Abstract
Human TNFAIP3 interacting protein 1 (TNIP1) has diverse functions including support of HIV replication through its interaction with viral Nef and matrix proteins, reduction of TNFα-induced signaling through its interaction with NF-κB pathway proteins, and corepression of agonist-bound retinoic acid receptors and peroxisome proliferator-activated receptors (PPAR). The wide tissue distribution of TNIP1 provides the opportunity to influence numerous cellular responses in these roles and defining control of TNIP1 expression would be central to improved understanding of its impact on cell function. We cloned 6kb of the human TNIP1 promoter and performed predictive and functional analyses to identify regulatory elements. The promoter region proximal to the transcription start site is GC-rich without a recognizable TATA box. In contrast to this proximal ~500bp region, 6kb of the promoter increased reporter construct constitutive activity over five-fold. Throughout the 6kb length, in silico analysis identified several potential binding sites for both constitutive and inducible transcription factors; among the latter were candidate NF-κB binding sequences and peroxisome proliferator response elements (PPREs). We tested NF-κB and PPAR regulation of the endogenous TNIP1 gene and cloned promoter by expression studies, electrophoretic mobility shift assays, and chromatin immunoprecipitations. We validated NF-κB sites in the TNIP1 promoter proximal and distal regions as well as one PPRE in the distal region. The ultimate control of the TNIP1 promoter is likely to be a combination of constitutive transcription factors and those subject to activation such as NF-κB and PPAR.
Keywords: PPAR, NF-κB, promoter, transcriptional regulation, response element
1. Introduction
The repertoire of possible roles for TNFAIP3 interacting protein 1 (TNIP1) is steadily increasing especially with its recent identification in genome-wide studies of gene-disease associations in lung cancer and psoriasis [1, 2]. TNIP1 is the NCBI designation for the protein alternatively known as NAF1, VAN, or ABIN-1. TNIP1 interacts with two HIV-encoded proteins, Nef and matrix; its isolation as the Nef-associated factor (NAF) was the initial eponymic report [3]. Later, it was also found to interact with HIV matrix and participate in the translocation of that protein from the cytoplasm to the nucleus, a step necessary for HIV replication. With this function identified [4], the protein was named virion-associated matrix-interacting nuclear shuttling protein (VAN). Over-expression of TNIP1 inhibits HIV replication [4] and increases cell-surface CD4 expression [3]. NF-κB activity is reduced by TNIP1 [5–9] and in these reports is referred to as ABIN-1, for A20-binding inhibitor of NF-κB. Mouse cells deficient for TNIP1 are hypersensitive to apoptosis following exposure to tumor necrosis factor α (TNFα) [10]. While these studies provide seminal advances in understanding TNIP1 protein function, little has been reported to date on characterization and control of its promoter, a deficit we sought to address in this study by cloning 6kb of the human TNIP1 promoter and identifying transcriptional regulatory elements within it.
Expression of TNIP1 and A20, one of its protein-protein interaction partners also known as TNF-α induced protein 3 (TNFAIP3), are up-regulated by NF-κB activation [11]. This association suggests a feedback system where a negative regulator of a signaling pathway is induced by that system. We recently identified TNIP1 as a corepressor of retinoic acid receptors (RARs) [12] and peroxisome proliferator-activated receptors (PPARs) (Flores et al, accepted). Its ligand requirement for interaction with RARs or PPARs but repression of their activity sets it apart from standard nuclear receptor (NR) corepressors. TNIP1 aligns with a growing group of ligand-dependent corepressors such as RIP140 and REA whose broad tissue distribution and impact on pharmacology, endocrinology and malignancy are demonstrating the importance of this atypical coregulator group [13 for review]. Like many of these atypical corepressors, amino acid motifs in TNIP1 required for ligand-dependent interaction with PPARs and RARs are actually characteristic of classical nuclear receptor coactivators and yet TNIP1 interaction leads to reduction in PPAR (Flores et al, accepted) and RAR [12] activity. Surprisingly, TNIP1 requires an intact activation function 2 (AF-2) domain in PPARs and RARs, a region in nuclear receptors otherwise expected to associate with coactivators.
Controlling the expression of NR coregulators has received much less attention than the protein-protein interactions of NRs and their coregulators. In this vein, it is important to note NR coregulator expression levels are frequently altered [14] during development, cell differentiation, tissue specialization or malignant conversion. Some are even governed by the ligand of the hormone receptor they coregulate [15, 16]. TNIP1 is expressed across different adult human cell and tissue types but at varying levels [3, 4]; what promoter regulatory elements could account for this were a focus of our investigation.
The diverse functions associated with TNIP1, participating in HIV replication, reducing NF-κB signaling, and repressing NR ligand-induced transcription, make understanding regulation of its promoter essential. We addressed this by cloning 6kb of the human TNIP1 gene upstream sequence and testing putative regulatory sites for transcriptional control of its expression. The nucleotide sequence of the TNIP1 promoter is increasingly GC-rich approaching the transcription start site region, characteristic of promoters without recognizable TATA boxes. Our novel findings include PPAR and NF-κB regulation of the TNIP1 promoter via elements within the distal region of the defined promoter; additionally, we confirmed NF-κB responsiveness in a smaller TNIP1 promoter clone as described previously [11]. We found binding of PPAR and NF-κB to synthetic TNIP1 promoter sequences in vitro as well as to the endogenous TNIP1 promoter in cells. We further found TNIP1 mRNA level to be increased in response to PPARγ. Positive control of TNIP1 expression by the transcription factors (NF-κB and PPAR) whose function is repressed by its protein may set the stage for regulatory feedback in normal cell physiology or disease states where NF-κB or PPAR signaling is altered.
2. Materials and methods
2.1. Identification, cloning, and in silico analysis of TNIP1 gene upstream sequence
Published and GenBank TNIP1 transcript sequences, a human chromosome 5 genomic contig, and transcripts from an Ensembl release (http://www.ensembl.org/Homo_sapiens/contigview?region=AC008641.7.1.176629) were used to identify bacterial artificial chromosomes containing relevant sequence upstream [11, 17] of the TNIP1 coding region. The 6kb, 3kb and 549bp promoter regions were generated via PCR from bacterial artificial chromosome CTB-35A8 (Invitrogen, Carlsbad, CA) with primers in Table 1 and sequenced at the UConn DNA Biotechnology Facility. Luciferase constructs were made by moving these three regions as Sal I – Xho I fragments into the Xho I site of promoterless pGL4.10 (Promega, Madison WI).
Table 1.
Primers for TNIP1 promoter cloning.
| Construct Name | Promoter Segment | Forward & Reverse | Cloning Sites |
|---|---|---|---|
| −549bp | −549 ~ +111 | TTTAAATCTAGAAATGCTTACGTGCCTTTTGG ATTTAAACTCGAGGGGAGCTTGGGGACACAG |
Xba I, Xho I |
|
|
|
|
|
| −3kb | −3106 ~ +111 | CAGCCTCCTTCTCCACTGTC ATTTAAACTCGAGGGGAGCTTGGGGACACAG |
Nhe I, Xho I |
|
|
|
|
|
| −6kb | −5887 ~ +111 | TTTAAAGTCGACCAGCCTCCTTCTCCACTGTC ATTTAAACTCGAGGGGAGCTTGGGGACACAG |
Sal I, Xho I |
|
|
|
|
|
Primers are all in 5′ → 3′ orientation. PCR products from the bacterial artificial chromosome template were digested with indicated restriction enzymes and cloned into the promoterless luciferase plasmid pGL-4.10 at the indicated or overhang compatible sites. Bolded nucleotides in primers indicate relevant restriction enzyme sites. The Nhe I site for cloning the 3kb fragment is internal to the amplicon.
The 6kb fragment was analyzed with the public domain programs MatInspector [18], Transfac [19], NUBIScan [20], and NHRScan [21]. Potential PPREs were compared against a consensus derived from human gene promoters [22–25] and visualized with WebLogo. The 3xC TNIP1 PPRE reporter was constructed as a three-times repeat of 5′-TCA CTT TGC CCT TTC CTC TCC TCC AGC TAG-3′ flanked by Spe I and Nhe I sites for cloning into pGL4.10 (TNIP1 PPRE element C in bold italics) with the thymidine kinase (tk) minimal promoter. Individual oligomer pairs, top and bottom strand, for self-multimerization, e.g., 3xA PPRE, or triplet multimerization, e.g., TNIP1 A-B-C PPRE, are described in Table 3. DNA sequencing confirmed number, sequence, and orientation of all oligomer inserts in the luciferase reporter. The distal NF-κB site and PPRE element C were subjected to site-directed mutagenesis using forward 5′-GGC CAC GTA TGT GAC GAG GCT ttt aat tga CCC CAG TGT CAC TTT GCC CTT TC-3′ and reverse 5′-GAA AGG GCA AAG TGA CAC TGG GGt caa tta aaA GCC TCG TCA CAT ACG TGG CC-3′, and forward 5′-GGA ATT TCC CCC AGT GTC ACT TCG CCt gTg tCT Cgc CTC CAG TGG TGC CCT AGA A-3′ and reverse 5′-TTC TAG GGC ACC ACT GGA Ggc GAG acA caG GCG AAG TGA CAC TGG GGG AAA TTC C-3′ primers (lower case letters denote mutant nucleotides), respectively, and QuikChange reagents (Stratagene, Agilent Technologies, Inc., Santa Clara, CA).
Table 3.
Oligomers containing candidate TNIP1 promoter PPREs.
| Predicted PPRE | Top & Bottom |
|---|---|
| A | ctagGAATGATGAAGAAAGGCCAGTTCATCCAAAg ctagcTTTGGATGAACTGGCCTTTCTTCATCATTC |
|
|
|
| B | ctaggCCTGGCTGACCTGTTCCCACTTCCAGGAGGg ctagcCCTCCTGGAAGTGGGAACAGGTCAGCCAGGc |
|
|
|
| C | ctagTCACTTTGCCCTTTCCTCTCCTCCAg ctagcTGGAGGAGAGGAAAGGGCAAAGTGA |
|
|
|
Oligomers are all in 5′ → 3′ orientation. Lower case nucleotides at oligomer ends indicate Nhe I compatible overhang, at 5′ end, and overhang and regeneration of Nhe I site, at 3′ end to allow for step-wise multimerization with subsequent PPRE oligomer. Nucleotides in capitals are from TNIP1 promoter sequence with DR1 core of predicted PPRE in bold type. The sequence for element C was also used in EMSA.
2.2. Electrophoretic Mobility Shift Assay (EMSA)
Bcl I (NEB, Beverly, MA) fragments of the TNIP1 distal 3kb promoter were prepared from cloned DNA propagated in GM2163 (Dam−) E. coli. Synthetic oligomers (Integrated DNA Technologies, Inc., Coralville, IA) were annealed and used as probes or competitors. Restriction fragments and double-stranded oligomers used as probes were labeled with α-32P dATP (GE Healthcare, Piscataway, NJ) at single-stranded overhangs by a T4 DNA polymerase (Promega) fill-in reaction. Unincorporated nucleotides were removed by column chromatography (Qiagen, Valencia, CA). For the variant EMSA, element Z, derived from the known PPRE in the ω-hydroxylase promoter, was used as a positive control [26]. It is present as a triple repeat in a Spe I to Nhe I fragment along with plasmid backbone sequence (702bp total) in the 3xZ probe EMSAs and a single repeat in the Z oligomer-based EMSAs. The top and bottom strands of the ω-hydroxylase Z element with single strand overhangs for labeling are 5′-CTA GGC-GCA AAC ACT GAA CTA GGG CAA AGT TGA GGG CAG-3′ and 5′-CTA GCT GCC CTC AAC TTT GCC CTA GTT CAG TGT TTG CGC-3′ [27]. The GL4 competitor is the analogous Spe I to Nhe I restriction fragment from pGL4.10 without PPRE inserts. For EMSA purposes, top and bottom strands for oligomers including the TNIP1 distal NF-κB site are 5′-CTA GCG AGG CTG GGA ATT TCC CCC AGT-3′ and 5′-CTA GAC TGG GGG AAA TTC CCA GCC TCG-3′.
Commercially available antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) for PPARγ (sc-7273), p65 RelA (sc-372), or control reactions (anti-Gal4 DNA binding domain, sc-510) were used for EMSA. Scrambled element sequences are same length and composition as the original but with the nucleotides in a random order generated by Shuffle DNA (http://eyegene.ophthy.med.umich.edu/shuffle/). Nuclear extract, protein:DNA binding and electrophoresis conditions have been described [28] with the reaction buffer adjusted to final concentrations of 0.25μg/μl BSA, 0.1μg/μl poly d(I-C) (Roche, Indianapolis, IN) and 0.5% Igepal (Sigma, St. Louis, MO).
