human development and disease has far-reaching implications, which would make immediate impacts in many fields ranging from stem cell biology to neuro science, and could serve as a novel means of disease-targeted therapeutic intervention. Therefore, as can be imagined, robust methods to identify and measure the abundance levels of epigenetic marks such as histone PTMs are of tremendous importance.
Why mass spectrometry-based proteomics for histone PTMs?
Mass spectrometry (MS) has played an increasingly important role for the analysis of a myriad of protein PTMs, including those from chromatin-associated proteins. The increased need for sensitivity and accurate identification of epigenetic determinants of disease is one of the many driving forces behind MS-based proteomics for understanding the role of histone PTMs. Traditionally, histone samples are tested for specific PTMs sites by immunoblotting assays for specific modifications. This approach serves a valuable role in chromatin analysis, especially when combined with targeted or large-scale gene expression analysis, such as chromatin immunoprecipitation (ChIP) coupled to PCR, DNA microarray (CHIP) or deep sequencing (Seq). However, there are several disadvantages associated with antibody-based methods. Most notable is the concern of antibodies cross-reacting with similar modifications on the same histone protein or on a different histone protein (i.e., H3K27me3 antibody binding H3K27me2 or H4K20me3), as well as antibodies recognizing unmodified histones and/or non-histone proteins with similar modifications and sequences. In one study profiling antibodies for histone PTMs in Drosophila melanogaster, 20–35% of the commercially available antibodies were deemed unsatisfactory, and other studies have shown similar results [20,21]. In addition, epitope occlusion, as well as cost and difficulty in manufacturing/validating antibodies, remain significant obstacles. Epitope occlusion, the ability of certain nonintended PTMs to block the intended recognition of a PTM by a site-specific antibody is particularly a major issue, especially on highly modified histones. Comparatively, MS provides a detailed, more unbiased method for discovering, screening and quantifying histone and non-histone proteins, as described in previous reviews [22,23]. Coupled with standard biological techniques, MS offers the unique capacity to compare protein and modification expression in the context of ‘normal’ cells versus altered cellular states, such as with infected or cancerous cells [24,25]. A myriad of cellular and tissue-specific conditions, such as mitotic regulation, tissue development and stem cell differentiation, can also be explored in detail. Although still a specialized approach, MS instrumentation and techniques are becoming more and more accessible to non-MS-trained scientists. In contrast to antibody-based investigations, MS also offers the ability to examine the combinatorial nature of PTMs in a high-throughput fashion. Therefore, MS-based methods are arguably stronger approaches than immunoassays, and serve as complementary experiments to genomic studies.
Discovery of novel histone PTMs by MS
Mass spectrometry has faithfully uncovered numerous novel PTMs that have been correlated with a rich transcriptional output. In earlier studies, MS was used to discover many new histone PTMs. For example, the arginine methylation of histone H4 (R3me1), a now well-studied modification that has been linked to transcriptional repression mediated by the protein arginine methyltransferase PRMT1, was first exposed via MS interrogation [26]. New lysine methylation sites found by proteomic investigations on other histones such as H2B and H1 have also recently been described [27,28]. Novel H2B modifications are particularly interesting finds, as this histone has been traditionally thought to contain a limited variety of PTMs and at a low abundance. These modifications found in yeast may be evolutionarily conserved, although the exact biological function remains to be determined. Previously unreported acetylation sites on histone H3 have also been catalyzed by MS detection. These include H3K56ac and H3K36ac, the former of which drives progression through the cell cycle in yeast, but in humans has been linked to the pluripotent transcriptional network in embryonic stem cells [29]. Histone H3K36ac is generated by the GCN5 acetyltransferase complex and has been found to occupy promoters of RNA polymerase II-transcribed genes, although the full effect of this modification remains unknown [30]. MS interrogation has also revealed the presence of new phosphorylation sites such as H3T45 phosphorylation. This mark has been shown to be involved in DNA replication in yeast [31] and associated with apoptosis in mammals [32]. Immobilized metal affinity chromatography in combination with MS was also more recently employed to demonstrate the existence of H3T6phos, a modification that influences binding of ING2 to its target H3K4me3 site [33]. This phenomenon of methylation binding by effector proteins being affected by nearby phosphorylation has been seen before when H3S10phos was shown to block heterochromatin protein 1 (HP1) from binding its target H3K9me3 site during mitosis [34,35].
More exciting, is the emergence of rarer and more exotic PTMs from histone screens by MS [36–39]. Recently and for the first time, histones have been described by both in vivo biological and mass spectrometric approaches to be modified by O-GlcNAcylation on H2A, H2B and H4, providing evidence of the existence of O-GlcNAc as part of the histone code [36]. MS analysis of histones from various eukaryotic sources has identified the in vivo existence of other atypical PTMs in addition to the O-GlcNAC PTM [36], such as propionylation, butyrylation, formylation and ADP-ribosylation [37–39]. Histone propionylation and butyrylation are very intriguing modifications, as they have been so far only detected on residues that are also known to be acetylated (i.e., H4K12), suggesting that a potential switch between modified forms on the same residue may exist and modulate biological function. This would be analogous to the opposing changes induced by histone H3K9 me3 or ac, the former mark being associated with HP1-binding and silenced genes, while the latter is known to reside on actively transcribed loci [40]. In addition, histone propionylation and butyrylation can be catalyzed by known histone acetyltransferases such as CREB-binding protein and p300, hinting at the role of these enzymes and PTMs in energy metabolism mediated through chromatin remodeling [39]. Subsequently, many non-histone proteins, such as p53, have also been detected to be propionylated; therefore, this modification may actually affect a much larger number of cellular pathways [41]. However, to date, the downstream transcriptional effects of these modifications are yet to be fully uncovered. MS has also been used to probe the uniqueness of histone PTMs and their abundances in specific organisms, cell and tissue types, although validation of these marks by orthogonal means, confirmation of specificity to biological material and deeper studies in potential function have not been fully attempted [42–45]. However, it is easy to see that in combination with classical molecular and biochemical techniques, proteomic approaches can provide the initial data to begin unraveling these new cellular mysteries.
Quantification of histone PTM abundance: comparative MS analyses
Precise identification of histone modifications is usually the first goal of any proteomic experiment. However, since the great majority of histones PTMs are not exclusive to any cellular condition or cell type, quantification of the abundance levels of the PTMs becomes an absolute necessity to find changes in histone PTMs from multiple physiological states. Histone PTMs can be quantified within diverse biological backgrounds based on their abundances relative to each other by a number of proteomic methods (i.e., isotopically labeled or label free). The biggest obstacle to gaining a robust quantification of histone PTMs is being able to generate a digestion of histones that will reproducibly contain the same peptides. Histones are highly basic proteins (rich in Arg and Lys residues) that when digested with common proteases used in proteomics research such as trypsin, will be cleaved into many short peptides. In addition, many of these peptides will be overlapping, containing the same residues, and thus quantification of specific modification sites becomes problematic. Therefore, strategies to minimize heterogeneity of peptides have been used by researchers in this area for a long time. A common approach is to employ the use of enzymes that cleave at only one amino acid, such as ArgC, or other proteases that target acidic residues, such as GluC. This was nicely demonstrated by McKittrick et al., while quantifying the amount of histone PTMs from the Drosophila histone H3.3 and H3.2 variants [46]. These experiments first pointed to H3.3 containing more PTMs linked to active genes than H3.2. More recent approaches have also included combining these targeted protease digestions with quantification using ‘multiple reaction monitoring’ on a triple quadrupole instrument, as well as other label-free methods [47]. While this has been a feasible tactic, often one finds that the proteases are not as specific as labeled and this can result in much longer, even more highly charged peptides that must be addressed by newer fragmentation approaches other than collisionally activated dissociation (CAD) (see the next section). In addition, we have observed that enzymes such as ArgC and and GluC can also cleave at other residues, such as at Lys (nonselective) and Gln (following deamidation) residues, and some sort of optimization of digestion conditions must be attained for quantitative and selective digestion. Therefore, chemical derivatization of samples followed by digestion with robust enzymes such as trypsin seem to be good alternative approaches.
