Summary
DNA injected into the C. elegans germline forms extrachromosomal arrays that segregate during cell division [1, 2]. The mechanisms underlying array formation and segregation are not known. Here, we show that extrachromosomal arrays form de novo centromeres at high frequency, providing unique access to a process that occurs with extremely low frequency in other systems [3–8]. De novo centromerized arrays recruit centromeric chromatin and kinetochore proteins and autonomously segregate on the spindle. Live imaging following DNA injection revealed that arrays form after oocyte fertilization via homologous recombination and non-homologous end joining. Individual arrays gradually transition from passive inheritance to active segregation during the early embryonic divisions. The Heterochromatin Protein 1 (HP1) family proteins HPL-1 and HPL-2 are dispensable for de novo centromerization even though arrays become strongly enriched for the heterochromatin-associated H3K9me3 modification over time. Partial inhibition of HP1 family proteins accelerates the acquisition of segregation competence. In addition to reporting the first direct visualization of new centromere formation in living cells, these findings reveal that naked DNA rapidly builds de novo centromeres in C. elegans embryos in an HP1-independent manner, and suggest that, rather than being a prerequisite, HP1-dependent heterochromatin antagonizes de novo centromerization.
Keywords: mitosis, centromere, kinetochore, heterochromatin, microtubule, tubulin, cell division, spindle, CenH3, CENP-A, histone variant, HP1
Results & Discussion
Extrachromosomal Arrays in C. elegans Form Centromeres and Segregate Autonomously
DNA injected into the C. elegans germline forms extrachromosomal arrays that segregate during cell division and can be transmitted across generations [1, 2]. To determine whether extrachromosomal arrays segregate using centromeres [9, 10] or employ an alternative mechanism, such as the “hitchhiking” of double-minute chromosomes and certain viral replicons [11], we constructed arrays by injecting a mixture of two plasmids (Fig. 1A). The first plasmid (p64xLacO) included 64 Lac operator repeats, allowing array visualization using Lac repressor (LacI). The second plasmid (pRF4) encoded the dominant mutant rol-6(su1006) which makes worms roll in a circular pattern (Roller phenotype) [1]. Three independent strains containing propagating arrays (each passed for >5 generations) were generated and arrays were visualized in fixed embryos using recombinant LacI. Typically, 1 or 2 copies of each array were observed per mitotic nucleus (Fig. 1C). Arrays were transmitted with >95% fidelity during embryonic cell divisions. Array inheritance across generations, which requires transmission through the mitotic proliferation and meiotic segregation events that generate the gametes [12, 13], occurred at a frequency of 20–50%. Array size was ~1 Mb based on DAPI staining using the endogenous chromosomes as standards (not shown). Arrays lacked extended telomeric repeats suggesting that they are either circular or have unstable ends (Fig. S1A).
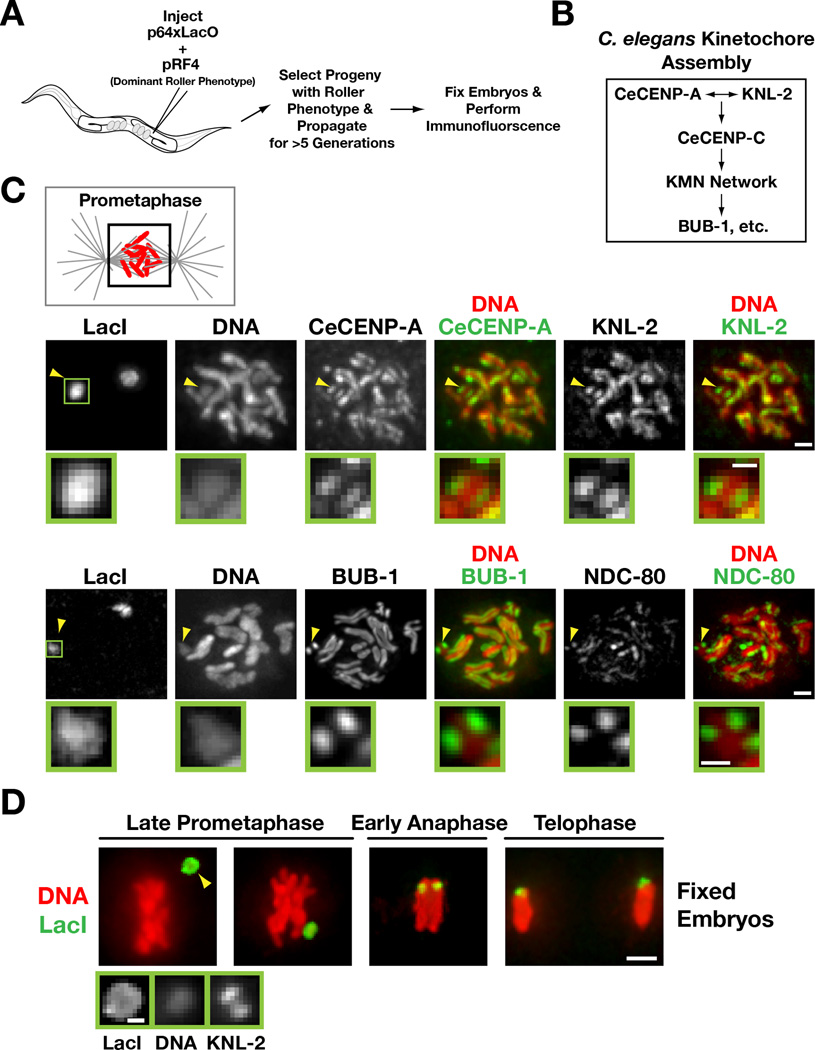
Figure 1. Kinetochores are present on extrachromosomal arrays that have been propagated for multiple generations.