2.3. Gene expression analysis
Twenty hours after seeding 1×104 cells per well of a 96 well plate, in triplicate, for each biological condition, HeLa cells were infected by adenovirus LacZ or adenovirus PPARγ, for 8h, followed by media supplementation with either vehicle (DMSO, 0.1% final concentration) or the PPARγ ligand troglitazone (5μM, final concentration). Cells were collected at 48 hr post-ligand treatment, using TaqMan Gene Expression Cells-to-CT Kit (ABI, Carlsbad, CA) for lysis and reverse transcription. Quantitative real-time PCR was performed using ABI TaqMan Gene Expression assays with the following human gene probes, TNIP1, Hs00374581_m1; ADRP, Hs00765634_m1; Human RPLP0 Endogenous Control, and run on an ABI 7500 Fast Real-Time PCR System with software 7500 Version 2.0. Analysis was performed from technical triplicates using the Comparative CT (ΔΔCT) protocol and represented as relative quantitation (RQ) with the RPLP0 as an endogenous control to normalize the amount of cDNA per reaction.
2.4. Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed with chromatin isolated from HeLa cells processed with a two-step fixation protocol utilizing first disuccinimidyl glutarate and then formaldehyde as described previously [29] and in consideration of prior publications regarding potential limits in detection of NF-κB [11] and nuclear receptor [30] binding to promoters. For ChIP of the candidate PPRE, HeLa cells were seeded at 5 × 106 per 10 cm plate and infected with adenovirus expressing either β-galactosidase (LacZ) as a negative control or the human cDNA for PPARγ, treated with 5 μM troglitazone 6 hr post-infection and cross-linked 42 hr later based on expression levels of PPARγ. For ChIP of candidate NF-κB binding sites, HeLa cells were seeded at 5 × 106 per 10cm plate, with 0.5% bovine serum albumin and no FBS, and treated 24 hr later with 30 ng/ml TNFα (Pierce, Rockford, IL) for one hour prior to the two-step fixation. Subsequent steps were as described [11, 29] with rabbit antibodies to the following: p65, sc-372X; PPARγ, sc-7196X; normal immunoglobulin, sc-2027, all from Santa Cruz. PCR cycling conditions and primers are listed in Table 2. Amplicons were resolved on an ethidium bromide stained 2% agarose gel.
Table 2.
ChIP assay primers.
| Target | Forward & Reverse | °C | Size |
|---|---|---|---|
| TNIP1 distal | GAGACCCAGGGCTCCTAGGAACTGT CCAACTATTTTCCCTGTGTCCCTG |
58 | 164 |
|
|
|
|
|
| TNIP1 proximal | ACAAGGAAGGGGCTGGGTGGTCT GAGGTAAACACCGGCTGCCTGCG |
64 | 226 |
|
|
|
|
|
| ADRP | GCAAAAAGAAGCTTGCTCAG TGTTGCCATCTTCAGTGTTT |
58 | 250 |
|
|
|
|
|
| GAPDH | ATGGTTGCCACTGGGGATCT TGCCAAAGCCTAGGGGAAGA |
58 | 174 |
|
|
|
|
|
| IκBα | GACGACCCCAATTCAAATCG TCAGGCTCGGGGAATTTCC |
58 | 300 |
Promoter regions of named genes are indicated as Target. Primers are all in 5′ → 3′ orientation with annealing temperatures indicated. PCR product base pair sizes are as predicted from UCSC in silico PCR using the human March 2006 assembly. Other conditions for 32 cycles were denaturation, 94°C, 20sec; annealing time, 30sec; extension temperature and time, 72°C, and 30sec; with a final extension of 72°C, 2min. Primers for TNIP1 distal and proximal are our design; primers for human ADRP, GAPDH [47], and IκBα [11], have been previously reported.
2.5. Cell culture and transfection
HeLa cells (ATCC, Manassas, VA) were maintained in a 3:1 mix of DMEM:F12, 10% FBS with 100units/ml penicillin and 100 μg/ml streptomycin; all culture reagents were from Invitrogen. Cells were seeded at 0.5×105 per well in 24-well plates but with 2.5% charcoal-stripped FBS (HyClone, Logan, UT), transfected 20 hr later with a total of 0.4 μg DNA per well using Fugene 6 (Roche) reagent:DNA ratios as per supplier’s directions. Transfections with differing length TNIP1 promoter constructs were normalized to have equal copies of TNIP1 promoter plasmid. Total DNA was standardized by carrier DNA or empty vector as appropriate. For PPAR activation, cells were re-fed vehicle control (0.1% DMSO) or ligand-containing media 10 μM WY14,643 (ChemSyn, Lenexa, KS), 1 μM L165,041 (Sigma) or 5 μM troglitazone (Cayman Chemical, Ann Arbor, MI) 18 hr post-transfection. RXRα activation was via 1 μM 9-cis retinoic acid (RA). Transfection with NF-κB refers to a 1:1 DNA combination of p50 and p65 CMV-driven expression constructs.
Relative light units (RLU) were determined on a LMaxII luminometer (Molecular Devices, Sunnyvale, CA) with commercial reagents (Promega) and normalized as previously described [31] with each transfection DNA combination performed in duplicate or triplicate. Transfections were conducted 2–3 times and showed consistent results across trials. The luciferase reporters with conalbumin minimal promoter alone or with the 3xNF-κB insert and the tk promoter alone or with the 3xZ element from ω-hydroxylase PPRE have been described [26, 32].
2.7. Statistical analyses
Statistical analysis was performed with Graphpad (La Jolla, CA) Prism software. Specific tests and p values are indicated in figure legends.
3. Results
3.1. Cloning, in silico analysis, and candidate transcription factor binding sites
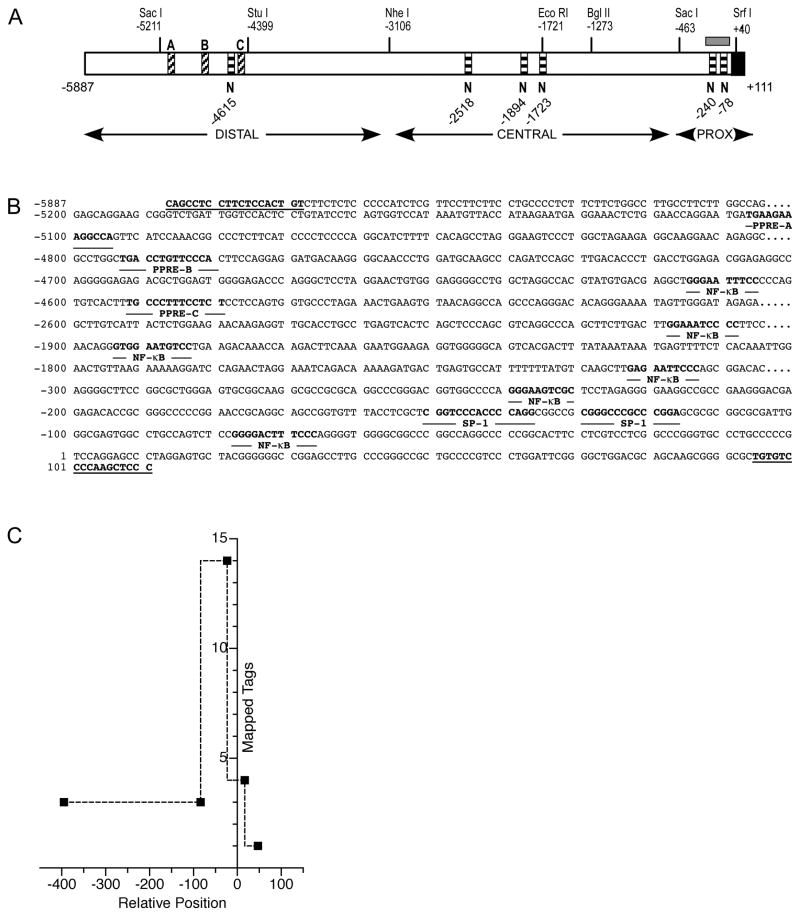
A 5998bp fragment (hereafter referred to as 6kb) upstream from the TNIP1 expected transcription start site was cloned from a bacterial artificial chromosome (see Materials and Methods), sequenced, and examined in silico for features characteristic of promoter function. TNIP1 promoter GC percentage strikingly rises approaching the transcription start site. Considering sequence from −1000 to +1, −600 to +1, and −200 to +1, the GC content is approximately 66, 74 and 79%, respectively, versus 50% throughout the total 6kb region. A CpG island overlaps the start site, extending from approximately −250 to −50 (Figure 1A). Two predicted SP1 sites within 200bp upstream of the transcription start site (−151 and −130) reflect the GC-rich nature of this region. The region lacks a recognizable consensus TATA box which is typical for GC-rich promoters [33].
Fig. 1. In silico identification of transcription factor binding sites in the human TNIP1 promoter.
(A) Schematic of 5′ region of human TNIP1 gene. Numbering is based on the ~600bp promoter isolated by Brasier et al [11]. Landmark restriction enzyme sites are shown. The Nhe I separates the distal region from the remainder of the promoter. Candidate NF-κB sites (indicated by letter N at nucleotide position and horizontal stripes) are marked under schematic. To avoid labels overlapping at this scale, all predicted NR response elements are indicated in Figure 3; candidate PPREs (indicated by letters A, B, and C above schematic and diagonal stripes) in the distal region are indicated. A CpG island (grey box) is shown over proximal region. Distal, Central and Proximal (PROX) refer to relative sections within the 6kb.
(B) Nucleotide sequence of predicted transcription factor binding sites and flanking regions: PPREs A, B, and C, NF-κB sites, and SP1 sites are bolded and named immediately under respective nucleotides. Terminal sequences (bolded, underlined) were used for PCR amplification.
(C) Plot of CAGE tag frequencies indicating transcription start sites for 5′ capped transcripts for the human TNIP1 promoter. The occurrence of mapped tags was plotted versus the relative promoter nucleotide position based on numbering in (A). The next most-frequent occurrence is of 7 tags is at relative position of approximately −6200.
We examined the 6kb promoter sequence for additional regulatory elements and found predicted sites with relevance to the expected roles of the TNIP1 gene product, namely TNIP1 inhibition of NF-κB signaling and corepression of NR activity. Within the 6kb TNIP1 promoter (Figure 1A and B) there are multiple potential NF-κB sites which are found in the distal region at −4615, the central region at −2518, −1894, and −1723, and the proximal region at −240 and −78. Several occurrences of NR binding half-sites in differing repeat organizations were also found. Three of these (Figure 1A and B) occurred in the distal promoter region (−5107, −4793, and −4593).
Cap-analysis of gene expression (CAGE) recognizes transcription start sites (TSS) [34]. Nucleotide sequences referred to as tags identify 5′-capped transcripts; tags can then be considered as i)distinct CAGE tag starting sites or ii)overlapping starting sites referred to as CAGE tag clusters. With this approach, we searched the CAGE database [35] for TNIP1 (Ensembl gene: ENSG00000145901) and 65 CAGE tags were returned. For TNIP1, the 5′-capped transcripts could be grouped into 49 CAGE tag starting sites (one or more of the 65 starting at the same nucleotide) and 32 tag clusters (representing start sites that overlap each other). Over half of the mapped transcription start sites localized within a stretch of ~600 nucleotides and included the most-frequently (14 tags) reported start region (Figure 1C). Other smaller tag clusters (≤4 tags) were found within the same region. This spread of start sites for TNIP1 is consistent with other CpG island-containing, TATA-less promoters [34]. About 10% of all TNIP1 transcription start sites localized ~5000 nucleotides upstream of this region, likely reflective of the alternative promoter which has been previously reported [17, 36]. The use of this alternative transcription start changes the sequence of a small, non-coding first exon, but does not change the coding region of the TNIP1 transcript. For consistency, we used the nucleotide numbering established by Brasier and colleagues [11] for their ~700 nucleotide TNIP1 promoter segment to assign the position numbering of the CAGE tag hits and of our 6kb promoter clone and its derivatives. Our cloned genomic region contains this segment and extends upstream of it to a relative nucleotide position of −5887 using their numbering.