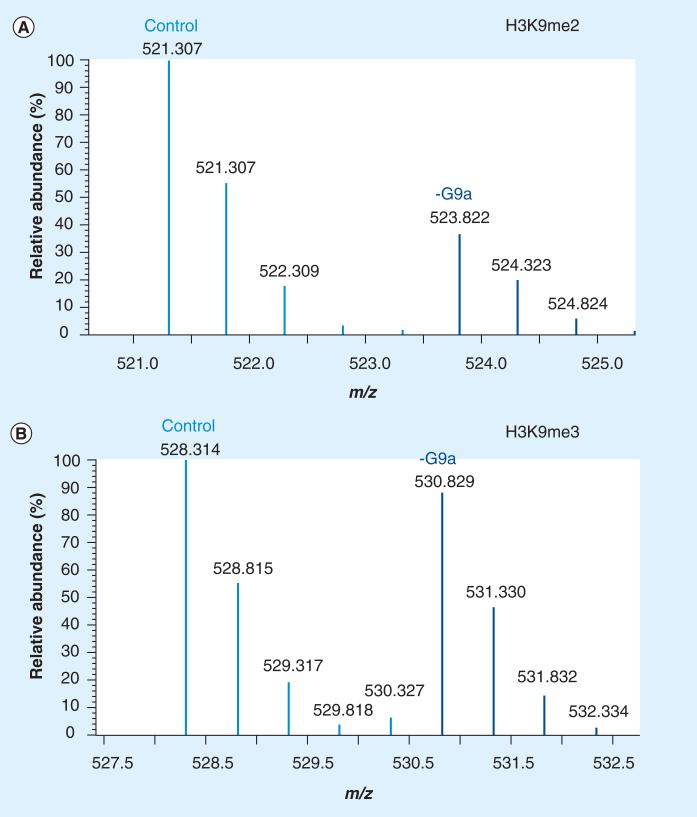
Using chemical derivatization as a means to modify cleavable residues has also been widely employed. Prior to proteolysis, the highly charged histones may be chemically derivatized using acetic or propionic anhydride [48,49]. Acetic anhydride derivatization is best utilized if a deuterated version is used, as shown previously to quantify the acetylation isomers on histone H4 and developmental changes of core histones from Drosophila melanogaster [50]. Propionylation of histones is also an attractive approach for preparing histones for MS analysis as a propionyl group is covalently added to unmodified lysines, thus blocking these sites from cleavage and resulting in slightly longer tryptic-like peptides ending in Arg residues that facilitate subsequent ionization, detection and fragmentation by MS. In a label-free approach, PTM abundances are determined by integrating the area under the curve for each charge state of the corresponding peptide. Intriguingly, methods have been refined to include the use of isotopically labeled propionic anhydride to compare PTM abundances between multiple sample types [51]. For example, histone samples from one cellular state can be labeled with deuterated propionic anhydride (D5) and mixed in 1:1 ratio with non-isotopically labeled (propionic anhydride; D0) histones from a different cellular state to quantify the histone PTMs. The propionylation methodology has been utilized by a large number of groups [52–55], which include quantification of histones from a variety of diverse cellular sources such as from the Trypanosoma brucei, the parasite responsible for African sleeping sickness [56], human and mouse melanoma cancer samples [57], and those displaying antimicrobial properties from shrimp hemocyte lysates [58], to just name a few. Last, this method ology has been shown to be effective for identifying changes in histone PTMs upon knockdown of specific histone-modifying enzymes [59]. As shown in Figure 1, knockdown of the methyltransferases G9a and G9a-like protein showed a large decrease in both H3K9me1 and H3K9me2, consistent with previous reports, but also demonstrated that the depletion of this methyltransferase complex also had a secondary effect, altering the levels of other nondirect sites such as H3K14ac and H3K79 methylation. In Figure 1A, the [M+2H]2+ ion peptides corresponding to the 9–17 fragment (prKme2STGGKprAPR) from a propionylated histone fragment possessing K9me2 can be seen to be decreased following siRNA knockdown of the methyltransferase G9a. The peptides are separated by 2.5 mass:charge ratio (m/z), which corresponds to a 5 Da overall mass shift, as one sample has been derivatized with D0 propionic anhydride and the other using D5 propionic anhydride. By contrast, the peptides corresponding to the 9–17 fragment (prKme3STGGKprAPR) containing K9me3 are not affected by depletion of G9a (Figure 1B), indicating that G9a is a specific H3K9me2 methyltransferase and does not affect the H3K9me3 chromatin pathway. Nevertheless, it should be noted that if detection of endogenous propionylation is the target of the studies, then these reagents should obviously be avoided.
Figure 1. Quantification of histone post-translational modifications by mass spectrometry.
(A) Quantitative stable isotope labeled analysis of the [M+2H]2+ 9–17 H3 peptide ion (prKme2STGGKprAPR) from control (scrambled siRNA) 521.307 m/z and G9a-depleted 523.822 m/z (siRNA directed toward the methyltransferase G9a). (B) The same analysis, this time of the [M+2H]2+ 9–17 H3 peptide ion (prKme3STGGKprAPR) from control (scrambled siRNA) 528.314 m/z and G9a-depleted 530.829 m/z (siRNA directed toward the methyltransferase G9a). The reduction of the methyltransferase G9a only shows an effect on the abundance levels of H3K9me2, but not H3K9me3.
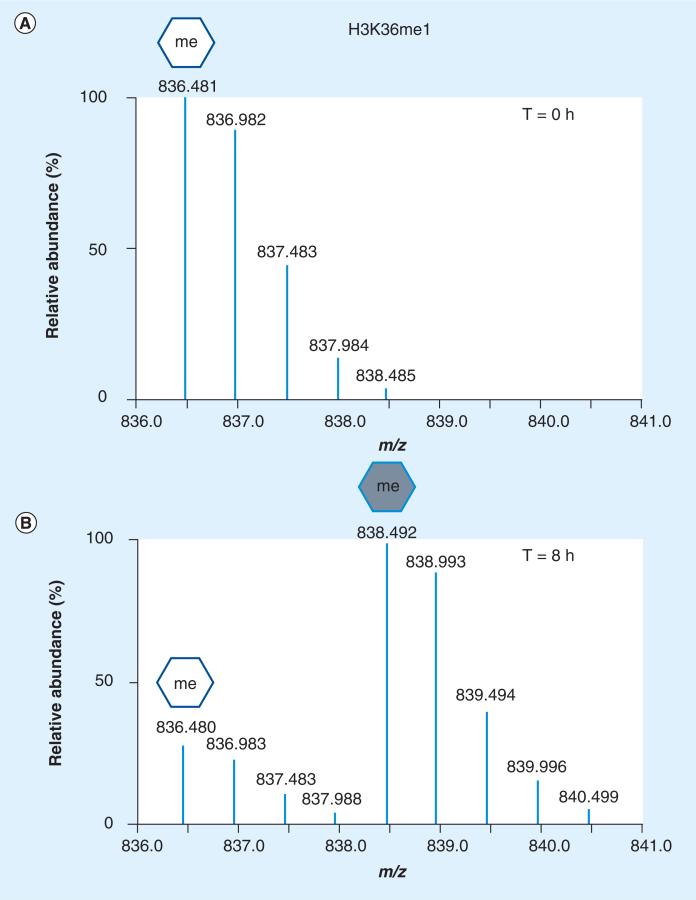
In vivo isotopic labeling of histones and their PTMs has also emerged as a key factor for quantifying abundance level changes. Stable isotopic labeling by amino acids in cell culture (SILAC) has become quite instrumental in capturing a glimpse of the cellular environment, especially when combined with specific induced or isolated biological conditions and MS readout [60,61]. In cell culture, isotopically ‘heavy’ amino acids are used to label one set of proteins with comparison to an unlabeled population. Examples of this methodology executed to find differentially expressed histone PTMs are becoming more prevalent. Bonenfant et al. used SILAC to monitor histone PTM dynamics during the cell cycle and determined that some marks were vastly changing during mitosis (histone H3 and H4 phosphorylation increased and H3K27/K36 methylation decreased) [62]. In similar studies, Mizzen and colleagues found by unlabeled and SILAC labeling that H4K20 methylation progression was tightly linked to cell cycle progression as well [63]. Another study used SILAC labeling and MS to point out the importance of the polycomb repressive complex Suz-12 in promoting the establishment of H3K27me2/me3 in mouse embryonic stem cells, and hinted at a switch between H3K27me3 and H3K27ac in this system [64]. Cuomo et al. also used SILAC approaches to determine potential breast cancer histone PTM signatures [65]. Here, the authors were able to confirm histone PTMs previously linked to cancer, such as a decrease of H4K20me3, but also found other novel histone PTM markers of breast cancer, such as decreased levels of H3K9me3. A slightly different version of SILAC has also been combined with MS experimentation to probe the turnover of histone PTMs and variants [66–68]. Zee et al. used this type of SILAC labeling to estimate the turnover of histone variants and modified forms from HeLa cells grown in standard media conditions and then transferred to ‘heavy’ lysine-containing media with time points collected every day post transfer. l-lysine-13C6 15N2 substituted in lysine-depleted media led to its incorporation in newly synthesized proteins. Here it was shown that certain histone variants such as H2A.Z do indeed have faster turnover rates than other canonical H2A variants [66]. In addition, large differences in half-lives were detected among PTM-modified peptides, with acetyl-containing peptides turning over much faster than methylated histone peptides. If 13CD3-methionine is added to methionine-depleted media, methyl-containing histone and non-histone peptides can be labeled. The cell is forced to use the ‘heavy’ labeled methionine to synthesize S-adenosyl methionine, which is utilized by methyltransferases as the sole