A) Schematic of experimental strategy to analyze array structure by immunofluorescence.
B) Simplified hierarchy of C. elegans kinetochore assembly. The KMN network is comprised of KNL-1, the MIS-12 complex, and the NDC-80 complex.
C) Chromatin-associated inner kinetochore components CeCENP-A and KNL-2 (top row), the microtubule-binding outer kinetochore protein NDC-80 and the spindle checkpoint kinase BUB-1 (bottom row) localize to opposing faces of LacO-containing extra-chromosomal arrays during prometaphase. Arrowheads point to the array; the boxed region is magnified below. Scale bar 1 µm (0.5 µm for magnified regions).
D) Immunofluorescence of the LacO-containing extra-chromosomal array (LacI) and DAPI staining during late prometaphase, early anaphase and telophase in embryos. Higher magnification view of the array (arrowhead) with KNL-2 staining is shown on the bottom. Scale bar 2 µm (0.5 µm for magnified regions).
To determine if arrays form centromeres, we performed immunofluorescence to localize conserved centromere/kinetochore proteins (Fig. 1B; [9, 10]). The centromeric histone CeCENP-A and its conserved assembly factor KNL-2 [14] localized on opposing faces of segregating arrays, in a pattern similar to that on endogenous chromosomes (Fig. 1C). The microtubule-binding kinetochore protein NDC-80, the checkpoint kinase BUB-1 also localized in a similar pattern (Fig. 1C), as did CENP-C and KNL-1 (Fig. S1B). We conclude that arrays build centromeres for segregation, rather than employ a hitchhiking mechanism. Consistent with this, array congression independently of endogenous chromosomes could be observed on the spindle (Fig. 1D). Thus, extrachromosomal arrays formed by DNA injection into the C. elegans germline are de novo centromerized and align and segregate autonomously on the mitotic spindle.
Extrachromosomal Arrays Form After Fertilization
By scoring for the Roller phenotype (Fig. S2A), we found that injected worms produce array-containing embryos for 24 hours beginning ~4 hours after injection, which is when the first oocytes containing injected DNA are fertilized (Fig. S2B). To directly visualize array formation, we imaged the gonad, oocytes, and embryos of worms expressing GFP∷LacI and mCherry∷H2b ~4–8 hours after injection of p64xLacO (Fig. 2A). Uninjected worms contained diffuse nuclear GFP∷LacI in the meiotic nuclei of the gonad, in oocytes, and in embryos (Fig. 2A). In injected worms, the GFP∷LacI signal in gonad nuclei and in oocytes was similar to that in uninjected worms, whereas fertilized embryos contained GFP∷LacI foci that resembled stable arrays. When embryos with transmitting arrays became adults, GFP∷LacI foci were detected in the meiotic and oocyte nuclei in their gonads, as well as in their embryos (Fig. 2A), demonstrating that GFP∷LacI is able to bind to LacO sequences in those tissues. As only p64xLacO was injected, array formation and transmission to progeny does not require endogenous C. elegans DNA (see also [2]).