3.2. Determination of multiple, functional NF-κB sites in the human TNIP1 promoter
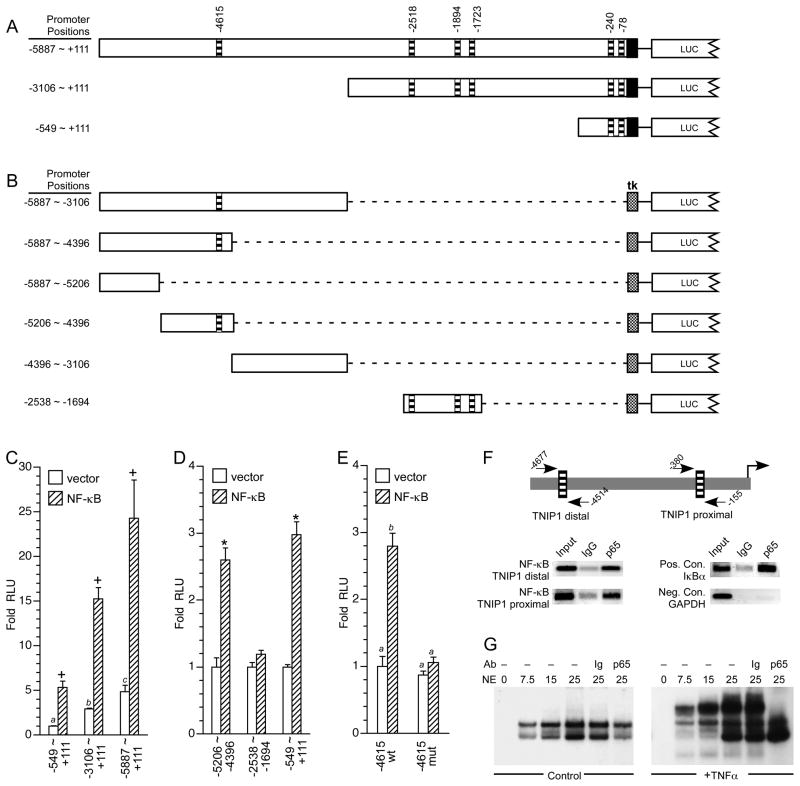
An initial analysis of the cloned TNIP1 promoter for constitutive transcriptional activity and regulation by NF-κB was performed with three constructs. Each starts at the same point and extends upstream to the proximal (−549 ~ +111, referred to hereafter as −549bp), central (−3106 ~ +111, referred to hereafter as −3kb) and distal regions (−5887 ~ +111, referred to hereafter as −6kb) (Figure 2A). Additional reporters were constructed to isolate candidate NF-kB sites (Figure 2B). We found reporter activity increased with increasing promoter length. The −3kb and −6kb constructs were approximately three- and five-fold more active than the −549bp proximal fragment (Figure 2C, open bars).
Fig. 2. TNIP1 promoter construct activity is regulated by proximal and distal NF-kB sites.
(A) TNIP1 promoter construct naming is based on position approximating their 5′-most nucleotide, −6kb (−5887 ~ +111), −3kb (−3106 ~ +111) and −549bp (−549 ~ +111), top to bottom in schematic. Horizontal striped bars, predicted NF-κB sites. Solid box, +1 to +111 of the cloned promoter as in Figure 1. LUC, 5′ end of luciferase coding region.
(B) Schematic of TNIP1 promoter upstream constructs named based on 5′ ~ 3′ nucleotides they include. Stippled box is tk minimal promoter. Distal promoter fragments were ligated (dashed line) to tk but are shown aligned to their native position in the 6kb construct in A for ease of comparison. LUC, as in A.
(C) HeLa cells transfected with equal copy number of TNIP1 promoter reporters 6kb (−5887 ~ +111), 3kb (−3106 ~ +111) or 549bp (−549 ~ +111). Promoter constructs were cotransfected with empty expression vector as control (open bars) or constructs expressing NF-κB subunits (p50 and p65, diagonal striped bars). Fold increase in normalized RLU’s are presented with signal from the −549 ~ +111-Luc construct in the absence of transfected NF-κB set at 1. Statistical significance indicated by +, p<0.005, Student’s t-test vector control compared to receipt of NF-κB; bars with different letters are values significantly different (p<0.01) from each other, one-way ANOVA, Tukey’s post-hoc test.
(D) HeLa cells transfected with equal copy number of TNIP1 promoter reporters −5206 ~ −4396, −2538 ~ −1694, or −549 ~ +111. Because the first two constructs contain the tk minimal promoter and the −549 ~ +111-Luc construct contains the native transcription elements, RLUs are presented for each reporter normalized to 1 in the presence of control empty expression vector (open bars) versus fold increase in activity due to coexpressed NF-κB subunits (p50 and p65, striped bars). Significance indicated by *, p<0.01, Student’s t-test for significantly different from corresponding empty vector control.
(E) HeLa cells transfected with equal copy number of TNIP1 promoter reporter −5206 ~ −4396 wild-type (wt) or with its NF-κB site mutated (mut). Conditions as in C with control empty expression vector (open bars, wt −5206 ~ −4396 set to 1) versus fold increase in activity due to coexpressed NF-κB subunits (p50 and p65, striped bars). Bars with different letters are values significantly different (p<0.001) from each other, one-way ANOVA, Tukey’s post-hoc test.
(F) ChIP analysis of endogenous NF-κB p65 subunit association with the HeLa genome TNIP1 promoter. Schematic shows relative position of PCR primers in the distal and proximal regions of the promoter. Lanes are PCR products from decross-linked chromatin without immunoprecipitation (Input), or chromatin immunoprecipitation with normal rabbit immunoglobulin (IgG), or anti-NF-κB p65 (p65). Indicative of p65 occupation of a known NF-κB site, a segment of the IκBα promoter was used as a positive control. GAPDH promoter was used as a negative control. Primer sequences and amplicon lengths are provided in Table 2.
(G) EMSA for protein binding to the TNIP1 distal NF-κB site. Specifics for binding reactions of increasing microgram amounts of HeLa nuclear extract (NE) protein, incubated without (−) antibody (Ab), or control immunoglobulin (Ig), or antibody directed against RelA (p65), are indicated at the top of individual lanes. NE were derived from control (no TNFα, left panel) or TNFα treated (right panel) cells. Disruption rather than supershift of the protein-probe complex has been previously noted in general and specifically for NF-κB [77, 78]. The TNFα-induced NF-κB-probe complex was disrupted by p65 antibody but not by control antibody (Ig).
Cotransfection with expression constructs for the p50 and p65 heterodimer subunits of NF-κB increased expression of each TNIP1 promoter reporter (Figure 2C, hatched bars) consistent with functionality of at least some of the in silico predicted NF-κB binding sites. Our results with the proximal −549 ~ +111 reporter (Figure 2C) are consistent with those of Brasier and colleagues [11] but our in silico analysis of the upstream regions suggested additional NF-κB sites. To assess which of these might contribute responsiveness to the p50/p65 subunits, central and distal promoter regions containing predicted sites were isolated and cloned into a reporter construct with the minimal tk promoter. The proximal −549 ~ +111 construct was used as an indicator of known p50/p65 responsiveness. We found the distal, but not the central, TNIP1 promoter region conferred NF-κB responsiveness (Figure 2D) which was lost when the −4615 NF-κB site was mutated (Figure 2E). For completeness and to assay for functional sites possibly missed by in silico analysis, we also tested constructs covering areas of no or poor scoring algorithm-predicted NF-κB sites, −5887 ~ −5206 and −4396 ~ −3106. Together, these constructs covered an additional ~2kb of the TNIP1 promoter but conferred no NF-κB responsiveness (not shown).
To test for binding of endogenous NF-κB to the TNIP1 promoter in the HeLa genome, chromatin immunoprecipitation assays were conducted on TNFα-stimulated cells. Primers were designed to flank and to be specific for the proximal and distal TNIP1 NF-κB sites. We found (Figure 2F) the p65 subunit (RelA) of endogenous NF-κB could be localized to distal and proximal regions of the TNIP1 promoter in TNFα-stimulated cells. Binding of p65 to the −4615 NF-κB sequence was confirmed by EMSA where a RelA-specific band from nuclear extracts of TNFα-stimulated cells was formed with double-stranded oligonucleotides representing this site (Figure 2G). These results confirm the previous [11] identification of NF-κB occupancy in the proximal (−549 ~ +111) TNIP1 promoter region and identify a new NF-κB site within the distal region (−5206 ~ −4396).
3.3. Regulation of the human TNIP1 promoter by PPARs
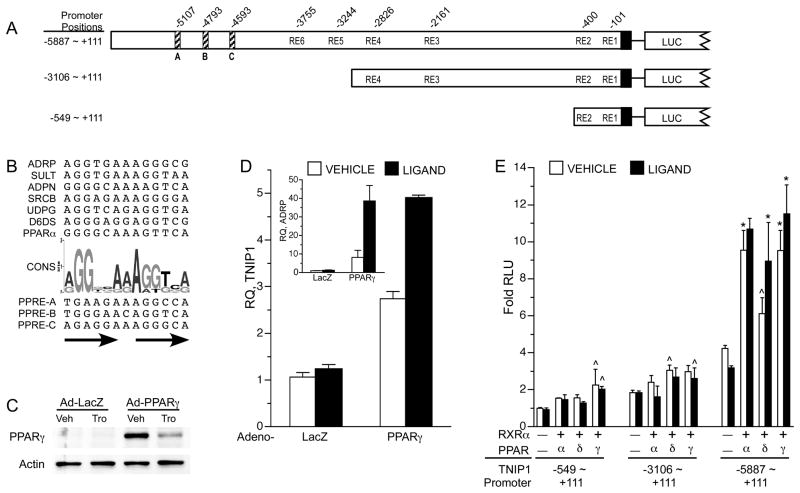
Several potential response elements (RE) conforming to the NR binding half-site 5′-A/GGG/TTCA −3′ consensus in differing repeat configurations were recognized throughout the 6kb TNIP1 promoter (Figure 3A). We focused on PPARs mediating transcriptional control of the TNIP1 promoter through these predicted REs for two reasons. First, three potential REs (labeled A, B, and C in Figure 3A) in the distal promoter region had the best matches to a consensus (Figure 3B) peroxisome proliferator response element (PPRE) derived from human gene promoters. Second, we have recently found TNIP1 protein can act as a corepressor of PPARs (Flores et al, accepted) suggesting that this NR could induce expression of a corepressor directed against it.
Fig. 3. Human TNIP1 expression is regulated by PPARs.
(A) TNIP1 promoter construct schematic. Diagonal stripe bars (A, B, and C), nuclear receptor response elements (RE) best-matching consensus PPRE. Other potential nuclear receptor REs of varying configurations labeled RE1 – 6 with nucleotide positions marked above promoter schematic. Solid box, +1 to +111 of cloned sequence as in Figure 1.
(B) Alignment of functional PPRE’s from human gene promoters with consensus sequence (CONS) given as a WebLogo graphical representation. Letter height reflects occurrence of nucleotide. ADRP, adipose differentiation-related protein. SULT, hepatic hydroxysteroid sulfotransferase 2A1. ADPN, adiponectin. SRCB, scavenger receptor class B, type I. UDPG, UDP-glucuronosyltransferase 1A9. D6DS, delta-6 desaturase. PPARα, peroxisome proliferator activated receptor alpha. Below graphic are the predicted TNIP1 PPRE’s (A, B, and C) from distal promoter, arrows indicate direct repeat.
(C) Western blot of protein lysates from HeLa cells infected with adenovirus expressing β-galactosidase (Ad-LacZ) as control or adenovirus expressing human PPARγ (Ad-PPARγ) 48 hours after infection and treatment with control media (0.1% DMSO vehicle, Veh) or 5μM troglitazone (Tro) for 28 hours before harvest. Actin signal from same blot shown as loading control.
(D) PPARγ regulation of endogenous TNIP1 gene expression. Expression of target genes, TNIP1 or ADRP, inset, measured by quantitative real-time PCR was normalized to RPLP0 from HeLa cells receiving either adenovirus LacZ as infection control or adenovirus PPARγ as in C. Levels are shown relative to the LacZ control receiving vehicle (0.1% DMSO, open bars). Ligand, 5μM troglitazone, filled bars. RQ, relative quantitation.
(E) HeLa cells transfected with equal copy number of TNIP1 promoter reporters −6kb (−5887 ~ +111), −3kb (−3106 ~ +111) or −549bp (−549 ~ +111). Promoter constructs were cotransfected with empty expression vector (labeled – under bars) as control or constructs expressing the indicated PPAR and RXRα as heterodimer partner. All open bars, vehicle, 0.1% DMSO. Fold increase in normalized RLU’s are presented with signal from −549 ~ +111-Luc construct in absence of transfected nuclear receptor heterodimer, vehicle control, set at 1 (first open bar). Solid bars PPAR ligands: PPARα, 10μM WY14643. PPARδ, 1μM L165041. PPARγ, 5μM troglitazone. In absence of transfected nuclear receptor heterodimer (first solid bar each promoter set), ligand was WY14643. For statistical tests, results were compared within a reporter construct transfection set and within vehicle or ligand treatments. Results significantly different from reporter with empty expression vector as determined by ANOVA, Dunnett’s post-hoc test, are marked ^, p<0.05; * p<0.01.