donor of methyl groups onto proteins. In this way, all methylation products are isotopically labeled and can be robustly and reproducibly quantified and traced by MS. This methodology was first demonstrated and termed ‘heavy-methyl’ SILAC and shown to be broadly applicable to any protein methylation (Lys or Arg) by the research group of Matthias Mann, and then first applied to histones by Jenuwein and co-workers to label ‘old’ versus ‘new’ histone H3K9me3 [69,70]. More recently, this type of heavy-methyl SILAC approach has been used with MS to determine the turnover of methylated histone peptides under steady-state and from synchronized cell populations [71]. This approach is demonstrated in Figure 2, where the full mass spectra of the doubly charged peptide ion spanning residues 27–40 of histone H3 possessing a monomethylation at K36 are shown. Here, at time 0 (Figure 2A), only the nonlabeled ‘old’ H3K36me1 peptide is detected, but following placement of cells in heavy methionine for 8 h we observe the presence of the ‘newly’ generated H3K36me1 species (Figure 2B) and a degradation of the H3K36me1 peptide, indicating turnover of the ‘old’ methyl form. This heavy-methyl SILAC labeling was used to determine that the dynamic turnover of histone lysine methylation depended on the degrees of methylation, with turnover rates in general following the trend me1 >me2 >me3, and the rates were slower if an acetylation PTM was also present on the peptide. In addition, methylation sites associated with active genes such as H3K36 turnover at more rapid rates than those associated with silenced genes such as H3K27 or H4K20. Combination of both SILAC approaches (protein and methyl labeling) from synchronized cells were used to monitor the progression of H3K79 methylation throughout the cell cycle, and led to the surprising conclusion that new and old histones were mono- and di-methylated at essentially equal rates, with little change among the three histone H3 variants [72].
Figure 2. Histone methylation turnover monitored by ‘heavy-methyl’ stable isotopic labeling by amino acids in cell culture and mass spectrometry.
(A) Full mass spectrum of the [M+2H]2+ 27–40 H3 peptide ion (prKprSAPATGGVKpr,me1KprPHR) monomethylated on K36 from cells growing at various time points in media supplemented with heavy methionine (13CD3). At time 0, only the unlabeled (white hexagon) isotopes are observed. (B) At 8 h, one can quantify other isotopes (gray hexagon) corresponding to the same peptide sequence containing a heavy methyl group (4 Da shift seen as 2 m/z increase), indicating turnover of this peptide.
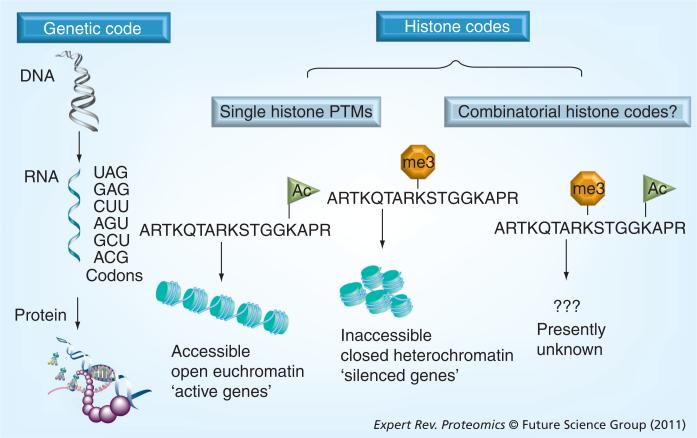
Combinatorial PTM histone codes?
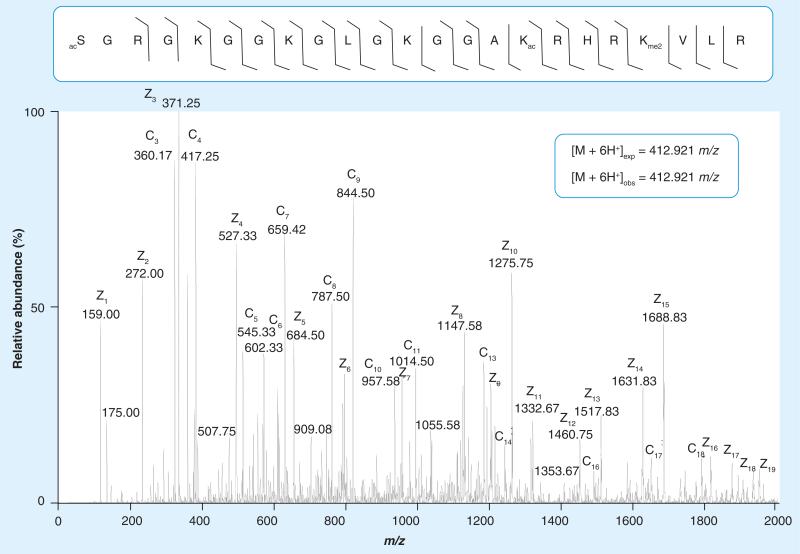
Being a very sensitive approach, bottom-up MS has led to the discovery and quantification of many novel histone PTMs; however, there are some limitations. Since in most bottom-up MS experiments short tryptic-like peptides are analyzed, the detection of simultaneously occurring long-distance PTMs cannot be achieved. Much effort has gone into research to determine how single histone modification sites are specifically linked to gene activation or silencing, but what type of effect unique combinations of simultaneously present modifications (histone codes) have on gene transcription remains unsolved (Figure 3). This challenge is first mainly due to the underdevelopment of tools to detect these combinatorial histone PTMs occurring on the same molecule. To address this problem, middle- and top-down approaches have been developed to examine combinatorial modifications on large polypeptides or intact proteins, respectively [73]. Electron capture dissociation (ECD)-based top-down MS analysis was first used to fragment intact whole histones, giving the first glimpses of the combinatorial PTM complexity of some of these proteins [74,75]. One of the more recent advances in the middle- and top-down approaches was the use of electron transfer dissociation (ETD), which is similar to ECD, but as opposed to CAD, allows more even fragmentation along the length of the peptide regardless of its modification state [76]. Although CAD works as a very effective fragmentation method for smaller peptides in a bottom-up MS workflow and has been used in middle-down experiments, it is somewhat more limited in its efficacy for fragmenting longer peptides, while ECD and ETD seemingly work better for larger peptides with higher charge states [77]. In addition, ETD has advantages over ECD of being able to be routinely performed outside of a Fourier transform ion cyclotron resonance mass spectrometer (FT-ICR-MS), and is extremely compatible with on-line liquid chromatography time-scales. An example of the advantage of using ETD fragmentation of a histone H4 modified form is shown in Figure 4. Here, a near complete fragment map of the 1–23 polypeptide of H4 containing combinatorial marks of N-terminal acetylation, K16ac and K20me2, was generated on an LTQ-Orbitrap instrument equipped with ETD fragmentation. These techniques (ECD and ETD) have allowed for the more precise capture of the combination of PTMs on a particular histone, especially when combined with some type of prefractionation of histone modified forms. For example, the use of ECD-based top-down MS with weak-cation exchange hydrophilic interaction liquid chromatography (WCX-HILIC) separation has shown that in HeLa cells there are 42 distinct types of histone H4 that are differentially methylated and acetylated only on five residues, K5, K8, K12, K16 and K20 [78], and these forms have been monitored through the cell cycle and found to be progressively increased, especially for H4K20 dimethylation. ETD fragmentation performed on an LTQ-Orbitrap instrument has also shown that there are 74 different forms of histone H4 in human embryonic stem cells, each containing a unique combinatorial code, with several changing abundance upon differentiation with 12-O-tetradecanoylphorbol-13-acetate [79]. Other reports have also detailed the combinatorial histone PTMs on histone H4 from other organisms and different physiological conditions. Histone H4 is arguably the easiest histone to work with, possessing the best ionization, separation and fragmentation properties of any histone at both the polypeptide and intact protein level. In addition, there is only one known sequence variant and H4 has a modest and simpler PTM profile than other histones. Hence, it is not surprising that the vast majority of middle- and top-down analyses of histones either focuses on or solely uses histone H4 as proof of principle [80].