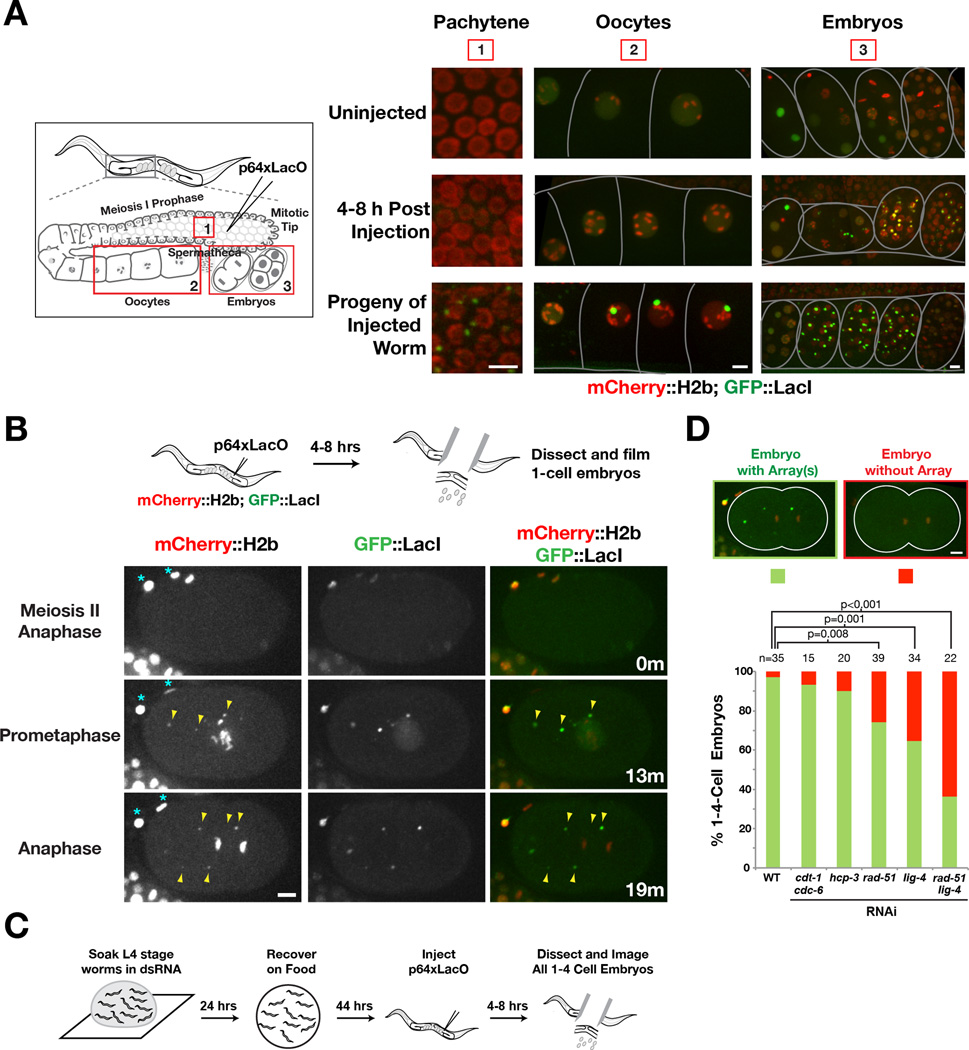
Figure 2. Timing of extrachromosomal array formation.
A) Direct imaging after injection of p64XLacO into the germline of a strain expressing mCherry∷H2b and GFP∷LacI. Worms were anesthetized prior to imaging. The C. elegans germline is a syncytium comprised primarily of nuclei in meiosis I prophase; after the turn, individual nuclear compartments expand and pinch off from the syncytium to form oocytes. Fertilization by sperm stored in the spermatheca generates embryos that are stored in the uterus and eventually released into the environment. Three different regions: 1) meiotic pachytene nuclei in the gonadal syncytium, 2) oocytes, and 3) embryos in the uterus, are shown for the indicated conditions. Scale bars 5 µm.
B) Time lapse images of a recently fertilized embryo dissected 4–8 hrs after p64xLacO injection, showing the appearance of GFP∷LacI foci that also contain mCherry∷H2b (arrowheads) after anaphase of meiosis II. Asterisks mark the polar bodies. Time (in minutes) relative to anaphase of meiosis II is shown. Scale bars 5 µm.
C) Schematic of the experimental approach used to analyze requirements for array formation.
D) Bar graph shows the percentage of 1 to 4-cell embryos with or without arrays in the indicated conditions. Representative images of a 1-cell embryo with or without an array(s) are shown on top. The number of embryos (n) analyzed for each condition is indicated. p values for the indicated comparisons were obtained using a 1-tail Z-test. Scale bars represent 5 µm.
The above results suggested that extrachromosomal arrays are formed after oocyte fertilization rather than in the gonad where the DNA is injected. To confirm this, we imaged embryos produced by injected worms starting around oocyte meiosis II, which occurs ~20 minutes after fertilization [15]; at this stage GFP∷LacI foci were not yet visible (Fig. 2B). Discrete GFP∷LacI foci began to appear in the cytoplasm around prophase of the first mitotic division (Fig. 2B). These foci contained mCherry∷H2b, suggesting that they are chromatinized. Numerous foci were detected per one-cell embryo (range 1–10; average 3; Fig. S2C,D); comparison with 2-cell and 4-cell embryos suggested that foci formation occurred primarily during the first division (Fig. S2C,D). Newly assembled foci did not align and segregate on the spindle. Thus, array formation occurs rapidly after fertilization but the ability to segregate is not acquired coincident with their formation.