We continued analysis of the promoter and its response to PPAR in HeLa cells given that these cells has been used for earlier TNIP1 promoter work describing a proximal [11] and distal NF-κB sites (Figure 2). This came with the experimental advantage that HeLa cells have been reported to have no or undetectable levels of PPARγ [37–39] providing a point of comparison for assessing TNIP1 expression in the absence of PPARγ versus its presence. To provide PPARγ to the HeLa system, we followed an approach similar to that used previously for the activation of PPAR genomic targets [40] and for study of candidate nuclear receptor-controlled elements [41]. HeLa cells were transduced with adenovirus expressing the β-galactosidase cDNA (LacZ) or the human PPARγ cDNA and cultured under media control (vehicle) and ligand (troglitazone) conditions and examined by western blot for PPARγ protein levels. Our results confirmed HeLa cells have no or undetectable levels of PPARγ protein. The viral expression system provided detectable levels of PPARγ and as expected [42], receptor protein levels decreased with ligand activation (Figure 3C). In this system, we found that the well-characterized and PPAR-regulatable target gene adipose differentiation-related protein (ADRP) [25] was unresponsive to the PPARγ ligand troglitazone in lacZ-transduced control cells but was induced by PPARγ expression and further increased by liganded receptor (Figure 3D, inset). Similarly, basal TNIP1 expression in the lacZ cells was not induced by PPARγ ligand. However, with the provision of PPARγ or PPARγ and ligand, expression of the endogenous TNIP1 gene was increased for both conditions compared to their corresponding LacZ controls (Figure 3D).
To localize the PPAR regulation of the TNIP1 promoter, we utilized the −6kb, −3kb and −549bp reporter constructs. Expression of PPARα, δ, or γ increased activity of the −6kb construct three- to four-fold with minimal increases in the −549bp or −3kb constructs (Figure 3E). This induction mapped to the distal 3kb promoter region containing the potential PPRE sites (A, B and C in Figure 3A). Therefore, our subsequent studies were directed to this region of the promoter.
3.4. PPARs target sites in the distal human TNIP1 promoter
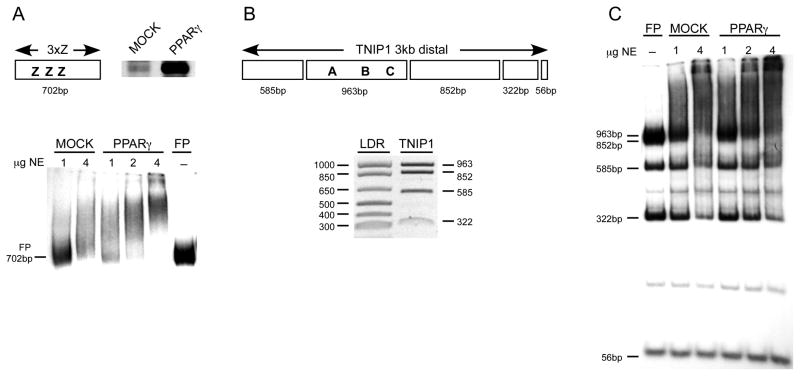
A combined physical and functional analysis of the 3kb distal portion of the TNIP1 promoter was undertaken to identify PPREs. A modified EMSA, suitable for scanning protein-binding sites in mixtures of large DNA fragments was employed in which a limited amount of probe is used with increasing amounts of nuclear extract protein [28, 43]. Interaction with DNA is assessed by decreased signal at the position of the free probe(s) as opposed to the formation of discrete band as in conventional EMSA. As a PPAR-relevant example, a 702bp fragment, containing three repeats of the element Z PPRE (Figure 4A, schematic) from the ω-hydroxylase gene promoter [26] was tested for proof of concept. In comparison to nuclear extracts from control COS7 cells which have low levels of endogenous PPARs (Figure 4A, western), the same amount of nuclear protein from COS7 cells transiently expressing human PPARγ is more effective in shifting the probe (Figure 4A, EMSA).
Fig. 4. Evidence for PPARγ interaction with the human TNIP1 distal promoter via a modified EMSA.
(A) top left, schematic of 3xZ EMSA probe, a 702bp fragment containing 3 repeats of the ω-hydroxylase Z (3xZ) PPRE. top right, western for endogenous and endogenous plus recombinant PPARγ, respectively, in equal microgram amounts of nuclear extracts from mock- and PPARγ-transfected COS7 cells. bottom, EMSA with 3xZ probe. Numbers at top of lanes are micrograms of nuclear extract (NE) proteins from mock and PPARγ-transfected cells. –, no nuclear protein added in right-most lane to establish position of free-probe (FP).
(B) top, schematic of predicted Bcl I fragments used as probes from the TNIP1 −3kb distal region. Relative positioning of predicted PPREs (A, B and C) best matching consensus sequence PPRE are shown. Fragments are in scale to each other. bottom, ethidium bromide stained gel of Bcl I digest of TNIP1 3kb distal promoter, LDR is DNA ladder with base pair sizes shown to left of gel. The TNIP1 963bp and 852bp fragments run as a doublet. The relatively short 56bp fifth fragment is not shown.
(C) Modified EMSA with TNIP1 distal 3kb probes illustrated in B. Mock and PPARγ-enriched nuclear proteins preferentially shift larger fragments from TNIP1 distal region. Lane labeling as in EMSA in A. Note decrease in any band’s intensity may reflect binding of other nuclear proteins in addition to endogenous and recombinant PPARγ.
The TNIP1 3kb distal promoter was Bcl I digested and the resulting five fragments (Figure 4B) were radiolabeled in bulk to produce a probe mixture covering the entire distal region. With this probe mixture we were able to assess the better consensus-matching A, B, and C candidate PPREs and lesser matching REs also present in the distal 3kb fragment in one EMSA. COS7 nuclear extracts with endogenous PPARs and extracts with enriched levels of PPARγ preferentially decreased the signal of the 963bp free probe band, which has three predicted PPREs, compared with other promoter fragments from the probe mixture (Figure 4C). Of note, this fragment contains predicted binding sites for additional proteins. The presence of those proteins in both the basal and PPARγ enriched extracts, from mock and PPARγ transfected cells, respectively, may lessen the difference in band intensity otherwise expected based strictly on the different in PPARγ levels. Other free probe band intensities, such as at 852bp and 585bp, decreased at increasing protein amounts, again, likely reflecting the binding of other nuclear proteins.
3.5. TNIP1 promoter responsiveness to PPARs maps to a direct repeat and is orientation independent
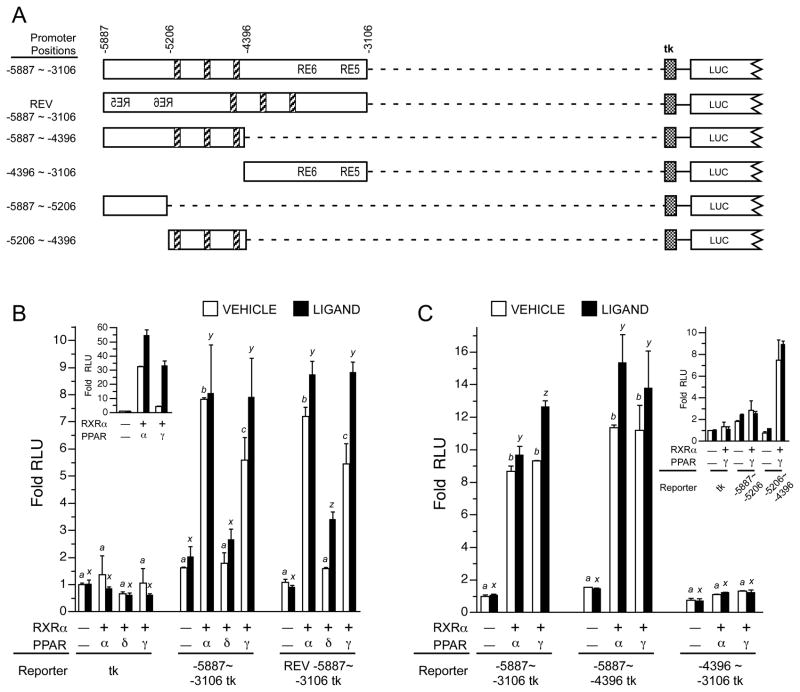
Reporter constructs with the TNIP1 distal 3kb fragment, −5887 ~ −3106, in forward or reverse orientations (Figure 5A) linked to the minimal tk promoter (Figure 5B) were induced by PPAR expression. The greatest increase came from PPARα and γ. The tk promoter alone was unaffected. Orientation independence of the 3kb fragment suggested the presence of an enhancer-like element possibly recognized by the receptor. As with the 6kb promoter reporter (Figure 3E), and the PPRE derived from the ω-hydroxylase gene (Figure 5B inset), extensive induction by PPAR occurred in the absence of experimentally added ligand. Promoter activation by PPARs without addition of exogenous ligand has also been seen with PPREs derived from other genes [44, 45].
Fig. 5. Isolation of TNIP1 distal promoter fragment positively regulated by PPAR.
(A) Schematic with numbering based on nucleotide position within intact promoter. Diagonal striped bars, nuclear receptor response elements (RE) best-matching consensus PPRE. RE5 and RE6 are as described in Figure 3. Stippled box, tk minimal promoter. LUC, 5′ end of luciferase gene. Distal promoter fragments were ligated (dashed line) to tk but are shown aligned to their native position in the −6kb construct for ease of comparison.
(B) Orientation independence of TNIP1 distal promoter PPAR responsiveness. HeLa cells were transfected with equal copy number of luciferase reporter constructs with the tk minimal promoter only or tk ligated to TNIP1 distal promoter fragment −5997 ~ −3106 in forward or reverse (REV) orientation. Transfections, expression and reporter constructs, vehicle (open bars) and ligands (solid bars), as in Figure 3. Fold increase in normalized RLU’s are presented with signal from tk-Luc construct in absence of transfected nuclear receptor heterodimer, vehicle control, set at 1 (first open bar). In absence of transfected nuclear receptor heterodimer (−/−, first solid bar each promoter set), ligand was WY14643. For statistical tests, results were compared within reporter constructs and either within vehicle (a, b, etc) or ligand treatments (x, y, etc). Different letters indicate different values, p<0.01, ANOVA, Tukey’s post-hoc test. Inset, PPAR induces 3xZ-tk-Luc with further activation in the presence of ligand. In absence of transfected nuclear receptor heterodimer ligand was WY14643. Transfections as in main graph.
(C) Localization of PPAR responsiveness to the −5206 to −4396bp fragment from TNIP1 distal promoter. HeLa cells transfected and cultured as in B. Normalized RLU’s are presented with signal from the distal −5887 ~ −3106-tk-Luc construct in absence of transfected nuclear receptor heterodimer, vehicle control, set at 1 (first open bar). In absence of transfected nuclear receptor heterodimer (−/−, first solid bar each promoter set), ligand was WY14643.
Inset, PPAR responsiveness can be further localized to region containing the three candidate PPREs in the right-hand half (−5206 ~ −4396bp fragment) of the −5887 ~ −4396bp region. Transfections as in main graph.
Given the better induction by α and γ, we used these two PPARs to narrow the response area. The left but not right half (−5887 ~ −4396 and −4396 ~ −3106, respectively, Figure 5C) of the distal 3kb fragment was induced by PPARα or γ to approximately the same extent as the entire region. A similar reductionist approach split this responsive fragment into approximate halves. The more-proximal ~800bp fragment (−5206 ~ −4396) which contained the three predicted PPREs, but not the more-distal fragment (−5887 ~ −5206) which had no predicted REs, retained activation by PPARγ (Figure 5C inset).
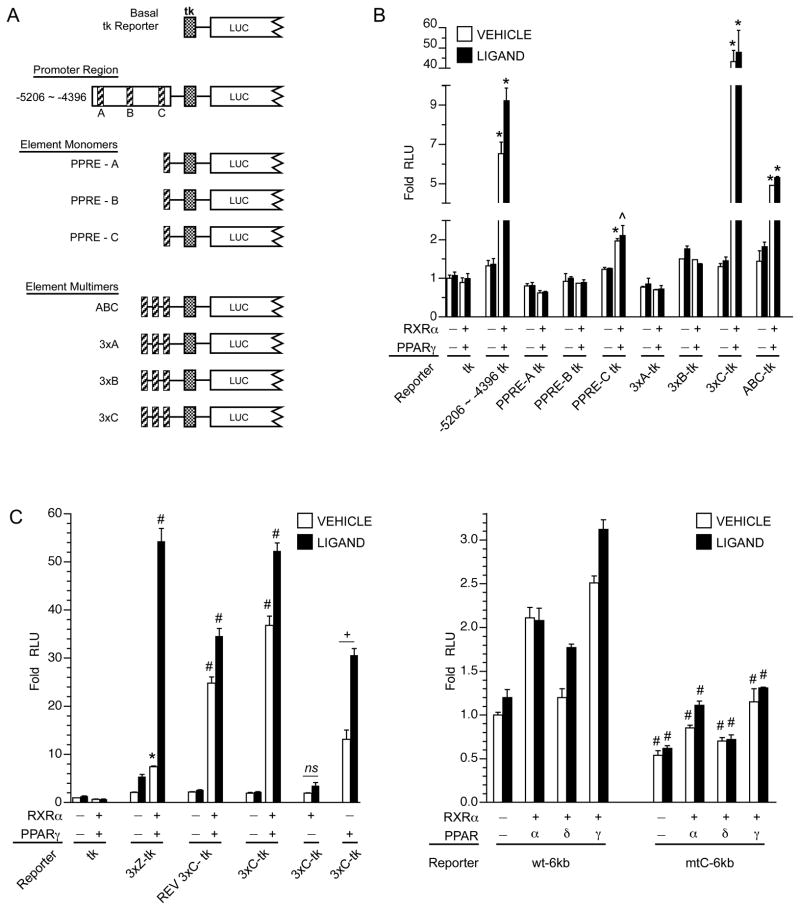
3.6. Identification of functional PPREs
The candidate TNIP1 PPREs A, B, and C, were prepared as synthetic oligomers, inserted as single or triple repeats into the tk minimal promoter reporter (Figure 6A), and compared against the −5206 ~ −4396 promoter fragment. In testing for regulation by PPAR, we chose to focus on PPARγ. There was evidence of its physical association with (Figure 4) and transcriptional activation of TNIP1 promoter-derived reporters with candidate PPREs (Figure 5C). Single as well as the repeat inserts of element C but not A or B showed positive regulation by coexpressed PPARγ (Figure 6B). This is similar to the effect seen with multimerization of previously described PPREs [26, 27]. In comparison, a construct containing one insert each of elements A, B, and C showed approximately one-third the induction of the triple C construct. As the candidate elements A and B showed no or only very minimal induction by PPARγ they were eliminated from further study.