Figure 3. Diagram showing the genetic code and potential histone codes.
In the genetic code (far left), genes (DNA sequence) are transcribed into mRNA. mRNA then uses a codon of three nucleotides and is translated on the ribosome into proteins. The histone code can be viewed from both a single PTM and combinatorial PTM point of view. Single PTMs, such as histone H3 Lys14ac, have been linked to active genes, while Lys9me3 on the same protein is found to be enriched in silenced genomic regions (middle). Nevertheless, the transcriptional outcome when both active and silencing PTMs are simultaneously present on the same histone molecule is not understood (combinatorial histone code, right).
PTM: Post-translational modification.
Figure 4. Middle-down electron transfer dissociation mass spectrum of the [M+6H]6+ 1–23 H4 peptide ion (SGRGKGGKGLGKGGAKRHRKVLR) containing N-terminal acetylation, acetylation on K16 and dimethylation on K20.
Lines between amino acid residues denote c and z electron transfer dissociation fragment ions successfully annotated from the spectrum, which were collected in a linear ion trap. Expected and observed precursor ion masses for the +6 charged peptide were collected in the higher resolution Orbitrap.
A greater challenge on PTM complexity lies on the landscape of histone H3, which has been reported to be decorated with a much larger number and variety of modifications than any of the other histones combined [73]. Continued refinement of liquid chromatography separation and fragmentation methods is heartening to the field of proteomics, as they provide more accurately and extensively the PTMs that are simultaneously present on a single protein. For instance, it has been shown that WCX-HILIC coupled to high-resolution MS can be used to determine the combinations of modifications present on single H3 forms [81]. WCX-HILIC separates modified histone forms by first separating out by charge state (or histone acetylation status) and then by hydrophilic content (or histone methylation degree). A middle-down MS platform using HILIC and ECD on an FT-ICR-MS found over 150 distinctly modified histone H3.2 forms [81]. More importantly, these data revealed an interplay between histone H3 acetylation and K4 methylation, with K4 methylation levels increasing robustly on highly acetylated forms. Similar information concerning PTM crosstalk has been indirectly extracted from some bottom-up experiments for histones H3 and H4 [82,83], although the direct measurement of the combinatorial PTM patterns is obtainable through middle-down studies. An online higher-throughput and MS-friendly version of this HILIC chromatography to create a nanoflow LC-MS/MS platform was coupled to an Orbitrap to reveal more than 200 forms of H3.2 and 70 forms of H4 from HeLa S3-derived histones [84]. This method also decreased the amount of starting material needed from over 100 μg to less than 0.5 μg, and sample analysis time from dozens of experiments and over 100 h of MS run time to one single 3-h run, thus potentially allowing for new biological applications of limited sample capacity. The beauty of WCX-HILIC is that it provides very high resolution to differences between modifications that have the same mass shift by MS, such as a trimethyl versus an acetyl group. Other ion exchange methods have been used previously, and allowed for the detection of over 40 modified species from Tetraheymena H3, and also resolved histone H1 phosphorylated isoforms [85,86]. HILIC chromatography alone has been applied to nearly all histones previously, so the potential for this type of separation to uncover new combinatorial PTM motifs and hier archies on these or related proteins remains high. It should be noted that advancements in bioinformatic capabilities have allowed for this complex combinatorial PTM data to be more readily analyzed (both identification and quantification) [87–89]. Continued improvements in computational approaches should allow for even more detailed data mining to be achieved, which will result in the identification of interdependent relationships between unique modified forms. We anticipate that in the near future, computational studies of the epigenetic landscape will become increasingly routine through development of these computational programs. In the advent of genome-wide studies, combinatorial PTMs are emerging as key signatures for cellular transcriptional outputs and should play major roles in defining a true histone code.
Distinguishing histone variants by MS
H1, H2A, H2B, H3 and H4 represent the canonical histones within the nucleoprotein structure of eukaryotic chromatin. However, within this family of proteins, each member has several variants, with the notable exception of histone H4 [90]. Differences within each subset of variants can vary from single amino acid changes to vast divergence among amino acid sequences. Intriguingly, mass shifts of multiples of 14 Da between variants are common and can be erroneously assigned as methylation PTMs, so care has to be made during assignment of variant-specific peptides. An array of mass spectrometric methods is currently available for isolating and separating these variants. Bottom-up MS, which can identify peptides unique to a particular protein, relies heavily on characterization of sequence differences among variants for efficient quantification. The lack of sequence variation among some histone variants, such as H2A.X and canonical H2As, however, make bottom-up analysis difficult. Nonetheless, there have been comprehensive and detailed peptide-centric workflows that have been shown to be effective for identification of histone variants from many organisms. Notably, a couple of bottom-up MS reports have detailed the H2B variants present in Arabidopsis thaliana, and also found a larger number of PTMs on these variants than expected, especially acetylated residues [91]. Other mass spectrometric bottom-up MS experiments have discovered novel histone variants, including two histone H3 variants (H3.Y and H3.X) in addition to the known H3 variants H3.1, H3.2 and H3.3 that may be primate specific [92]. H3.Y depletion affects cellular proliferation and modulates genes involved in cell cycle progression. In addition, bottom-up MS has been used to annotate the PTMs and variants of mammalian H2B, H2A (i.e., macroH2A) or linker histone H1 variants from divergent species and physiological states [93–95].