To delineate the processes that contribute to array formation, we used RNA interference to inhibit replication initiation (CDT-1 and CDC-6), centromeric chromatin assembly (CeCENP-A/HCP-3), homologous recombination (RAD-51) and non-homologous end joining (LIG-4) (Fig. 2C). Inhibiting replication or centromeric chromatin assembly had no effect (Fig. 2D; Fig. S2E,F). In contrast, inhibiting homologous recombination or non-homologous end joining reduced array formation, consistent with prior analysis of array structure [2]; an additive effect was seen when both pathways were inhibited (Fig. 2D). These results indicate that homologous recombination and non-homologous end joining concatemerize injected DNA in the cytoplasm of the one-cell embryo to form arrays. Why array formation occurs after fertilization and not in the germline where the DNA is injected is currently unclear—this timing may reflect need for the injected DNA to access components restricted to the nuclear compartment.
Extrachromosomal Arrays Acquire Segregation Competency Over Multiple Cell Cycles
Arrays selected over multiple generations show robust segregation in one-cell and later stage embryos (Fig. 1). In contrast, from 30 one-cell embryos imaged 4–8 h after injection (total of ~100 arrays) only one array appeared to segregate (Fig. 2B, 3A; Fig. S3A). These results suggest that arrays must mature to segregate. Consistent with this idea, a higher percentage of arrays in cells from later stage embryos are segregation competent compared to arrays in early embryos (Fig. 3A, S3A).
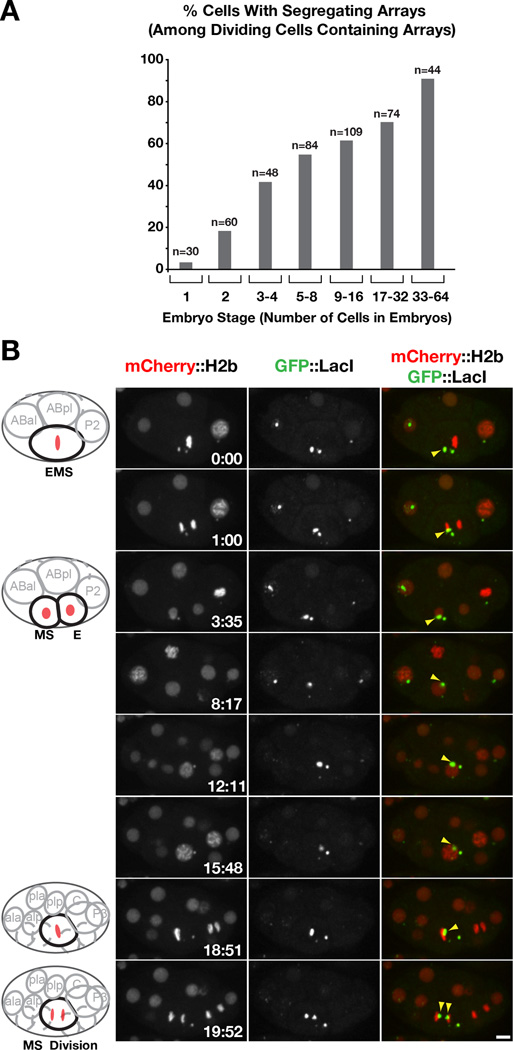
Figure 3. Extrachromosomal arrays acquire segregation competency during early embryonic divisions.
A) Graph showing the percentage of cells with a segregating array among all cells that contained arrays and underwent mitosis during the imaging period; the X-axis indicates the embryo stage. The number of cells (n) analyzed at each stage is indicated.
B) Timelapse images following an array that failed to segregate in the EMS cell (in a 6-cell stage embryo) but 16 minutes later segregated in the daughter MS cell (in a 12-cell stage embryo). See also Fig. S3C. Scale bar 5 µm.
By following individual arrays over multiple cell cycles, we observed six examples of arrays acquiring segregation competence. In each example, the newly formed array initially failed to segregate, passively remaining in one of the two daughter cells during each division, and was then observed to align at the metaphase plate and segregate (Fig. 3B). Once segregation competency was acquired, arrays continued to segregate in subsequent divisions (Fig. S3C). One possibility is that nuclear access by the cytoplasmically formed arrays is required to establish segregation competency. Consistent with this idea, perturbing nuclear envelope disassembly in the early embryonic divisions slowed the acquisition of segregation competence (Fig. S3D–F). Although we attempted to directly visualize centromere/kinetochore assembly, we were unable to directly correlate array maturation with the loading of centromere/kinetochore proteins due to close proximity of segregating arrays to endogenous chromosomes and limitations in imaging centromere/kinetochore proteins in living cells (not shown).