Fig. 6. Identification of TNIP1 promoter element C as a functional PPRE.
(A) Schematic of reporter constructs. Candidate PPREs (diagonal striped bars) A, B, and C were introduced as single or triple inserts in basal reporter construct. Stippled box, tk minimal promoter. LUC, 5′ end of luciferase coding region.
(B) TNIP1 promoter element C provides PPAR responsiveness. HeLa cells transfected with equal copy number luciferase reporter constructs with tk minimal promoter only or tk promoter ligated to TNIP1 distal promoter fragment −5206 ~ −4396, or single, or three times repeats of candidate elements A, B, or C, or a mini-gene version of the candidate PPREs in a contiguous stretch of sequence (see schematics in A). Transfections, expression and reporter constructs, vehicle (open bars) and ligands (solid bars), as in Figure 3. Normalized RLU’s are presented from representative experiment with signal from tk-Luc construct in the presence of vehicle set at 1; note breaks in y-axis and changes in scale. Fold increase in normalized RLU’s are presented with signal from the tk-Luc construct in absence of transfected nuclear receptor heterodimer, vehicle control, set at 1 (first open bar). Error bars are SD. Statistically significant differences indicated by ^, p<0.05, or *, p<0.01, from Student’s t-test for reporter activity without (−/−) versus with (+/+) expression of PPARγ/RXRα, within vehicle or ligand treatments. ns, not significant difference.
(C) TNIP1 promoter element C functions as an orientation-independent PPRE. HeLa cells transfected and cultured as in B. Normalized RLU’s are presented with signal from tk-Luc construct in presence of vehicle set at 1. Ligand for transfections with the PPARγ/RXRα heterodimer pair or PPARγ alone was troglitazone as in Figure 3. Ligand for RXRα alone transfection was 1μM 9-cis RA. Error bars are SEM. REV 3xC is a three times repeat of element C but in an orientation reverse that occurring in the native promoter. Statistically significant differences indicated by *, p<0.01, or #, p<0.001, from Student’s t-test for reporter activity without (−/−) versus with (+/+) expression of PPARγ/RXRα. ns, not significant difference. +, p<0.005 for troglitazone induction.
(D) Mutation of element C in the 6kb promoter construct reduces activation by PPAR. HeLa cells transfected and cultured as in B. Normalized RLU’s are presented with signal from the wild-type 6kb-Luc construct (wt-6kb) without transfected receptors, in presence of vehicle, set at 1. Ligands as in Figure 3E. Statistically significant differences indicated by #, p<0.001, from Student’s t-test for the 6kb-Luc construct with a mutated element C (mtC-6kb) compared against the corresponding condition for wt-6kb.
Potential synergy amongst concatenates of the same DR element (such as 3xC) has been previously described including for the ω-hydroxylase PPRE [27] which we have used here for comparison (3xZ-tk, Figure 6C). Orientation independence for the triple C insert (REV 3xC-tk, Figure 6C) control by PPARγ was also detected paralleling our earlier results with the distal 3kb TNIP1 promoter fragment (REV −5887 ~ −3106, Figure 5B). In the forward orientation, the TNIP1 3xC was activated to an extent similar to the 3xZ derived from the known ω-hydroxylase PPRE. Transfections for responsiveness of element C to PPARγ had coexpressed the heterodimer partner RXRα. When PPARγ was transfected alone, there was about half the induction obtained with the heterodimer. However, transfection of RXRα alone provided no induction (Figure 6C) even with 9-cis RA treatment suggesting element C was acting as a PPRE and not a binding site for RXR homodimers.
With the PPAR-responsiveness of element C established as a monomer and multimer repeat, we extended its analysis back to the context of the 6kb promoter. Mutagenesis of element C caused a loss in reporter induction associated with expression of any PPAR subtype (Figure 6D). Retention of some PPAR induction is consistent with our in silico prediction of possible response elements, albeit those with lesser matches to consensus sequences, within the proximal 3kb promoter (Figure 3A) and some increase in reporter activity for that construct coexpressed with PPAR (Figure 3E). Compared to the wild-type sequence, there was also decrease in promoter activity of the element C mutant (vehicle control and ligand conditions) in the absence of transfected receptors. This may reflect loss of activity from disruption of some site for an additional transcription factor overlapping the element C PPRE (see Discussion).
3.7. PPARγ binds the TNIP1 promoter in vitro and in vivo
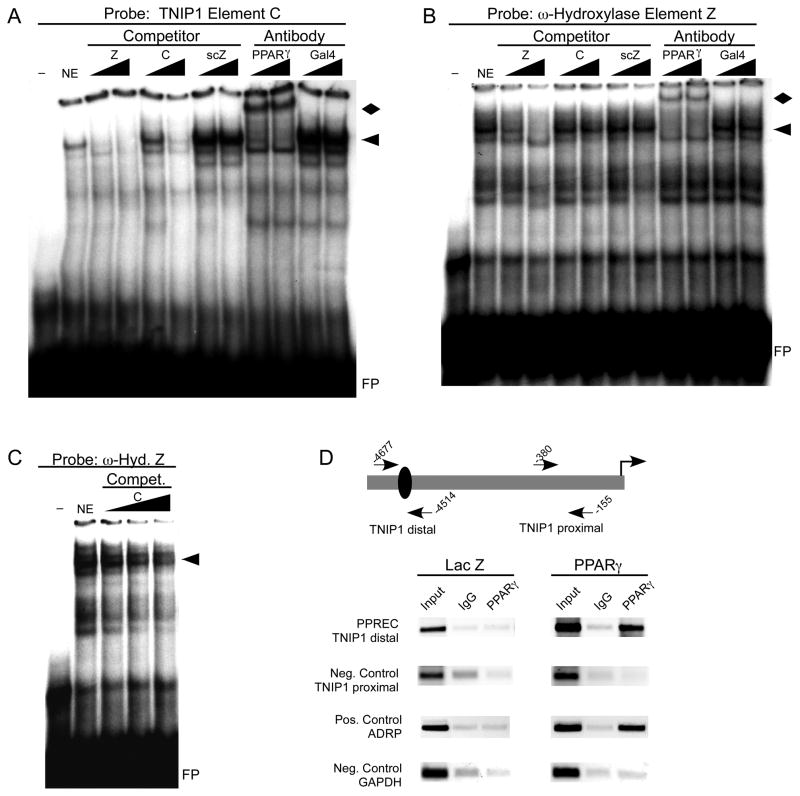
To complement the above functional analysis of the TNIP1 element C PPRE, we examined its ability to physically interact with PPARγ. PPARγ bound element C PPRE in an EMSA (Figure 7A). Increasing amounts of unlabeled element C as well as the ω-hydroxylase element Z PPRE competed for binding when element C was used as probe. Antibody against PPARγ successfully super-shifted the TNIP1 element C PPRE-protein complex but no super-shift occurred with the negative control antibody. Parallel results were obtained when the ω-hydroxylase element Z PPRE was used as probe (Figure 7B). In this case TNIP1 element C was a less effective competitor against Z than when Z was used as competitor against the C probe. This may have to do with the Z element having nucleotides other than those contained within the direct repeat that promote interaction with PPARs [27]. A titration with additional amounts of element C did show further competition with the Z PPRE for binding PPARγ (Figure 7C) consistent with it being a valid binding element for PPARγ protein.
Fig. 7. TNIP1 promoter element C is a PPARγ-binding site.
(A) EMSA with double-stranded oligomer representing TNIP1 promoter element C. First lane, no nuclear protein ( − ). Remaining lanes with 10μg of PPARγ-enriched COS7 nuclear extract (NE) protein. Triangles indicate increasing amounts of cold competitor or antibody. Competitors: Z, PPRE from Z element of ω-hydroxylase promoter. C, C element from TNIP1 promoter. scZ, scrambled version of Z element PPRE of same length and composition but with nucleotides in random order. Antibodies: PPARγ or Gal4 DNA binding domain (Gal4-BD). arrowhead, PPARγ-containing complex. diamond, PPARγ-containing super-shifted complex. FP, free probe.
(B) EMSA with double-stranded Z element of ω-hydroxylase promoter. Binding conditions, competitors and antibodies as in A.
(C) EMSA with double-stranded Z element of ω-hydroxylase promoter. Binding conditions as in B with increased titration of TNIP1 element C as cold competitor.
(D) ChIP analysis of PPARγ association with the endogenous TNIP1 promoter. Schematic shows relative position of PCR primers surrounding the TNIP1 PPREC in the distal region and a downstream proximal region of the promoter as a negative control. Lanes are PCR products from decross-linked chromatin without immunoprecipitation (Input), or immunoprecipitation with normal rabbit immunoglobulin (IgG), or anti-PPARγ (PPARγ). Indicative of PPARγ occupation of a known PPRE, a segment of the ADRP promoter was used as a positive control. GAPDH promoter was used as a negative control. Primer sequences and amplicon lengths are provided in Table 2.
To test for binding of PPARγ to the TNIP1 promoter in the HeLa genome, ChIP assays were conducted (Figure 7D). HeLa cells have been reported to have no or undetectable levels of PPARγ by Glass [39] and Lazar [38]. Our receptor activity studies (Figure 5B, inset) are consistent with this. No increase in the PPRE-containing 3xZ-TK-Luc reporter occurred in HeLa cells treated with troglitazone in the absence of transfected PPARγ. Thus HeLa cells can be used for comparison of chromatin derived from control cultures (transduced with adenovirus carrying the β-galactosidase cDNA) versus those experimentally expressing PPARγ (transduced with adenovirus carrying the human PPARγ cDNA) similar to our qPCR experiments above and previous work by Mandrup and colleagues [40] and Aikawa and colleagues [46]. The PPRE characterized for the human ADRP [25, 47] was used as a positive control and point of reference for this approach. Chromatin from the control β-galactosidase-expressing HeLa cells incubated with PPARγ antibody did not give any signal over the normal IgG control. Recombinant expression of PPARγ was necessary for positive ChIP results from both the TNIP1 PPRE as well as the previously established ADRP PPRE. Parallel to this, the recombinant PPARγ expression did not generate off-target binding as demonstrated by two negative controls, a TNIP1 promoter region without PPREs or a separate gene, GAPDH, with no known PPRE. These ChIP results are consistent with the above in silico, transfection, and EMSA findings of a PPRE in the human TNIP1 distal promoter.