Nevertheless, top-down and middle-down MS capture the m/z of whole intact proteins and substantially larger polypeptides, respectively, and hold huge promise for histone variant analysis. Before MS detection, the proteins/polypeptides are usually first separated by some type of liquid chromatography. Offline reverse-phase HPLC has been widely used to separate out bulk histone variants, particularly in the cases of H2As, H3s and H1s, which often can resolve into separate peaks. Commonly, these separations have been coupled on-line to MS analysis, making intact protein profiling of histones a rapid and facile experiment [96]. Thorough characterization of histone variants by top-down MS has been performed on all histones to date. At the forefront of these studies is the Kelleher research group, which has published a series of manuscripts, that examine histone variants using ECD and CAD fragmentation in top-down MS mode [97–99]. These reports showed that H2A and H2B families contains a large number of gene products expressed in human cells, the majority of which are not modified and do not vary in abundance through the cell cycle. H3 variants from human, rat and yeast have also been the subject of focus by top and middle-down MS techniques and found to have interestingly minor differences in overall PTM profiles in the mammalian cells [45,98,100]. Specifically, it was found that H3.3 seems to be slightly enriched in PTMs associated with active genes (previously seen by bottom-up MS), and that the major sites of modification were at K9me2, K14ac and K23ac [98]. Histones isolated from Saccharomyces cerevisiae deletion mutants targeting the major histone acetylation complexes such as the SAGA complex revealed that H2B and H3 seemed to be the most-affected histones with much reduced acetylation levels, especially in a GCN5 deletion strain [100]. No information on individual forms could be made by these analyses, as the H3 isoforms were fragmented ‘en masse’; however, these results have been useful to infer some general ‘bulk’ relationships and patterns among the modified residues.
Towards gene-specific histone signatures
To date, nearly all of the MS analyses of histones have been performed from bulk isolated histones (i.e., global histones). However, the value of isolating out a specific gene or genomic region with its associated chromatin protein components would allow for investigations of the local chromatin environment (including histones) from the specific genomic location. This has incredible implications in many fields, most excitingly the specific chromatin analysis of disease-associated genes, and could provide glimpses into the epigenetic components of the disease state. In recent years, both chromatin biologists and proteomic groups have shown in a few independent studies that proteins can be purified with specific genomic loci. Regions of differential transcriptional activity are demarcated by unique chromatin boundaries whose integrity is maintained by binding of chromatin-associated remodeling proteins. Tackett et al. showed that these chromatin boundary complexes could be isolated by targeted affinity purification of the boundary-associated proteins [101]. In particular, the proteins Pol2 and Dpb4 were tagged with protein A and chromatin sub-complexes isolated both with and without their cognate DNA and histones attached. Results showed that disruption of these boundary complexes tested using deletion strains does indeed affect gene expression levels of target genes. Interestingly, these experiments also showed for the first time that minute amounts of histones (here specifically H4 acetylation) isolated from sub-genome-wide populations could be quantified by MS-based proteomics. The addition of SILAC labeling to other isolation approaches have also been useful for identifying gene-specific protein interactions (DNA–protein interactions). Mittler et al. described the use of a DNA affinity probe to determine the specificity of transcription factor binding to various functional DNA sequence elements, and adaptations of this technique could prove useful for isolation of loci-specific histones [102]. Most recently, a technique called ‘proteomics of isolated chromatin segments’ has been demonstrated to be a useful tool for isolation of distinct genomic loci [103]. This method utilizes sequence-specific nucleic acid probes to affinity isolate genomic DNA with its associated chromatin proteins, and was successfully used to map the protein networks of telomeric DNA regions. Methods like these and others ultimately seek to differentiate between distinct genomic sequences that are specific to varying cellular conditions, such as disease states, tumorigenesis and developmental regulation. Techniques for analyzing gene-specific signatures are becoming increasingly sensitive and, if coupled to MS analysis, could be a powerful approach for understanding the molecular mechanism behind specific DNA templated events at both the histone and non-histone level. In addition, ChIP approaches that have been coupled to microarray (ChIP-CHIP) or deep sequencing (ChIP-Seq) are also providing a means of enriching for a more concentrated specific genomic population for downstream MS (ChIP-MS) assessment [21].
Identification of histone-binding proteins
One of the most significant discoveries connecting histone-binding proteins to transcription came when Allis and colleagues identified a Tetrahymena histone acetyltransferase as homologous to the well-known yeast GCN5 transcriptional regulator [104]. Since that time, researchers have used elegant experiments to identify other proteins that modify or bind to very specific histone PTMs [1–3]. As mentioned earlier, the histone code hypothesis postulates that the histone PTMs act as a recruitment platform for proteins that contain specialized domains that bind these modification sites. In support, several proteins that bind specific histone methylation and acetylation sites have been discovered [105]. For instance, HP1 possesses a chromodomain that binds H3K9me3 and this interaction has been shown to be important for maintaining heterochromatic regions by further recruitment of the methyltransferase SuVar39-h1 and other factors [106]. In addition, the double bromodomain-containing proteins are known to bind acetylated histones, and couple histone acetylation to transcription by promoting RNA polymerase II transcription through nucleosomes [107]. However, until recently, these discoveries had been performed by relatively low-throughput through biological experiments. In recent years, MS has been used to identify potential histone PTM binding proteins following affinity purification of the binding protein using synthetically modified histone peptides as baits [108]. However, the large-scale and quantitative nature of proteomics experiments have allowed for rapid screening of potential binding proteins in an unbiased manner. For example, Vermeulen et al. employed SILAC labeling and MS to show that the plant homeodomain finger of Taf3, a component of the TFIID transcription factor, binds H3K4me3 [109]. Disruption of this interaction resulted in loss of transcription and binding of the TFIID complex at a subset of promoters. This work was the first to describe the molecular mechanisms coupling the promoter-specific H3K4me3 mark with the transcription initiation machinery. This methodology has also been repeated by the same group to identify potential binding proteins to several other histone PTMs associated with gene activation or repression, such as H3K36me3 and H4K20me3 [110]. The sensitivity and robustness of LC-MS/MS combined with immunoaffinity have continued to provide a means to identify PTM-specific binding proteins [111]. In addition, histone peptide array proteomic approaches have also emerged as a different means to identify histone PTM binding proteins[112]. We expect that continued optimization of these methods and advances in bioinformatics will progress to large-scale discovery methods, capable of determining the role of multiple histone PTMs and their binding complexes in a systematic fashion.
Expert commentary
Currently, histone PTMs encompass an area of chromatin biology that has far-reaching functional implications in transcription, DNA damage and other nuclear events. In more recent years, MS has become the method of choice for analysis of protein PTMs, and has already made key contributions to the histone field. The advent of combinatorial histone PTM MS has offered the promise of access to gene-specific epigenetic signatures that combine histone PTMs, recruited proteins and chromatin-associated enzymes. In recent years, with the increasing prevalence of ‘method-blending’, ChIP-Seq and ChIP-MS continue to provide critical insight into the chromatin environment. Technical advancements and steadily decreasing costs of these techniques have provided critical access to the molecular interactions between histone PTMs and specific gene targets. These advances allow us to refine our techniques to capture the most transient epigenetic responses to cellular perturbations. However, with the constant enhancement and optimization of mass spectrometric functions, it is clear that we have only now uncovered the proverbial ‘tip of the iceberg’.
Five-year view
Histone PTMs are part of the epigenetic mechanisms that are now being linked to several human disorders and diseases. These PTMs may drive altered gene-expression patterns that in turn affect progression of disease. Therefore, as can be imagined, it will become of increasing importance to quantify histone PTM levels occurring on disease-related genes. Nevertheless, the ability to detect small amounts of sub-genome-wide histone PTMs has so far proven to be a technical challenge. While a few proof-of-principle examples have been highlighted in this article, this type of selective MS analysis of histones remains far from routine. Advances in several areas will be needed to address this challenge, including continued improvements in dynamic range and sensitivity of MS instruments to continue to lower the limit of detection of substoichiometric histone PTMs present in a sample. Concurrent with increased sensitivity for proteomic analyses, the development of oligonucleotide-specific probes that can remove the targeted genomic region with its intact chromatin profile will likely be integral to studying gene-specific histone modifications. Another area of rapid growth in the proteomic community has been adaptation of middle-and top-down MS, and this methodology has made its way to the histone PTM realm in full force. The ability to detect combinatorial histone PTMs is now much easier than it has been before, but the most difficult issue with these analyses still remains: deconvolution of the data. In particular, despite the chromatographic and mass/charge separation of modified histone peptides and proteins provided in the mass spectrum, one still encounters many co-eluting and isobaric analytes that lead to highly complex tandem mass spectrum with multiple-fragment ion series shared and unique to the multiple forms. The parsing of those unique fragment ions is an enormous computational problem that must be solved for deep sequencing of highly modified histone peptides and proteins. The bioinformatics being developed in the post-genome era is transforming our ability to analyze the extremely large data sets that can be a limiting factor in the frequent use of middle-down and top-down MS, which give us the best estimations of gene-specific global histone PTMs and their downstream transcriptional effects. In order for complex relationships and crosstalk between histone PTMs to be deciphered, methods to cluster and mine these data must be created. These methods have been used heavily in the genomics fields, so crossover into the histone proteomics should be accomplished. Histone modifications continue to emerge as a significant epigenetic mechanism potentially affecting both human physiology and disease and, as such, proteomics-based investigations are ideally suited for quantitative exploration of the combinatorial histone PTM space, which may prove vital for precisely determining the existence and rules governing an in vivo histone code.