Cumulatively, our results establish that array maturation lags behind array formation likely due to the kinetics of de novo centromerization, which may require nuclear access.
Inhibition of Heterochromatin Protein 1 (HPL-1 and HPL-2) Does Not Prevent De Novo Centromerization of Arrays
Previous work has led to conflicting views on the relationship between heterochromatin and de novo centromere formation. Studies in fission yeast and Drosophila have indicated that new centromere formation requires heterochromatin or a heterochromatin-euchromatin boundary [3, 4, 16–18]. By contrast, studies in mammalian cells have shown that neocentromeres can lack substantial associated heterochromatin domains [19, 20] and have suggested that heterochromatin assembly antagonizes de novo centromerization [21–23]. We therefore used extrachromosomal arrays in C. elegans to investigate the relationship between heterochromatin and de novo centromerization. Single deletion mutations in the genes encoding the two HP1 family proteins HPL-1 (hpl-1(tm1624)) and HPL-2 (hpl-2(tm1489)) are viable, although both mutants exhibit a reduced brood size [24, 25]. The percentage of progeny of pRF4- injected worms exhibiting the Roller phenotype was not affected by either the single or double deletions (Fig. 4A), suggesting that HPL-1 and HPL-2 are not required for array formation or transmission. To analyze de novo centromere formation, we propagated Roller worms generated by injection of a p64xLacO/pRF4 mixture into the hpl-1(tm1624);hpl-2(tm1489) double mutant for multiple generations and performed immunofluorescence. Arrays in the double mutant had CeCENP-A signals on opposing sides (Fig. 4B), similar to arrays in wild-type worms (Fig. 1C). Thus, robust de novo centromerization is observed when both C. elegans HP1 proteins are absent.
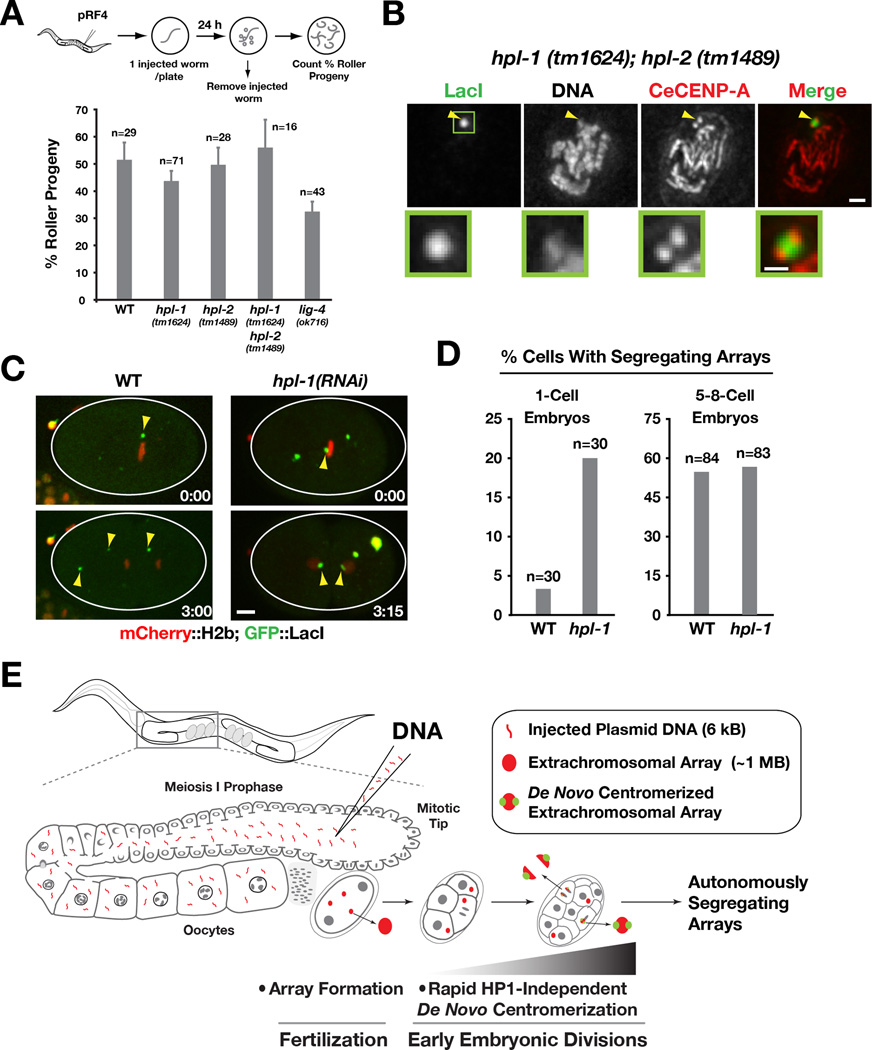
Figure 4. Heterochromatin protein 1 (HPL-1/HPL-2) mutants do not affect array formation or de novo centromerization.