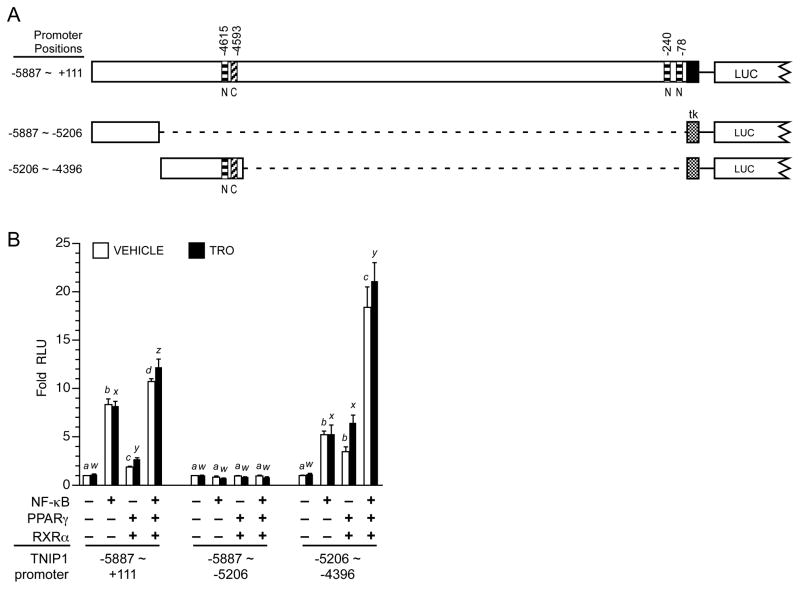
3.8. Evidence for combined NF-κB/PPAR activation of the TNIP1 promoter
Activity of the TNIP1 promoter is up-regulated by NF-κB (Figure 2) and PPARs (Figure 3) through sites localized to specific regions within the promoter (Figure 8A for summary schematic and Figures 5, 6, 7 for data). In these analyses, up-regulation was achieved with expression of the individual respective transcription factor, NF-κB or PPAR. To extend these studies, we compared (Figure 8B) individual and combined expression of NF-κB and PPARγ on the full length TNIP1 6kb promoter (−5887 ~ +111), a distal promoter fragment (−5887 ~ −5206), and a second distal promoter fragment contiguous to this one (−5206 ~ −4396). The three reporters were selected based on the 6kb construct having the most promoter sequence available and the −5206 to −4396 fragment containing NF-κB and PPAR responsiveness. The −5887 to −5206 construct was included for comparison; the in silico analysis had predicted no NF-κB or PPAR binding sites within it. Individually, NF-κB or PPARγ activated transcription from the 6kb construct (Figure 8B, left bar set) with the induction from NF-κB being greater perhaps because of the multiple NF-κB binding sites (see schematic Figure 8A) versus the one PPRE within it. When NF-κB and PPARγ were coexpressed there was an additive effect on the 6kb promoter (last pair of left bar set). In agreement with the in silico analysis of no predicted binding sites for NF-κB or PPAR, the −5887 to −5206 distal promoter fragment had no responsiveness to these factors whether provided individually or in combination (Figure 8B, middle bar set). In contrast, the −5206 to −4396 distal TNIP1 promoter fragment with the NF-κB site and one PPRE had a ~5–6-fold induction from either of these transcription factors (Figure 8B, right bar set). Coexpressing NF-κB and PPARγ produced an apparent synergistic effect (last pair of right bar set). For both the 6kb promoter and the −5206 ~ −4396 distal fragment, the increases obtained by combined NF-κB and PPARγ continued in the presence of the PPARγ ligand troglitazone.
Fig. 8. Individually and together, NF-κB and PPARγ stimulate TNIP1 promoter reporter activity.
(A) Schematic of TNIP1 promoter constructs. Diagonal striped bar, element C PPRE. Horizontal striped bars, NF-κB. Stippled box, tk minimal promoter. LUC, 5′ end of luciferase gene. Distal promoter fragments were ligated (dashed line) to tk but are shown aligned to their native position in the 6kb construct for ease of comparison.
(B) Combined NF-κB and PPARγ activation of TNIP1 promoter. HeLa cells were transfected with equal copy number of the indicated TNIP1 promoter constructs. The two distal fragments utilized the tk minimal promoter (see panel A). Cells were cotransfected with empty expression vector as control (first bar pair, each set) or constructs expressing NF-κB (p50 and p65), or PPARγ and RXRα, or both. Normalized RLU from each TNIP1 promoter construct for control empty expression vector transfection, vehicle-treated media culture condition, was set at one for each reporter. Change in expression for each reporter is expressed as fold of its respective empty vector, vehicle control. Error bars are SEM. Open bars: vehicle control. Solid bars: PPARγ ligand, 1μM troglitazone (TRO). For statistical tests, results were compared within reporter constructs and either within vehicle (a, b, etc) or ligand treatments (w, x, etc). Different letters indicate different values, p<0.05, ANOVA, Newman-Keuls multiple comparison test.
4. Discussion
This study’s initial goal was to define regulation of the TNIP1 promoter given its wide tissue expression and our other findings (Flores et al, accepted) of TNIP1 ability to repress PPAR signaling. Overall, the functional contribution of atypical corepressors such as TNIP1, LCoR and RIP140 [48, 49] may provide a transcriptional regulatory system more capable of fine adjustments rather than maximal off/on responses by typical corepressors and coactivators alone. They may also contribute to early but negative ligand regulation of genes. An example of this was recently documented following retinoic acid treatment of keratinocytes; at one hour 134 genes were induced and 181 were suppressed [50]. It would be reasonable to expect similar results with PP/PPAR in relevant cell types and that these reductions could involve corepressors of ligand-bound nuclear receptors such as TNIP1 among other possible mechanisms. Beyond this, TNIP1 has a role in reducing NF-κB activity [5, 6, 51, 52] and yet its expression is increased in response to TNFα [11, 29, 53] and in inflammatory disease states such as psoriasis [2]. To determine under what conditions TNIP1 expression levels might change and thus its repression of NF-κB and/or PPAR, it became important to understand what regulated TNIP1 promoter activity both near and distant from its transcription start site as repeats of control elements or distinct transcription factor binding site could augment or provide new control opportunites. A previous report on an ~700bp fragment of the TNIP1 promoter demonstrated regulation by NF-κB near the transcriptional start site [11]. To more fully examine what might regulate TNIP1 expression, we cloned ~6kb of the promoter. Our functional analysis demonstrated previously unknown NF-κB and PPRE sites in the promoter distal region highlighting the potential of elements outside of the proximal region. Predominance of NF-κB or PPARs in regulation of TNIP1 expression will likely vary with the differing levels of these transcription factors in different cell types [54]. Clearly however, no one of these factors, even NF-κB with binding sites in the distal and proximal regions of the TNIP1 promoter, is likely to be the sole factor driving expression. We found ~40% of the 6kb promoter activity was resistant to increasing amounts of a dominant-negative IκB (not shown). Ultimate control of TNIP1 promoter activity will likely be a combinatorial interplay of inducible transcription factors described here and constitutive factors, such as Sp1, currently under study in our laboratory.
In contrast to some earlier described PPREs, such as the motifs from the ω-hydroxylase or acyl coA oxidase promoters, the TNIP1 element C PPRE has a lesser degree of induction due to receptor and ligand after its activation by receptor under vehicle conditions. However, agonist-independent NR activity or only marginal enhancement of activity in response to agonist (which we observe in our studies) have previously been conclusively demonstrated. In particular, for PPARs, it has been shown that all three subtypes can associate with coactivators in a ligand-independent manner due in part to the coactivator-compatible conformation of the receptors’ AF-2 domain [55]. A very recent, extensive PPAR-responsiveness study classifying different modes of transcriptional regulation by PPARs (in this case delta) at PPREs included those elements in genes otherwise positively regulated by PPAR binding in their promoter with no or weak further induction by a receptor-specific agonist [56]. Separately, Shimizu et al [57] showed PPARα responsible for a 2.5–3-fold increase in reporter constructs based on ~3kb of the PEX11alpha gene with only ~30% further increase from ligand.
Other functionality of element C, such as apparent fold induction greater with PPARγ alone than with the PPARγ/RXRα heterodimer may be due to the relative proportions of receptors achieved experimentally. Although of a different sequence than element C, others have addressed a similar PPAR/RXR result in the description of PPAR/RXR heterodimers and PPAR homodimers binding to a PPRE presumably based on relative amounts of either dimer partner [58–60]. Should such homodimers be formed, they may recruit different ratios of receptor coactivators than the PPAR/RXR heterodimer and be dependent on the element sequence or surrounding nucleotides. Finally, as in other genes controlled by other NRs, there may be additional PPREs active with the endogenous promoter but upstream of our cloned region that could account for the greater response of the endogenous TNIP1 gene to ligand and receptor than seen with the promoter reporter constructs. The recent work of Brown and colleagues [61] illustrates this for estrogen response elements with the identification of an estrogen receptor binding region 67 kilobases upstream of the c-myc gene conferring estrogen-responsiveness to its transcription.
Partial reduction of reporter activity when element C was mutated in the absence of exogenous receptor and ligand (Figure 6D) may be attributed to loss of activity from disruption of some site for an additional transcription factor overlapping the element C PPRE. DR1 motifs such as element C also function as binding sites for RXR homodimers as well as RXR heterodimer partners beyond PPAR such as RAR [62], COUP and HNF4 [63 for review]. While the results (Figure 6C) with transfected RXR argue against element C activation by RXR homodimers, it is possible that basal activity of the TNIP1 promoter is in part from occupation by other RXR heterodimer partners such as RAR found in HeLa cells [64, 65 and not shown].
The coexistence within the same promoter of NF-κB and nuclear receptor binding sites has been previously reported. For instance, the iNOS promoter is NF-κB inducible [66 for review] and is also regulated through the RXR/RAR heterodimer binding a retinoid response element [67]. In addition to the potential combinatorial regulation by NF-kB and RXR/RAR, this promoter has an element bound and activated by other RXR heterodimers where CAR, PXR, TR or VDR serve as the partners [68]. Parallel to TNIP1, the promoter of hyaluronan synthase 2 gene [69] contains multiple NF-κB sites and one nuclear receptor site, in this case for RAR, and is activated by both factors depending whether the cell is exposed to TNFα or retinoic acid. We suggest that genes such as iNOS, HAS2, and TNIP1 are subject to integrated regulation by the elements within their promoters, the transcription factor repertoire present in a given cell type, and the presence of particular chemical or ligand stimuli which together will lead to gene activation. Thus the multi-factorial regulation of TNIP1 expression is consistent with the well-established concept of context-dependent regulation of gene expression [70].
The presence of both NF-κB and nuclear receptor binding sites in the promoters of genes such as TNIP1 is different from other promoter studies where cross-talk of NF-κB and nuclear receptors has been investigated and antagonism reported [71, 72]. In these cases, where no PPARγ binding site is described, expression is inducible by NF-κB, and reduced by PPARγ. Decreased expression may occur through sequestration of cofactors by PPARγ or binding of the two proteins to each other [71, 72].
TNIP1 associates with agonist-bound PPAR; it represses transcription targeting a region of the receptor typically involved in physical association with transcriptional coactivators (Flores et al, accepted) placing it in an atypical and intriguing subgroup of nuclear receptor coregulatory proteins [13 for review]. Overall, the functional contribution of atypical corepressors such as TNIP1, LCoR and RIP140 [48, 49] may provide a transcriptional regulatory system capable of fine adjustments to ligand-induced transcription rather than maximal off or on responses respectively, by typical corepressors and coactivators alone. Nuclear receptor regulation of gene expression involves dynamic [73] exchange of transcription factors providing entry for ligand-dependent corepressors as well as coactivators. Interestingly, another atypical NR coregulator, hairless, also has a complex relationship with one of the NRs it targets, the vitamin D receptor. Recently, Engelhard and colleagues [74] reported ligand-independent activation of the hairless promoter constructs by the vitamin D receptor. This was cell type-dependent occurring in 3T3 fibroblasts but not in a keratinocyte cell line. Similar to our investigations for NF-κB and PPAR interaction with the TNIP1 promoter, comprehensive regulation of these promoters is likely to be influenced by cell-dependent amounts of constitutive and inducible transcription factors. While only a few corepressors of ligand-bound nuclear receptors have been identified to date, they are likely to have wide relevance in control of metabolism and differentiation given their control of NRs integral to these processes. Among these, receptor interacting protein (RIP) 140 has had over a dozen years of research clarifying its function as a ligand-dependent nuclear receptor corepressor [75 for review] and more recently in control of its own expression. Certain parallels can be drawn between it and TNIP1. Both RIP140 and TNIP1 proteins rely on leucine rich motifs that are otherwise characteristic of coactivators for their interaction with NRs. Yet these proteins ultimately corepress liganded NRs. At the promoter level, both are positively regulated by NRs that they ultimately repress. RIP140 expression is positively regulated by the estrogen and retinoic acid receptors [15, 16] and it in turn represses the activity of these NRs. PPARs promote TNIP1 promoter activity and yet TNIP1 represses PPARs (Flores et al accepted). Similarly, activated NF-κB increases TNIP1 promoter activity but TNIP1 protein inhibits NF-κB function [9 and references therin, 11]. The results of this study are consistent with a self- and trans-limiting feedback loop where PPAR or NF-κB stimulation of TNIP1 expression restricts ongoing activity of either or both factors until such a time when their contribution to TNIP1 expression has been tempered. Negative feedback loops involving signal pathway coregulators and their target proteins may buffer against extremes of signaling by functioning as suggested by Werner and colleagues as a “tunable rheostat” [76] responsible for a finer regulation response rather than an absolute on/off.
