Abstract
Analyzing the massive amounts of neural data collected using microelectrodes to extract biologically relevant information is a major challenge. Many scientific findings rest on the ability to overcome these challenges and to standardize experimental analysis across labs. This can be facilitated in part through comprehensive, efficient and practical software tools disseminated to the community at large. We have developed a comprehensive, MATLAB-based software package - entitled NeuroQuest - that bundles together a number of advanced neural signal processing algorithms in a user-friendly environment. Results demonstrate the efficiency and reliability of the software compared to other software packages, and versatility over a wide range of experimental conditions.
Keywords: neural data analysis, neural signal processing, spike train analysis, spike detection, spike sorting
1. Introduction
Ensemble recording of neural signals provides a window of opportunity to understand how the brain represents and interprets the physical world in the coordinated spiking patterns of its neurons. Spike data have been extensively collected to study how neuronal ensembles represent sensory inputs (Panzeri et al., 2001; DeWeese et al., 2003; Tiesinga et al., 2008), plan motor actions (Velliste et al., 2008; Reimer and Hatsopoulos, 2010) and perform complex cognitive tasks such as decision making (Harris et al., 2003; Fujisawa et al., 2008; Beck et al., 2008). Prior to interpreting the information they contain, however, spike data recorded with microelectrodes have to undergo many processing steps that range from simple detection of spike presence in noisy traces to fitting advanced mathematical models to distinct patterns in neural spike trains.
Developing software packages that can efficiently help researchers in processing and analyzing neural signals is therefore of utmost importance and many have been and continue to be developed in recent years. Generally speaking, these can be categorized into two groups: Processing and Analysis software packages. Processing packages are designed to extract spike trains from the extracellular recordings, and mainly consist of filtering, spike detection, and spike sorting (Fee et al., 1996; Hulata et al., 2002; Hermle et al., 2004; Quiroga et al., 2004; Rutishauser et al., 2006; Hazan et al., 2006; Takekawa et al., 2010). Analysis software packages are designed to extract information from spike train data and perform statistical inference about some underlying hypotheses being tested (Brown et al., 2004). Very few software tools, if any, exist to date that integrate both types in one comprehensive package. As a result, most research labs rely on in-house analysis tools in one form or another. When coupled with a significant lack of community–wide standardized set of tools that enables replicating experimental data analysis across labs, objective comparison between scientific findings becomes very challenging and time consuming. With a significant trend towards very large scale ensemble recording (Buzsáki, 2006), a potential need to automate and streamline the processing of large scale datasets would inevitably arise to alleviate the burden of continued user supervision typically performed in today’s experiments.
In this paper, we present a detailed description of NeuroQuest - a MATLAB-based software package for analyzing extracellular neural data acquired using penetrating microelectrodes (Kwon et al., 2009). The software is packaged in a simple Graphical User Interface (GUI) exclusively designed for ensemble neural data analysis and statistical inference, with a non-expert user in mind. It can be distinguished from other software packages in several aspects: First, the package includes algorithms for spike detection and sorting, as well as spike train analysis. This creates a unified processing environment that is efficient and time saving. Second, the spike detection algorithm implemented in NeuroQuest outperform those in other software packages such as KlustaKwik, WAVE_CLUS, and OSort (Harris et al., 2000; Quiroga et al., 2004; Rutishauser et al., 2006), especially in low Signal to Noise Ratios (SNRs) experiments. Robust spike detection and sorting are crucial because all-subsequent data analysis depends largely on the outcome of these steps. Third, NeuroQuest is designed to cope with distinct data characteristics that may be observed in different brain areas, such as bursting versus sparse spiking patterns. Fourth, NeuroQuest provides a variety of graphical tools to help the user with automated or semi-automated parameter selection by instantaneously illustrating the effect of a particular selection on the analysis results, thereby reducing the user time during the learning phase. Individual modules are designed as independent GUIs that allow developers to easily integrate their own processing routines within the software. Finally, NeuroQuest provides advanced spike train analysis tools that enable identifying the functional and effective connectivity between simultaneously recorded neurons. This is an important step, particularly when studying network dynamics to elucidate the potential role of correlation coding, if any, of behavioral covariates.