A) Bar graph showing the percentage of Roller progeny produced during the first 24 hrs post-injection of pRF4 in wild-type, hpl-1(tm1624), hpl-2(tm1489), hpl-1(tm1624);hpl-2(tm1489) and lig-4(ok716) mutants. The number of worms analyzed in each condition (n) is indicated. Error bar represents 95% confidence interval for the mean.
B) CeCENP-A localizes to opposing faces of LacO-containing extrachromosomal arrays that have been generated and propagated in the hpl-1(tm1624);hpl-2(tm1489) double mutant. Arrowheads point to the array; the boxed region is magnified below. Scale bar 1 µm (0.5 µm for magnified regions).
C) Examples of 1-cell embryos in wild-type and hpl-1(RNAi), dissected and imaged 4–8 h after p64xLacO injection. hpl-1 RNAi was performed as in Fig. 3B, except that worms were recovered for 20h prior to p64xLacO injection. The wild-type embryo image is the same as in Fig. 2B. Scale bar 5 µm.
D) Bar graph showing the percentage of cells with a segregating array in wild-type and hpl-1 inhibited (using either the mutant or RNAi) at the1-cell stage and at the 5 to 8-cell stage. The number of cells analyzed at each stage (n) is indicated. Note that the Y-axis is modified to facilitate comparison of the two conditions at the two different embryo stages.
E) Model summarizing the key findings. Array formation occurs immediately after fertilization but array centromerization occurs over a longer time scale. HP1 family proteins are dispensable for both array formation and centromerization.
We next analyzed H3K9 methylation (H3K9me3), the modification recognized by HP1 family proteins that is representative of heterochromatin. Arrays selected over multiple generations in wild-type worms exhibited strong H3K9me3 staining (Fig. S4A) [26], indicating compatibility of this heterochromatic mark with centromerization and segregation. In the hpl-1(tm1624);hpl-2(tm1489) double mutant, H3K9me3 staining on arrays was reduced by >90% (signal in the double mutant was 8.5 ± 5.7% (n=11) of that in controls; Fig. S4A). Thus, H3K9me3 is strongly enriched on the propagated repetitive arrays in an HP1-dependent manner but neither this accumulation, nor the HP1 family proteins that bind to it, are essential for de novo centromerization.
Partial Inhibition of Heterochromatin Protein 1 Accelerates Acquisition of Segregation Competence
We next analyzed the consequence of HP1 family protein inhibitions immediately after DNA injection. Inhibiting both HPL-1 and HPL-2 has pleiotropic deleterious effects; therefore, for the acute imaging assays we analyzed single inhibitions. Array formation was not affected by inhibition of either protein by mutation or RNAi (Fig. S4B). To analyze segregation, we chose the single HPL-1 inhibition because it exhibits fewer defects compared to the HPL-2 inhibition, suggesting that it is a weak perturbation of heterochromatin.
In one-cell stage hpl-1(RNAi) embryos, we observed a 6-fold increase in the frequency of segregating arrays compared to controls (Fig. 4C,D); this is a modest effect as only 1 array (from a total of ~100 arrays in 30 embryos) was observed to segregate in controls. This observed increase is not due to increased time spent in the first division or to an increased number of arrays formed following HPL-1 inhibition (not shown). By the 5–8 cell stage, the frequency of segregating arrays was not significantly different between HPL-1-inhibited and wild-type embryos. Thus, weak heterochromatin inhibition accelerates acquisition of segregation competence but does not affect the percentage of arrays ultimately able to segregate. We did not detect significant levels of H3K9me3 on newly formed arrays in the early divisions (Fig. S4C), or even in later stage embryos, suggesting that the strong enrichment observed after propagation for multiple generations (Fig. S4A) occurs on a substantially longer time scale than centromerization.
Cumulatively, the results above show that heterochromatin assembly is not required for extrachromosomal array formation or de novo centromerization in C. elegans embryos and suggest that weakening heterochromatin may accelerate centromerization. These conclusions contrast with the conclusions derived from studies in fission yeast and cultured Drosophila cells. One potential explanation for the difference in the relationship between heterochromatin and de novo centromerization between systems could be the transcriptional status of their genomes. Fission yeast/Drosophila cells in culture are actively transcribing their genomes and experiencing transcription-coupled histone turnover; heterochromatic regions in these cells are silenced and may therefore provide a neighborhood permissive for new CENP-A deposition. In C. elegans, transcription is inhibited during early embryonic divisions [27] and hence heterochromatin may not be important for CENP-A domains to form.