5. Conclusions
Approximately 6000bp of the human TNIP1 promoter have been isolated and characterized. The TNIP1 distal promoter, approximately 4600 nucleotides upstream of a frequently utilized transcription start site region, contains PPARγ and NF-κB sites that bind the respective transcription factors in vivo. These binding sites drive transcription via their respective protein factors. The varied activity of PPAR and NF-κB across different cell types may contribute to differences in TNIP1 expression in those cells.
Highlights.
Six kilobases of the human TNIP1 promoter were cloned and examined in silico
Functional NF-κB and PPAR sites have been identified in the human TNIP1 promoter
NF-κB and PPARγ bind to the endogenous TNIP1 promoter
The TNIP1 promoter PPRE functions in an orientation-independent manner
TNIP1 expression is likely to vary as abundance and activity of NF-κB and PPAR change
Acknowledgments
We thank D. Goldhamer for GM2163 E. coli, R. Hay and E. Zandi for conalbumin-luc and NF-κB-conalbumin-luc reporters, C. Giardina for discussions on NF-κB and expression constructs for dominant negative IκB, p50, and p65, and E. Barry and T. Rasmussen for advice on ChIP assays. The work was supported by a National Institute of Arthritis and Musculoskeletal and Skin Diseases grant (AR048660 to BJA). Partial support was provided by a pre-doctoral fellowship from the American Foundation for Pharmaceutical Education, the Boehringer Ingelheim Graduate Fellowship in Toxicology, and the Edward A. Khairallah Summer Graduate Fellowship (IG), a UConn Multicultural Program pre-doctoral fellowship and Boehringer Ingelheim Summer Fellowship (PCE), a Neiforth PharmD Studentship from UConn School of Pharmacy (CPS), and a UConn Presidential Scholars Enrichment Award for undergraduates (SEL).
Abbreviations used
- Nef
negative factor of HIV
- NR
nuclear receptor
- PP
peroxisome proliferator
- PPRE
PP response element
- RIP
receptor interacting protein
- RQ
relative quantitation
- scZ
scrambled sequence of Z element
- TNIP1
TNFAIP3-interacting protein 1
- TNFAIP3
TNF-α-induced protein 3
- TRO
troglitazone
Footnotes
Conflict of interest
The authors declare that they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Igor Gurevich, Email: igor.gurevich@huskymail.uconn.edu.
Carmen Zhang, Email: carmen.c.zhang@uconn.edu.
Priscilla C. Encarnacao, Email: priscilla.encarnacao@uconn.edu.
Charles P. Struzynski, Email: charles.struzynski@huskymail.uconn.edu.
Brian J. Aneskievich, Email: brian.aneskievich@uconn.edu.
References
- 1.Hosgood HD, 3rd, Menashe I, Shen M, Yeager M, Yuenger J, Rajaraman P, He X, Chatterjee N, Caporaso NE, Zhu Y, Chanock SJ, Zheng T, Lan Q. Pathway-based evaluation of 380 candidate genes and lung cancer susceptibility suggests the importance of the cell cycle pathway. Carcinogenesis. 2008;29:1938–1943. doi: 10.1093/carcin/bgn178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, Gudjonsson JE, Li Y, Tejasvi T, Feng BJ, Ruether A, Schreiber S, Weichenthal M, Gladman D, Rahman P, Schrodi SJ, Prahalad S, Guthery SL, Fischer J, Liao W, Kwok PY, Menter A, Lathrop GM, Wise CA, Begovich AB, Voorhees JJ, Elder JT, Krueger GG, Bowcock AM, Abecasis GR. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukushi M, Dixon J, Kimura T, Tsurutani N, Dixon MJ, Yamamoto N. Identification and cloning of a novel cellular protein Naf1, Nef-associated factor 1, that increases cell surface CD4 expression. FEBS Lett. 1999;442:83–88. doi: 10.1016/s0014-5793(98)01631-7. [DOI] [PubMed] [Google Scholar]
- 4.Gupta K, Ott D, Hope TJ, Siliciano RF, Boeke JD. A human nuclear shuttling protein that interacts with human immunodeficiency virus type 1 matrix is packaged into virions. J Virol. 2000;74:11811–11824. doi: 10.1128/jvi.74.24.11811-11824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heyninck K, Kreike MM, Beyaert R. Structure-function analysis of the A20-binding inhibitor of NF-kappa B activation, ABIN-1. FEBS Lett. 2003;536:135–140. doi: 10.1016/s0014-5793(03)00041-3. [DOI] [PubMed] [Google Scholar]
- 6.Heyninck K, De Valck D, Vanden Berghe W, Van Criekinge W, Contreras R, Fiers W, Haegeman G, Beyaert R. The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. J Cell Biol. 1999;145:1471–1482. doi: 10.1083/jcb.145.7.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beyaert R, Heyninck K, Van Huffel S. A20 and A20-binding proteins as cellular inhibitors of nuclear factor-kappa B-dependent gene expression and apoptosis. Biochem Pharmacol. 2000;60:1143–1151. doi: 10.1016/s0006-2952(00)00404-4. [DOI] [PubMed] [Google Scholar]
- 8.Mauro C, Pacifico F, Lavorgna A, Mellone S, Iannetti A, Acquaviva R, Formisano S, Vito P, Leonardi A. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J Biol Chem. 2006;281:18482–18488. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- 9.Wagner S, Carpentier I, Rogov V, Kreike M, Ikeda F, Lohr F, Wu CJ, Ashwell JD, Dotsch V, Dikic I, Beyaert R. Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene. 2008;27:3739–3745. doi: 10.1038/sj.onc.1211042. [DOI] [PubMed] [Google Scholar]
- 10.Oshima S, Turer EE, Callahan JA, Chai S, Advincula R, Barrera J, Shifrin N, Lee B, Yen B, Woo T, Malynn BA, Ma A. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature. 2009;457:906–909. doi: 10.1038/nature07575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian B, Nowak DE, Jamaluddin M, Wang S, Brasier AR. Identification of direct genomic targets downstream of the nuclear factor-kappaB transcription factor mediating tumor necrosis factor signaling. J Biol Chem. 2005;280:17435–17448. doi: 10.1074/jbc.M500437200. [DOI] [PubMed] [Google Scholar]
- 12.Gurevich I, Aneskievich BJ. Liganded RARalpha and RARgamma interact with but are repressed by TNIP1. Biochem Biophys Res Commun. 2009;389:409–414. doi: 10.1016/j.bbrc.2009.08.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurevich I, Flores AM, Aneskievich BJ. Corepressors of agonist-bound nuclear receptors. Toxicol Appl Pharmacol. 2007;223:288–298. doi: 10.1016/j.taap.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lonard DM, Lanz RB, O’Malley BW. Nuclear receptor coregulators and human disease. Endocr Rev. 2007;28:575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- 15.Augereau P, Badia E, Balaguer P, Carascossa S, Castet A, Jalaguier S, Cavailles V. Negative regulation of hormone signaling by RIP140. J Steroid Biochem Mol Biol. 2006;102:51–59. doi: 10.1016/j.jsbmb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 16.White KA, Yore MM, Warburton SL, Vaseva AV, Rieder E, Freemantle SJ, Spinella MJ. Negative feedback at the level of nuclear receptor coregulation. J Biol Chem. 2003;278:43889–43892. doi: 10.1074/jbc.C300374200. [DOI] [PubMed] [Google Scholar]
- 17.Shiote Y, Ouchida M, Jitsumori Y, Ogama Y, Matsuo Y, Ishimaru F, Tanimoto M, Shimizu K. Multiple splicing variants of Naf1/ABIN-1 transcripts and their alterations in hematopoietic tumors. Int J Mol Med. 2006;18:917–923. [PubMed] [Google Scholar]
- 18.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 19.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Podvinec M, Kaufmann MR, Handschin C, Meyer UA. NUBIScan, an in silico approach for prediction of nuclear receptor response elements. Mol Endocrinol. 2002;16:1269–1279. doi: 10.1210/mend.16.6.0851. [DOI] [PubMed] [Google Scholar]
- 21.Sandelin A, Wasserman WW. Prediction of nuclear hormone receptor response elements. Mol Endocrinol. 2005;19:595–606. doi: 10.1210/me.2004-0101. [DOI] [PubMed] [Google Scholar]
- 22.Fang HL, Strom SC, Cai H, Falany CN, Kocarek TA, Runge-Morris M. Regulation of human hepatic hydroxysteroid sulfotransferase gene expression by the peroxisome proliferator-activated receptor alpha transcription factor. Mol Pharmacol. 2005;67:1257–1267. doi: 10.1124/mol.104.005389. [DOI] [PubMed] [Google Scholar]
- 23.Pineda Torra I, Jamshidi Y, Flavell DM, Fruchart JC, Staels B. Characterization of the human PPARalpha promoter: identification of a functional nuclear receptor response element. Mol Endocrinol. 2002;16:1013–1028. doi: 10.1210/mend.16.5.0833. [DOI] [PubMed] [Google Scholar]
- 24.Tang C, Cho HP, Nakamura MT, Clarke SD. Regulation of human delta-6 desaturase gene transcription: identification of a functional direct repeat-1 element. J Lipid Res. 2003;44:686–695. doi: 10.1194/jlr.M200195-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Targett-Adams P, McElwee MJ, Ehrenborg E, Gustafsson MC, Palmer CN, McLauchlan J. A PPAR response element regulates transcription of the gene for human adipose differentiation-related protein. Biochim Biophys Acta. 2005;1728:95–104. doi: 10.1016/j.bbaexp.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 26.Sher T, Yi HF, McBride OW, Gonzalez FJ. cDNA cloning, chromosomal mapping, and functional characterization of the human peroxisome proliferator activated receptor. Biochemistry. 1993;32:5598–5604. doi: 10.1021/bi00072a015. [DOI] [PubMed] [Google Scholar]
- 27.Palmer CNA, Hsu MH, Griffin KJ, Johnson EF. Novel sequence determinants in peroxisome proliferator signaling. J Biol Chem. 1995;270:16,114–116,121. doi: 10.1074/jbc.270.27.16114. [DOI] [PubMed] [Google Scholar]
- 28.Gurevich I, Zhang C, Aneskievich BJ. Scanning for transcription factor binding by a variant EMSA. Methods Mol Biol. 2010;585:147–158. doi: 10.1007/978-1-60761-380-0_11. [DOI] [PubMed] [Google Scholar]
- 29.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 30.Gillespie RF, Gudas LJ. Retinoid regulated association of transcriptional co-regulators and the polycomb group protein SUZ12 with the retinoic acid response elements of Hoxa1, RARbeta(2), and Cyp26A1 in F9 embryonal carcinoma cells. J Mol Biol. 2007;372:298–316. doi: 10.1016/j.jmb.2007.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munoz E, Courtois G, Veschambre P, Jalinot P, Israel A. Tax induces nuclear translocation of NF-kappa B through dissociation of cytoplasmic complexes containing p105 or p100 but does not induce degradation of I kappa B alpha/MAD3. J Virol. 1994;68:8035–8044. doi: 10.1128/jvi.68.12.8035-8044.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 34.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, Forrest AR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kitazume Y, Kawaji H, Kai C, Nakamura M, Konno H, Nakano K, Mottagui-Tabar S, Arner P, Chesi A, Gustincich S, Persichetti F, Suzuki H, Grimmond SM, Wells CA, Orlando V, Wahlestedt C, Liu ET, Harbers M, Kawai J, Bajic VB, Hume DA, Hayashizaki Y. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 35.Kawaji H, Kasukawa T, Fukuda S, Katayama S, Kai C, Kawai J, Carninci P, Hayashizaki Y. CAGE Basic/Analysis Databases: the CAGE resource for comprehensive promoter analysis. Nucleic Acids Res. 2006;34:D632–636. doi: 10.1093/nar/gkj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Favre M, Butticaz C, Stevenson B, Jongeneel CV, Telenti A. High frequency of alternative splicing of human genes participating in the HIV-1 life cycle: a model using TSG101, betaTrCP, PPIA, INI1, NAF1, and PML. J Acquir Immune Defic Syndr. 2003;34:127–133. doi: 10.1097/00126334-200310010-00002. [DOI] [PubMed] [Google Scholar]
- 37.Hayes MM, Lane BR, King SR, Markovitz DM, Coffey MJ. Peroxisome proliferator-activated receptor gamma agonists inhibit HIV-1 replication in macrophages by transcriptional and post-transcriptional effects. J Biol Chem. 2002;277:16913–16919. doi: 10.1074/jbc.M200875200. [DOI] [PubMed] [Google Scholar]