Key issues.
Histone post-translational modifications (PTMs) affect many nuclear processes, including gene activation and silencing, by recruitment of protein factors that recognize the PTMs with specialized domains.
Site-specific antibodies are the main tool used for analysis of histone PTMs, but suffer from many limitations, including cross-reactivity and epitope occlusion and are not quantitative.
Mass spectrometry has become the most powerful method for the large-scale discovery and quantification of histone PTMs, and has already produced a rich catalog of novel modifications, many of which are the focus of current biological research.
Top- and middle-down mass spectrometry methods are essentially the only tools available to characterize combinatorial histone PTMs simultaneously occurring on the same molecule (the histone code).
Future proteomics research is heading towards interrogation of histones from defined genomic loci. This will require continued improvements in mass spectrometry instrumentation paired with continued development of genomic methods to accurately affinity purify such material and analyze sparse amounts of protein.
Acknowledgments
Benjamin A Garcia gratefully acknowledges support from the American Society for Mass Spectrometry Research award sponsored by the Waters Corporation, NSF Early Faculty CAREER award, NSF grant (CBET-0941143) and an NIH Innovator award (DP2OD007447) from the Office of the Director, NIH. Barry M Zee is supported by an NSF Graduate research fellowship.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Campos EI, Reinberg D. Histones: annotating chromatin. Annu. Rev. Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 4.Magenis RE, Brown MG, Lacy DA, Budden S, LaFranchi S. Is Angelman syndrome an alternate result of del(15) (q11q13)? Am. J. Med. Genet. 1987;28(4):829–838. doi: 10.1002/ajmg.1320280407. [DOI] [PubMed] [Google Scholar]
- 5.Nicholls RD, Knoll JH, Butler MG, Karam S, Lalande M. Genetic imprinting suggested by maternal heterodisomy in nondeletion Prader–Willi syndrome. Nature. 1989;342(6247):281–285. doi: 10.1038/342281a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hasegawa T, Hara M, Ando M, et al. Cytogenetic studies of familial Prader–Willi syndrome. Hum. Genet. 1984;65(4):325–330. doi: 10.1007/BF00291556. [DOI] [PubMed] [Google Scholar]
- 7.Smeets DF, Hamel BC, Nelen MR, et al. Prader–Willi syndrome and Angelman syndrome in cousins from a family with a translocation between chromosomes 6 and 15. N. Engl. J. Med. 1992;326(12):807–811. doi: 10.1056/NEJM199203193261206. [DOI] [PubMed] [Google Scholar]
- 8.Weksberg R, Nishikawa J, Caluseriu O, et al. Tumor development in the Beckwith–Wiedemann syndrome is associated with a variety of constitutional molecular 11p15 alterations including imprinting defects of KCNQ1OT1. Hum. Mol. Genet. 2001;10(26):2989–3000. doi: 10.1093/hmg/10.26.2989. [DOI] [PubMed] [Google Scholar]
- 9.DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am. J. Hum. Genet. 2003;72(1):156–160. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prawitt D, Enklaar T, Gartner-Rupprecht B, et al. Microdeletion of target sites for insulator protein CTCF in a chromosome 11p15 imprinting center in Beckwith–Wiedemann syndrome and Wilms’ tumor. Proc. Natl. Acad. Sci. USA. 2005;102(11):4085–4090. doi: 10.1073/pnas.0500037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amir RE, Van den Veyver IB, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 12.Carney RM, Wolpert CM, Ravan SA, et al. Identification of MeCP2 mutations in a series of females with autistic disorder. Pediatr. Neurol. 2003;28(3):205–211. doi: 10.1016/s0887-8994(02)00624-0. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Freitas MA, Wickham J, et al. Differential expression of histone post-translational modifications in acute myeloid and chronic lymphocytic leukemia determined by high-pressure liquid chromatography and mass spectrometry. J. Am. Soc. Mass Spectrom. 2004;15(1):77–86. doi: 10.1016/j.jasms.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Linggi BE, Brandt SJ, Sun ZW, Hiebert SW. Translating the histone code into leukemia. J. Cell. Biochem. 2005;96(5):938–950. doi: 10.1002/jcb.20604. [DOI] [PubMed] [Google Scholar]
- 15.Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005;37(4):391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 16.Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435(7046):1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 17.Wang GG, Song J, Wang Z, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding. finger. Nature. 2009;459(7248):847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer. 2006;6(1):38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 19.Moving AHEAD with an international human epigenome project. Nature. 2008;454(7205):711–715. doi: 10.1038/454711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10(10):669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr. Biol. 2011;21(1):53–58. doi: 10.1016/j.cub.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brumbaugh J, Phanstiel D, Coon JJ. Unraveling the histone's potential: a proteomics perspective. Epigenetics. 2008;3(5):254–257. doi: 10.4161/epi.3.5.7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trelle MB, Jensen ON. Functional proteomics in histone research and epigenetics. Expert Rev. Proteomics. 2007;4(4):491–503. doi: 10.1586/14789450.4.4.491. [DOI] [PubMed] [Google Scholar]
- 24.Evertts AG, Zee BM, Garcia BA. Modern approaches for investigating epigenetic signaling pathways. J. Appl. Physiol. 2010;109(3):927–933. doi: 10.1152/japplphysiol.00007.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eberl HC, Mann M, Vermeulen M. Quantitative proteomics for epigenetics. ChemBioChem. 2010;12(2):224–234. doi: 10.1002/cbic.201000429. [DOI] [PubMed] [Google Scholar]
- 26.Strahl BD, Briggs SD, Brame CJ, et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 2001;11(12):996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 27.Weiss T, Hergeth S, Zeissler U, et al. Histone H1 variant-specific lysine methylation by G9a/KMT1C and Glp1/KMT1D. Epigenetics Chromatin. 2010;3(1):7. doi: 10.1186/1756-8935-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardner KE, Zhou L, Parra MA, Chen X, Strahl BD. Identification of lysine 37 of histone H2B as a novel site of methylation. PLoS One. 2011;6(1):e16244. doi: 10.1371/journal.pone.0016244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie W, Song C, Young NL, et al. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol. Cell. 2009;33(4):417–427. doi: 10.1016/j.molcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris SA, Rao B, Garcia BA, et al. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J. Biol. Chem. 2007;282(10):7632–7640. doi: 10.1074/jbc.M607909200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker SP, Phillips J, Anderson S, et al. Histone H3 Thr 45 phosphorylation is a replication-associated post-translational modification in S. cerevisiae. Nat. Cell. Biol. 2010;12(3):294–298. doi: 10.1038/ncb2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurd PJ, Bannister AJ, Halls K, et al. Phosphorylation of histone H3 Thr-45 is linked to apoptosis. J. Biol. Chem. 2009;284(24):16575–16583. doi: 10.1074/jbc.M109.005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garske AL, Oliver SS, Wagner EK, et al. Combinatorial profiling of chromatin binding modules reveals multisite discrimination. Nat. Chem. Biol. 2010;6(4):283–290. doi: 10.1038/nchembio.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischle W, Tseng BS, Dormann HL, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438(7071):1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 35.Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438(7071):1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 36.Sakabe K, Wang Z, Hart GW. β-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl Acad. Sci. USA. 2010;107(46):19915–19920. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wisniewski JR, Zougman A, Mann M. Nepsilon-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res. 2008;36(2):570–577. doi: 10.1093/nar/gkm1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Messner S, Altmeyer M, Zhao H, et al. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;38(19):6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, Sprung R, Tang Y, et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics. 2007;6(5):812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng Z, Tang Y, Chen Y, et al. Molecular characterization of propionyllysines in non-histone proteins. Mol. Cell. Proteomics. 2009;8(1):45–52. doi: 10.1074/mcp.M800224-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiong L, Adhvaryu KK, Selker EU, Wang Y. Mapping of lysine methylation and acetylation in core histones of Neurospora crassa. Biochemistry. 2010;49(25):5236–5243. doi: 10.1021/bi1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jufvas A, Stralfors P, Vener AV. Histone variants and their post-translational modifications in primary human fat cells. PLoS One. 2011;6(1):e15960. doi: 10.1371/journal.pone.0015960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tweedie-Cullen RY, Reck JM, Mansuy IM. Comprehensive mapping of post-translational modifications on synaptic, nuclear, and histone proteins in the adult mouse brain. J. Proteome Res. 2009;8(11):4966–4982. doi: 10.1021/pr9003739. [DOI] [PubMed] [Google Scholar]
- 45.Garcia BA, Thomas CE, Kelleher NL, Mizzen CA. Tissue-specific expression and post-translational modification of histone H3 variants. J. Proteome Res. 2008;7(10):4225–4236. doi: 10.1021/pr800044q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl Acad. Sci. USA. 2004;101(6):1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darwanto A, Curtis MP, Schrag M, et al. A modified ‘cross-talk’ between histone H2B Lys-120 ubiquitination and H3 Lys-79 methylation. J. Biol. Chem. 2010;285(28):21868–21876. doi: 10.1074/jbc.M110.126813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith CM, Haimberger ZW, Johnson CO, et al. Heritable chromatin structure: mapping ‘memory’ in histones H3 and H4. Proc. Natl Acad. Sci. USA. 2002;99(Suppl. 4):16454–16461. doi: 10.1073/pnas.182424999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia BA, Mollah S, Ueberheide BM, et al. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat. Protoc. 2007;2(4):933–938. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonaldi T, Imhof A, Regula JT. A combination of different mass spectroscopic techniques for the analysis of dynamic changes of histone modifications. Proteomics. 2004;4(5):1382–1396. doi: 10.1002/pmic.200300743. [DOI] [PubMed] [Google Scholar]
- 51.Plazas-Mayorca MD, Zee BM, Young NL, et al. One-pot shotgun quantitative mass spectrometry characterization of histones. J. Proteome Res. 2009;8(11):5367–5374. doi: 10.1021/pr900777e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robin P, Fritsch L, Philipot O, Svinarchuk F, Ait-Si-Ali S. Post-translational modifications of histones H3 and H4 associated with the histone methyltransferases Suv39h1 and G9a. Genome Biol. 2007;8(12):R270. doi: 10.1186/gb-2007-8-12-r270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drogaris P, Wurtele H, Masumoto H, Verreault A, Thibault P. Comprehensive profiling of histone modifications using a label-free approach and its applications in determining structure–function relationships. Anal. Chem. 2008;80(17):6698–6707. doi: 10.1021/ac800739d. [DOI] [PubMed] [Google Scholar]
- 54.Peters AH, Kubicek S, Mechtler K, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 2003;12(6):1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 55.Chen X, Xiong J, Xu M, Chen S, Zhu B. Symmetrical modification within a nucleosome is not required globally for histone lysine methylation. EMBO Rep. 2011;12(3):244–251. doi: 10.1038/embor.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mandava V, Fernandez JP, Deng H, Janzen CJ, Hake SB, Cross GA. Histone modifications in Trypanosoma brucei. Mol. Biochem. Parasitol. 2007;156(1):41–50. doi: 10.1016/j.molbiopara.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kapoor A, Goldberg MS, Cumberland LK, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468(7327):1105–1109. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ouvry-Patat SA, Schey KL. Characterization of antimicrobial histone sequences and posttranslational modifications by mass spectrometry. J. Mass Spectrom. 2007;42(5):664–674. doi: 10.1002/jms.1200. [DOI] [PubMed] [Google Scholar]
- 59.Plazas-Mayorca MD, Bloom JS, Zeissler U, et al. Quantitative proteomics reveals direct and indirect alterations in the histone code following methyltransferase knockdown. Mol. Biosyst. 2010;6(9):1719–1729. doi: 10.1039/c003307c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ong SE, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 61.Pimienta G, Chaerkady R, Pandey A. SILAC for global phosphoproteomic analysis. Methods Mol. Biol. 2009;527:107–116. x. doi: 10.1007/978-1-60327-834-8_9. [DOI] [PubMed] [Google Scholar]
- 62.Bonenfant D, Towbin H, Coulot M, Schindler P, Mueller DR, van Oostrum J. Analysis of dynamic changes in post-translational modifications of human histones during cell cycle by mass spectrometry. Mol. Cell. Proteomics. 2007;6(11):1917–1932. doi: 10.1074/mcp.M700070-MCP200. [DOI] [PubMed] [Google Scholar]
- 63.Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol. Cell. Biol. 2008;28(1):468–486. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jung HR, Pasini D, Helin K, Jensen ON. Quantitative mass spectrometry of histones H3.2 and H3.3 in Suz12-deficient mouse embryonic stem cells reveals distinct, dynamic post-translational modifications at Lys-27 and Lys-36. Mol. Cell. Proteomics. 2010;9(5):838–850. doi: 10.1074/mcp.M900489-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cuomo A, Moretti S, Minucci S, Bonaldi T. SILAC-based proteomic ana lysis to dissect the ‘histone modification signature’ of human breast cancer cells. Amino Acids. 2011;41(2):387–399. doi: 10.1007/s00726-010-0668-2. [DOI] [PubMed] [Google Scholar]
- 66.Zee BM, Levin RS, Dimaggio PA, Garcia BA. Global turnover of histone post-translational modifications and variants in human cells. Epigenetics Chromatin. 2010;3(1):22. doi: 10.1186/1756-8935-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scharf AN, Barth TK, Imhof A. Establishment of histone modifications after chromatin assembly. Nucleic Acids Res. 2009;37(15):5032–5040. doi: 10.1093/nar/gkp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328(5974):94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 69.Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods. 2004;1(2):119–126. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- 70.Fodor BD, Kubicek S, Yonezawa M, et al. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20(12):1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In vivo residue-specific histone methylation dynamics. J. Biol. Chem. 2010;285(5):3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sweet SM, Li M, Thomas PM, Durbin KR, Kelleher NL. Kinetics of re-establishing H3K79 methylation marks in global human chromatin. J. Biol. Chem. 2010;285(43):32778–32786. doi: 10.1074/jbc.M110.145094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Young NL, Plazas-Mayorca MD, Garcia BA. Systems-wide proteomic characterization of combinatorial post-translational modification patterns. Expert Rev. Proteomics. 2010;7(1):79–92. doi: 10.1586/epr.09.100. [DOI] [PubMed] [Google Scholar]
- 74.Pesavento JJ, Kim YB, Taylor GK, Kelleher NL. Shotgun annotation of histone modifications: a new approach for streamlined characterization of proteins by top down mass spectrometry. J. Am. Chem. Soc. 2004;126(11):3386–3387. doi: 10.1021/ja039748i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Medzihradszky KF, Zhang X, Chalkley RJ, et al. Characterization of Tetrahymena histone H2B variants and posttranslational populations by electron capture dissociation (ECD) Fourier transform ion cyclotron mass spectrometry (FT-ICR MS). Mol. Cell. Proteomics. 2004;3(9):872–886. doi: 10.1074/mcp.M400041-MCP200. [DOI] [PubMed] [Google Scholar]