Conclusion
The results described here establish extrachromosomal arrays in C. elegans as a robust model for de novo centromerization. Arrays form soon after fertilization but take additional time to mature for autonomous segregation. Both array formation and transmission can occur in the absence of HP1 family proteins (Fig. 4E). While C. elegans is holocentric, it employs conserved machinery involved in CENP-A targeting and chromosome segregation [14, 28]. Thus, investigation of de novo centromerization in C. elegans has the potential to inform efforts on artificial chromosome engineering in human cells, especially the mechanisms that self-organize CENP-A chromatin to form a platform for kinetochore assembly. In particular, two of the attributes of extrachromosomal arrays—the robust ability to build an autonomous segregating unit independent of DNA sequence and maintain transgene expression from the introduced sequence in somatic cells—are key goals of artificial chromosome engineering for therapeutic delivery of genetic material in humans [29].
Materials and Methods
Worm strains, RNA Interference, DNA Injection
C. elegans strains used in this study are listed in Table S1. All strains were maintained at 20°C, except for PFR40, PFR61, OD568 and OD569, which were maintained at 16°C. Double-stranded RNAs were prepared as described using primers containing T3 and T7 promoters to amplify 500–1000 bp corresponding to the gene regions from genomic DNA or cDNA as templates [30], and RNA interference was performed by soaking larval L4 worms in 5 µl 1µg/ml dsRNAs (Table S2) for 24 hrs in a humidified chamber, and recovering soaked worms on NGM plates seeded with E. coli OP50 [31] for 24 hrs for array segregation experiments or 40 hrs for array formation experiments. 100 ng/µl purified plasmid DNA was injected into gonads of young adult worms using standard methods.
Immunofluorescence
Immunofluorescence microscopy was performed as described previously (Oegema, 2001) using a 20 min cold methanol fixation. Antibodies used against CeCENP-A, KNL-2, CeCENP-C, KNL-1, NDC-80 and BUB-1 were directly labeled with fluorescent dyes (Cy2, Cy3 or Cy5) and used at 1 µg/ml [30, 32]. For Fig. 1C, CeCENP-A and KNL-2 and Fig. 1D, recombinant LacI purified from E. coli was added to the fixed embryos for 90 mins, and then crosslinked in 3% formaldehyde for 15 mins as described (Monen, 2007). Other IF experiments were performed in a strain expressing GFP∷LacI, so no recombinant LacI was added. Antibody against LacI (mouse monoclonal, UPSTATE, 05-503) and antibodies against H3K9Me3 (rabbit polyclonal, Abcam, Ab8898) were used with fluorescent dye-conjugated secondary antibodies. Images were acquired using a 100X, 1.35 NA Olympus U-Plan Apo oil objective and a CoolSnap CCD camera (Roper Scientific) mounted on a Delta Vision deconvolution microscope system (Applied Precision). All fixed images are projections of wide-field Z-planes acquired every 0.2 µm, and deconvolved using Softworx software (Applied Precision).
Live imaging
DNA injected worms were imaged 4–8 hrs after injection. Worms were anesthetized in 1 mg/ml Tricane (ethyl 3-aminobenzoate methanesulfonate salt) and 0.1 mg/ml of tetramisole hydrochloride (TMHC) dissolved in M9 then were transferred to an agarose pad for imaging as described [31]. Gonads and in utero embryos were imaged in 80 × 0.5 µm Z-series. Embryos were dissected in M9 medium and imaged on agar pads, or dissected in meiosis medium and mounted on a metal holder, and imaged in 9 × 1 µm Z-series at 1 min time interval with Yokogawa spinning disk confocal head (CSU-X1) mounted on Zeiss Axio Observer Z1 inverted microscope system equipped with a 63×1.4NA Plan-Apochromat objective and a QuantEM:512SC EMCCD camera (Photometrics). Acquisition parameters, shutters, and focus were controlled by AxioVision software (Zeiss).
Highlights.
DNA injected in germline transforms into extrachromosomal arrays after fertilization
Extrachromosomal arrays form de novo centromeres and acquire segregation competency
HP1 family proteins are dispensable for de novo centromere formation
Partial inhibition of HP1 accelerates de novo centromere formation
Supplementary Material
Acknowledgements
We thank Anne Villenueve, in whose lab the GFP∷LacI strain was generated, Diego Folco for useful discussions, Becky Green and other members of the Oegema/Desai labs for comments on the manuscript. This work was supported by a grant from the NIH to A.D. (GM074215) and a fellowship to K.W.Y.Y. from the Croucher Foundation (Hong Kong). K.O. and A.D. receive salary and additional support from the Ludwig Institute for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: K.W.Y.Y. designed and conducted all of the experiments with input from A.D. and K.O. K.N. generated the GFP∷LacI strain. K.W.Y.Y., A.D., and K.O. wrote the manuscript.