- 38.Wang C, Fu M, D’Amico M, Albanese C, Zhou JN, Brownlee M, Lisanti MP, Chatterjee VK, Lazar MA, Pestell RG. Inhibition of cellular proliferation through IkappaB kinase-independent and peroxisome proliferator-activated receptor gamma-dependent repression of cyclin D1. Mol Cell Biol. 2001;21:3057–3070. doi: 10.1128/MCB.21.9.3057-3070.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Straus DS, Pascual G, Li M, Welch JS, Ricote M, Hsiang CH, Sengchanthalangsy LL, Ghosh G, Glass CK. 15-deoxy-delta 12,14-prostaglandin J2 inhibits multiple steps in the NF-kappa B signaling pathway. Proc Natl Acad Sci, USA. 2000;97:4844–4849. doi: 10.1073/pnas.97.9.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen R, Grontved L, Stunnenberg HG, Mandrup S. Peroxisome proliferator-activated receptor subtype- and cell-type-specific activation of genomic target genes upon adenoviral transgene delivery. Mol Cell Biol. 2006;26:5698–5714. doi: 10.1128/MCB.02266-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song S, Attia RR, Connaughton S, Niesen MI, Ness GC, Elam MB, Hori RT, Cook GA, Park EA. Peroxisome proliferator activated receptor alpha (PPARalpha) and PPAR gamma coactivator (PGC-1alpha) induce carnitine palmitoyltransferase IA (CPT-1A) via independent gene elements. Mol Cell Endocrinol. 2010;325:54–63. doi: 10.1016/j.mce.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hauser S, Adelmant G, Sarraf P, Wright HM, Mueller E, Spiegelman BM. Degradation of the peroxisome proliferator-activated receptor gamma is linked to ligand-dependent activation. J Biol Chem. 2000;275:18527–18533. doi: 10.1074/jbc.M001297200. [DOI] [PubMed] [Google Scholar]
- 43.Delacroix L, Begon D, Chatel G, Jackers P, Winkler R. Distal ERBB2 promoter fragment displays specific transcriptional and nuclear binding activities in ERBB2 overexpressing breast cancer cells. DNA Cell Biol. 2005;24:582–594. doi: 10.1089/dna.2005.24.582. [DOI] [PubMed] [Google Scholar]
- 44.Helledie T, Grontved L, Jensen SS, Kiilerich P, Rietveld L, Albrektsen T, Boysen MS, Nohr J, Larsen LK, Fleckner J, Stunnenberg HG, Kristiansen K, Mandrup S. The gene encoding the Acyl-CoA-binding protein is activated by peroxisome proliferator-activated receptor gamma through an intronic response element functionally conserved between humans and rodents. J Biol Chem. 2002;277:26821–26830. doi: 10.1074/jbc.M111295200. [DOI] [PubMed] [Google Scholar]
- 45.Basu-Modak S, Braissant O, Escher P, Desvergne B, Honegger P, Wahli W. Peroxisome proliferator-activated receptor beta regulates acyl-CoA synthetase 2 in reaggregated rat brain cell cultures. J Biol Chem. 1999;274:35881–35888. doi: 10.1074/jbc.274.50.35881. [DOI] [PubMed] [Google Scholar]
- 46.Dean J, Plante J, Huggins GS, Snyder RO, Aikawa R. Role of cyclic AMP-dependent kinase response element-binding protein in recombinant adeno-associated virus-mediated transduction of heart muscle cells. Hum Gene Ther. 2009;20:1005–1012. doi: 10.1089/hum.2009.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tachibana K, Kobayashi Y, Tanaka T, Tagami M, Sugiyama A, Katayama T, Ueda C, Yamasaki D, Ishimoto K, Sumitomo M, Uchiyama Y, Kohro T, Sakai J, Hamakubo T, Kodama T, Doi T. Gene expression profiling of potential peroxisome proliferator-activated receptor (PPAR) target genes in human hepatoblastoma cell lines inducibly expressing different PPAR isoforms. Nucl Recept. 2005;3:3. doi: 10.1186/1478-1336-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernandes I, Bastien Y, Wai T, Nygard K, Lin R, Cormier O, Lee HS, Eng F, Bertos NR, Pelletier N, Mader S, Han VK, Yang XJ, White JH. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol Cell. 2003;11:139–150. doi: 10.1016/s1097-2765(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 49.Fritah A, Christian M, Parker MG. The metabolic coregulator RIP140: an update. Am J Physiol Endocrinol Metab. 2010;299:E335–E340. doi: 10.1152/ajpendo.00243.2010. [DOI] [PubMed] [Google Scholar]
- 50.Lee DD, Stojadinovic O, Krzyzanowska A, Vouthounis C, Blumenberg M, Tomic-Canic M. Retinoid-responsive transcriptional changes in epidermal keratinocytes. J Cell Physiol. 2009;220:427–439. doi: 10.1002/jcp.21784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wullaert A, Wielockx B, Van Huffel S, Bogaert V, De Geest B, Papeleu P, Schotte P, El Bakkouri K, Heyninck K, Libert C, Beyaert R. Adenoviral gene transfer of ABIN-1 protects mice from TNF/galactosamine-induced acute liver failure and lethality. Hepatology. 2005;42:381–389. doi: 10.1002/hep.20785. [DOI] [PubMed] [Google Scholar]
- 52.El Bakkouri K, Wullaert A, Haegman M, Heyninck K, Beyaert R. Adenoviral gene transfer of the NF-kappa B inhibitory protein ABIN-1 decreases allergic airway inflammation in a murine asthma model. J Biol Chem. 2005;280:17938–17944. doi: 10.1074/jbc.M413588200. [DOI] [PubMed] [Google Scholar]
- 53.Gallagher J, Howlin J, McCarthy C, Murphy EP, Bresnihan B, FitzGerald O, Godson C, Brady HR, Martin F. Identification of Naf1/ABIN-1 among TNF-alpha-induced expressed genes in human synoviocytes using oligonucleotide microarrays. FEBS Lett. 2003;551:8–12. doi: 10.1016/s0014-5793(03)00823-8. [DOI] [PubMed] [Google Scholar]
- 54.Kostadinova R, Wahli W, Michalik L. PPARs in diseases: control mechanisms of inflammation. Curr Med Chem. 2005;12:2995–3009. doi: 10.2174/092986705774462905. [DOI] [PubMed] [Google Scholar]
- 55.Molnar F, Matilainen M, Carlberg C. Structural determinants of the agonist-independent association of human peroxisome proliferator-activated receptors with coactivators. J Biol Chem. 2005;280:26543–26556. doi: 10.1074/jbc.M502463200. [DOI] [PubMed] [Google Scholar]
- 56.Adhikary T, Kaddatz K, Finkernagel F, Schonbauer A, Meissner W, Scharfe M, Jarek M, Blocker H, Muller-Brusselbach S, Muller R. Genomewide analyses define different modes of transcriptional regulation by peroxisome proliferator-activated receptor-beta/delta (PPARbeta/delta) PLoS One. 2011;6:e16344. doi: 10.1371/journal.pone.0016344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimizu M, Yamashita D, Yamaguchi T, Hirose F, Osumi T. Aspects of the regulatory mechanisms of PPAR functions: analysis of a bidirectional response element and regulation by sumoylation. Mol Cell Biochem. 2006;286:33–42. doi: 10.1007/s11010-005-9052-z. [DOI] [PubMed] [Google Scholar]
- 58.Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 59.Todorov VT, Desch M, Schmitt-Nilson N, Todorova A, Kurtz A. Peroxisome proliferator-activated receptor-gamma is involved in the control of renin gene expression. Hypertension. 2007;50:939–944. doi: 10.1161/HYPERTENSIONAHA.107.092817. [DOI] [PubMed] [Google Scholar]
- 60.Okuno M, Arimoto E, Ikenobu Y, Nishihara T, Imagawa M. Dual DNA-binding specificity of peroxisome-proliferator-activated receptor gamma controlled by heterodimer formation with retinoid X receptor alpha. Biochem J. 2001;353:193–198. doi: 10.1042/0264-6021:3530193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang C, Mayer JA, Mazumdar A, Fertuck K, Kim H, Brown M, Brown PH. Estrogen Induces c-myc Gene Expression via an Upstream Enhancer Activated by the Estrogen Receptor and the AP-1 Transcription Factor. Mol Endocrinol. 2011;25:1527–1538. doi: 10.1210/me.2011-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Durand B, Saunders M, Leroy P, Leid M, Chambon P. All-trans and 9-cis retinoic acid induction of CRABPII transcription is mediated by RAR-RXR heterodimers bound to DR1 and DR2 repeated motifs. Cell. 1992;71:73–85. doi: 10.1016/0092-8674(92)90267-g. [DOI] [PubMed] [Google Scholar]
- 63.Chan LS, Wells RA. Cross-Talk between PPARs and the Partners of RXR: A Molecular Perspective. PPAR Res. 2009;2009:925309. doi: 10.1155/2009/925309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee HK, Park UH, Kim EJ, Um SJ. MED25 is distinct from TRAP220/MED1 in cooperating with CBP for retinoid receptor activation. Embo J. 2007;26:3545–3557. doi: 10.1038/sj.emboj.7601797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keriel A, Stary A, Sarasin A, Rochette-Egly C, Egly JM. XPD mutations prevent TFIIH-dependent transactivation by nuclear receptors and phosphorylation of RARalpha. Cell. 2002;109:125–135. doi: 10.1016/s0092-8674(02)00692-x. [DOI] [PubMed] [Google Scholar]
- 66.De Bosscher K, Vanden Berghe W, Haegeman G. Cross-talk between nuclear receptors and nuclear factor kappaB. Oncogene. 2006;25:6868–6886. doi: 10.1038/sj.onc.1209935. [DOI] [PubMed] [Google Scholar]
- 67.Zou F, Liu Y, Liu L, Wu K, Wei W, Zhu Y, Wu J. Retinoic acid activates human inducible nitric oxide synthase gene through binding of RARalpha/RXRalpha heterodimer to a novel retinoic acid response element in the promoter. Biochem Biophys Res Commun. 2007;355:494–500. doi: 10.1016/j.bbrc.2007.01.178. [DOI] [PubMed] [Google Scholar]
- 68.Toell A, Kroncke KD, Kleinert H, Carlberg C. Orphan nuclear receptor binding site in the human inducible nitric oxide synthase promoter mediates responsiveness to steroid and xenobiotic ligands. J Cell Biochem. 2002;85:72–82. [PubMed] [Google Scholar]
- 69.Saavalainen K, Tammi MI, Bowen T, Schmitz ML, Carlberg Cm. Integration of the activation of the human hyaluronan synthase 2 gene promoter by common cofactors of the transcription factors retinoic acid receptor and nuclear factor kappaB. J Biol Chem. 2007;282:11530–11539. doi: 10.1074/jbc.M607871200. [DOI] [PubMed] [Google Scholar]
- 70.Alvarez M, Rhodes SJ, Bidwell JP. Context-dependent transcription: all politics is local. Gene. 2003;313:43–57. doi: 10.1016/s0378-1119(03)00627-9. [DOI] [PubMed] [Google Scholar]
- 71.Li M, Pascual G, Glass CK. Peroxisome proliferator-activated receptor gamma-dependent repression of the inducible nitric oxide synthase gene. Mol Cell Biol. 2000;20:4699–4707. doi: 10.1128/mcb.20.13.4699-4707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chung SW, Kang BY, Kim SH, Pak YK, Cho D, Trinchieri G, Kim TS. Oxidized low density lipoprotein inhibits interleukin-12 production in lipopolysaccharide-activated mouse macrophages via direct interactions between peroxisome proliferator-activated receptor-gamma and nuclear factor-kappa B. J Biol Chem. 2000;275:32681–32687. doi: 10.1074/jbc.M002577200. [DOI] [PubMed] [Google Scholar]
- 73.Rochette-Egly C. Dynamic combinatorial networks in nuclear receptor-mediated transcription. J Biol Chem. 2005;280:32565–32568. doi: 10.1074/jbc.R500008200. [DOI] [PubMed] [Google Scholar]
- 74.Engelhard A, Bauer RC, Casta A, Djabali K, Christiano AM. Ligand-independent regulation of the hairless promoter by vitamin D receptor. Photochem Photobiol. 2008;84:515–521. doi: 10.1111/j.1751-1097.2008.00301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.White R, Morganstein D, Christian M, Seth A, Herzog B, Parker MG. Role of RIP140 in metabolic tissues: connections to disease. FEBS Lett. 2008;582:39–45. doi: 10.1016/j.febslet.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 76.Werner SL, Kearns JD, Zadorozhnaya V, Lynch C, O’Dea E, Boldin MP, Ma A, Baltimore D, Hoffmann A. Encoding NF-kappaB temporal control in response to TNF: distinct roles for the negative regulators IkappaBalpha and A20. Genes Dev. 2008;22:2093–2101. doi: 10.1101/gad.1680708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. J. Wiley; 2004. [Google Scholar]
- 78.Bruggeman LA, Adler SH, Klotman PE. Nuclear factor-kappa B binding to the HIV-1 LTR in kidney: implications for HIV-associated nephropathy. Kidney Int. 2001;59:2174–2181. doi: 10.1046/j.1523-1755.2001.00732.x. [DOI] [PubMed] [Google Scholar]