- 76.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence ana lysis by electron transfer dissociation mass spectrometry. Proc. Natl Acad. Sci. USA. 2004;101(26):9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Coon JJ, Ueberheide B, Syka JE, et al. Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc. Natl Acad. Sci. USA. 2005;102(27):9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pesavento JJ, Bullock CR, LeDuc RD, Mizzen CA, Kelleher NL. Combinatorial modification of human histone H4 quantitated by two-dimensional liquid chromatography coupled with top down mass spectrometry. J. Biol. Chem. 2008;283(22):14927–14937. doi: 10.1074/jbc.M709796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Phanstiel D, Brumbaugh J, Berggren WT, et al. Mass spectrometry identifies and quantifies 74 unique histone H4 isoforms in differentiating human embryonic stem cells. Proc. Natl Acad. Sci. USA. 2008;105(11):4093–4098. doi: 10.1073/pnas.0710515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garcia BA. What does the future hold for top down mass spectrometry? J. Am. Soc. Mass Spectrom. 2010;21(2):193–202. doi: 10.1016/j.jasms.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 81.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nat. Methods. 2007;4(6):487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 82.Zhang K, Williams KE, Huang L, et al. Histone acetylation and deacetylation: identification of acetylation and methylation sites of HeLa histone H4 by mass spectrometry. Mol. Cell. Proteomics. 2002;1(7):500–508. doi: 10.1074/mcp.m200031-mcp200. [DOI] [PubMed] [Google Scholar]
- 83.Zhang K, Siino JS, Jones PR, Yau PM, Bradbury EM. A mass spectrometric ‘Western blot’ to evaluate the correlations between histone methylation and histone acetylation. Proteomics. 2004;4(12):3765–3775. doi: 10.1002/pmic.200400819. [DOI] [PubMed] [Google Scholar]
- 84.Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. High throughput characterization of combinatorial histone codes. Mol. Cell. Proteomics. 2009;8(10):2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Taverna SD, Ueberheide BM, Liu Y, et al. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc. Natl Acad. Sci. USA. 2007;104(7):2086–2091. doi: 10.1073/pnas.0610993104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Garcia BA, Joshi S, Thomas CE, et al. Comprehensive phosphoprotein ana lysis of linker histone H1 from Tetrahymena thermophila. Mol. Cell. Proteomics. 2006;5(9):1593–1609. doi: 10.1074/mcp.M600086-MCP200. [DOI] [PubMed] [Google Scholar]
- 87.DiMaggio PA, Jr, Young NL, Baliban RC, Garcia BA, Floudas CA. A mixed integer linear optimization framework for the identification and quantification of targeted post-translational modifications of highly modified proteins using multiplexed electron transfer dissociation tandem mass spectrometry. Mol. Cell. Proteomics. 2009;8(11):2527–2543. doi: 10.1074/mcp.M900144-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guan S, Burlingame AL. Data processing algorithms for ana lysis of high resolution MSMS spectra of peptides with complex patterns of posttranslational modifications. Mol. Cell. Proteomics. 2010;9(5):804–810. doi: 10.1074/mcp.M900431-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Frank AM, Pesavento JJ, Mizzen CA, Kelleher NL, Pevzner PA. Interpreting top-down mass spectra using spectral alignment. Anal. Chem. 2008;80(7):2499–2505. doi: 10.1021/ac702324u. [DOI] [PubMed] [Google Scholar]
- 90.Talbert PB, Henikoff S. Histone variants – ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 2010;11(4):264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 91.Bergmuller E, Gehrig PM, Gruissem W. Characterization of post-translational modifications of histone H2B-variants isolated from Arabidopsis thaliana. J. Proteome Res. 2007;6(9):3655–3668. doi: 10.1021/pr0702159. [DOI] [PubMed] [Google Scholar]
- 92.Wiedemann SM, Mildner SN, Bonisch C, et al. Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J. Cell. Biol. 2010;190(5):777–791. doi: 10.1083/jcb.201002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bonenfant D, Coulot M, Towbin H, Schindler P, van Oostrum J. Characterization of histone H2A and H2B variants and their post-translational modifications by mass spectrometry. Mol. Cell. Proteomics. 2006;5(3):541–552. doi: 10.1074/mcp.M500288-MCP200. [DOI] [PubMed] [Google Scholar]
- 94.Chu F, Nusinow DA, Chalkley RJ, Plath K, Panning B, Burlingame AL. Mapping post-translational modifications of the histone variant macroH2A1 using tandem mass spectrometry. Mol. Cell. Proteomics. 2006;5(1):194–203. doi: 10.1074/mcp.M500285-MCP200. [DOI] [PubMed] [Google Scholar]
- 95.Wisniewski JR, Zougman A, Kruger S, Mann M. Mass spectrometric mapping of linker histone H1 variants reveals multiple acetylations, methylations, and phosphorylation as well as differences between cell culture and tissue. Mol. Cell. Proteomics. 2007;6(1):72–87. doi: 10.1074/mcp.M600255-MCP200. [DOI] [PubMed] [Google Scholar]
- 96.Su X, Lucas DM, Zhang L, et al. Validation of an LC-MS based approach for profiling histones in chronic lymphocytic leukemia. Proteomics. 2009;9(5):1197–1206. doi: 10.1002/pmic.200800333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Siuti N, Roth MJ, Mizzen CA, Kelleher NL, Pesavento JJ. Gene-specific characterization of human histone H2B by electron capture dissociation. J. Proteome Res. 2006;5(2):233–239. doi: 10.1021/pr050268v. [DOI] [PubMed] [Google Scholar]
- 98.Thomas CE, Kelleher NL, Mizzen CA. Mass spectrometric characterization of human histone H3: a bird's eye view. J. Proteome Res. 2006;5(2):240–247. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 99.Boyne MT, 2nd, Pesavento JJ, Mizzen CA, Kelleher NL. Precise characterization of human histones in the H2A gene family by top down mass spectrometry. J. Proteome Res. 2006;5(2):248–253. doi: 10.1021/pr050269n. [DOI] [PubMed] [Google Scholar]
- 100.Jiang L, Smith JN, Anderson SL, Ma P, Mizzen CA, Kelleher NL. Global assessment of combinatorial post-translational modification of core histones in yeast using contemporary mass spectrometry. Lys4 trimethylation correlates with degree of acetylation on the same H3 tail. J. Biol. Chem. 2007;282(38):27923–27934. doi: 10.1074/jbc.M704194200. [DOI] [PubMed] [Google Scholar]
- 101.Tackett AJ, Dilworth DJ, Davey MJ, et al. Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol. 2005;169(1):35–47. doi: 10.1083/jcb.200502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mittler G, Butter F, Mann M. A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome Res. 2009;19(2):284–293. doi: 10.1101/gr.081711.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dejardin J, Kingston RE. Purification of proteins associated with specific genomic loci. Cell. 2009;136(1):175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brownell JE, Zhou J, Ranalli T, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84(6):843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 105.Daniel JA, Pray-Grant MG, Grant PA. Effector proteins for methylated histones: an expanding family. Cell Cycle. 2005;4(7):919–926. doi: 10.4161/cc.4.7.1824. [DOI] [PubMed] [Google Scholar]
- 106.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410(6824):116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 107.LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol. Cell. 2008;30(1):51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wysocka J, Swigut T, Milne TA, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121(6):859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 109.Vermeulen M, Mulder KW, Denissov S, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131(1):58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 110.Vermeulen M, Eberl HC, Matarese F, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142(6):967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 111.Chan DW, Wang Y, Wu M, Wong J, Qin J, Zhao Y. Unbiased proteomic screen for binding proteins to modified lysines on histone H3. Proteomics. 2009;9(9):2343–2354. doi: 10.1002/pmic.200800600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bua DJ, Kuo AJ, Cheung P, et al. Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PLoS One. 2009;4(8):e6789. doi: 10.1371/journal.pone.0006789. [DOI] [PMC free article] [PubMed] [Google Scholar]