References
- 1.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stinchcomb DT, Shaw JE, Carr SH, Hirsh D. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol Cell Biol. 1985;5:3484–3496. doi: 10.1128/mcb.5.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- 5.Ketel C, Wang HS, McClellan M, Bouchonville K, Selmecki A, Lahav T, Gerami-Nejad M, Berman J. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 2009;5:e1000400. doi: 10.1371/journal.pgen.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mejia JE, Alazami A, Willmott A, Marschall P, Levy E, Earnshaw WC, Larin Z. Efficiency of de novo centromere formation in human artificial chromosomes. Genomics. 2002;79:297–304. doi: 10.1006/geno.2002.6704. [DOI] [PubMed] [Google Scholar]
- 7.Nakashima H, Nakano M, Ohnishi R, Hiraoka Y, Kaneda Y, Sugino A, Masumoto H. Assembly of additional heterochromatin distinct from centromere-kinetochore chromatin is required for de novo formation of human artificial chromosome. J Cell Sci. 2005;118:5885–5898. doi: 10.1242/jcs.02702. [DOI] [PubMed] [Google Scholar]
- 8.Williams BC, Murphy TD, Goldberg ML, Karpen GH. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat Genet. 1998;18:30–37. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]
- 9.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 10.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanda T, Otter M, Wahl GM. Mitotic segregation of viral and cellular acentric extrachromosomal molecules by chromosome tethering. J Cell Sci. 2001;114:49–58. doi: 10.1242/jcs.114.1.49. [DOI] [PubMed] [Google Scholar]
- 12.Hubbard EJ, Greenstein D. Introduction to the germ line. WormBook. 2005:1–4. doi: 10.1895/wormbook.1.18.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schvarzstein M, Wignall SM, Villeneuve AM. Coordinating cohesion, co-orientation, and congression during meiosis: lessons from holocentric chromosomes. Genes Dev. 24:219–228. doi: 10.1101/gad.1863610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNally KL, McNally FJ. Fertilization initiates the transition from anaphase I to metaphase II during female meiosis in C. elegans. Dev Biol. 2005;282:218–230. doi: 10.1016/j.ydbio.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, Simmer F, Urano T, Hamilton GL, Allshire RC. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science. 2009;234:1716–1719. doi: 10.1126/science.1172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olszak AM, van Essen D, Pereira AJ, Diehl S, Manke T, Maiato H, Saccani S, Heun P. Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat Cell Biol. doi: 10.1038/ncb2272. [DOI] [PubMed] [Google Scholar]
- 19.Alonso A, Hasson D, Cheung F, Warburton PE. A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin. 3:6. doi: 10.1186/1756-8935-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimes BR, Babcock J, Rudd MK, Chadwick B, Willard HF. Assembly and characterization of heterochromatin and euchromatin on human artificial chromosomes. Genome Biol. 2004;5:R89. doi: 10.1186/gb-2004-5-11-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 23.Okamoto Y, Nakano M, Ohzeki J, Larionov V, Masumoto H. A minimal CENP-A core is required for nucleation and maintenance of a functional human centromere. EMBO J. 2007;26:1279–1291. doi: 10.1038/sj.emboj.7601584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Couteau F, Guerry F, Muller F, Palladino F. A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep. 2002;3:235–241. doi: 10.1093/embo-reports/kvf051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schott S, Coustham V, Simonet T, Bedet C, Palladino F. Unique and redundant functions of C. elegans HP1 proteins in post-embryonic development. Dev Biol. 2006;298:176–187. doi: 10.1016/j.ydbio.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 26.Bean CJ, Schaner CE, Kelly WG. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat Genet. 2004;36:100–105. doi: 10.1038/ng1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar LG, Wolf N, Wood WB. Early transcription in Caenorhabditis elegans embryos. Development. 1994;120:443–451. doi: 10.1242/dev.120.2.443. [DOI] [PubMed] [Google Scholar]
- 28.Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, 3rd, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macnab S, Whitehouse A. Progress and prospects: human artificial chromosomes. Gene Ther. 2009;16:1180–1188. doi: 10.1038/gt.2009.102. [DOI] [PubMed] [Google Scholar]
- 30.Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green RA, Kao HL, Audhya A, Arur S, Mayers JR, Fridolfsson HN, Schulman M, Schloissnig S, Niessen S, Laband K, et al. A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell. 2011;145:470–482. doi: 10.1016/j.cell.2011.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Hyman A, Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C.elegans. Genes Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.