2. Methods
2.1. Structure and Organization of NeuroQuest
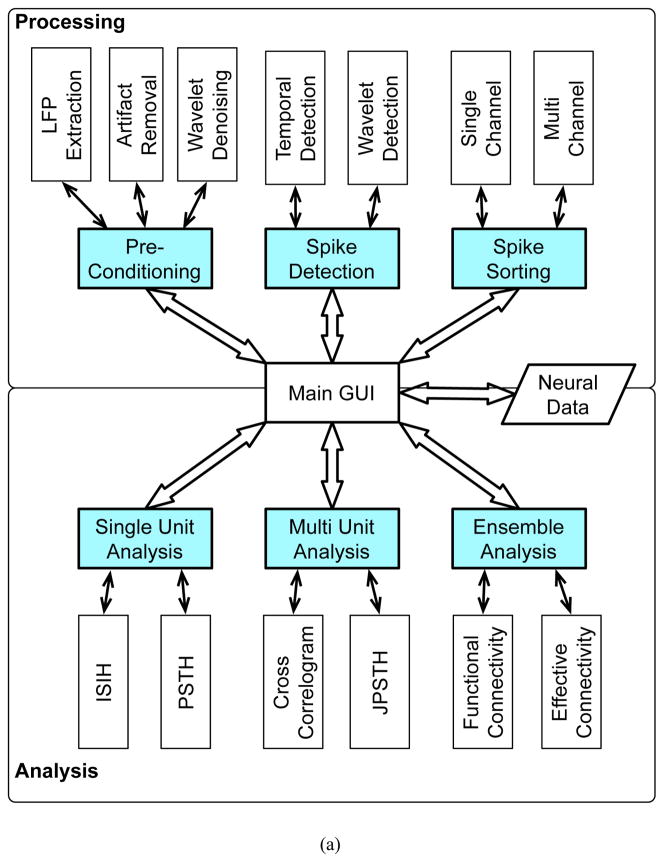
NeuroQuest bundles six processing modules as illustrated in Figure 1.a. These are classified into Processing and Analysis modules. The Processing modules provide a number of tools that extract spikes from raw or pre-processed extracellular recording and perform spike sorting, while the Analysis modules perform statistical analysis of the extracted spike trains. Based on the format of the input data, a corresponding group of modules becomes available in the main menu.
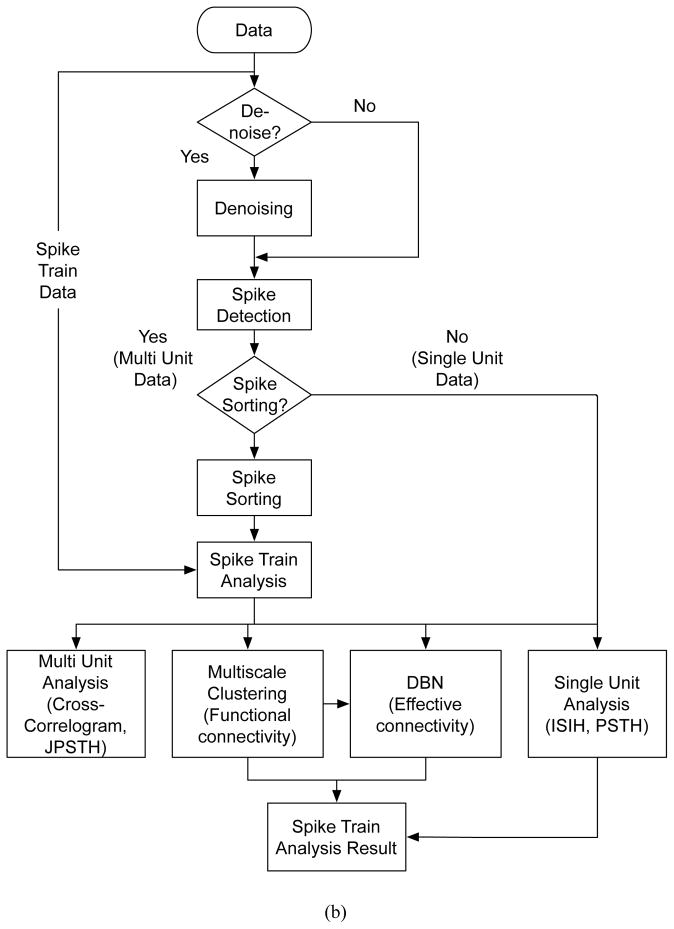
Figure 1. NeuroQuest v1.0 architecture and Flowchart.
(a) Architechture: A total of 6 processing modules (colored blocks) provided in the software are classified into two groups: Processing group and Analysis group. Individual modules are connected through the main GUI which has an access to the input and output data. Spike sorting modules require the raw or preprocessed extracellular recordings, while spike analysis tools handle single or multiple spike trains. Once the input data is loaded, the corresponding group of modules becomes available in the main menu. Each module contains sub-modules that assist to yield more accurate analysis results. (b) Flowchart of NeuroQuest: After the extracellular recording data is loaded, spike sorting tools are activated for further processing. The first stage is to denoise the data to enhance the neural yield. After denoising, spikes are detected and subsequently sent to the spike sorting algorithm to obtain the spike trains. These are further analyzed using the primary spike train analysis tools such as Interspike Interval Histogram (ISIH), Peristimulus Time Histogram (PSTH), Joint Peristimulus Time Histogram (JPSTH), Cross-Correlogram (CC)(Oweiss and Anderson, 2002b; Oweiss, 2010), and the ensemble analysis tools such as functional and effective connectivity estimation.
Figure 1.b illustrates the complete processing and analysis steps in NeuroQuest. Extracellular recordings are first pre-conditioned to remove noise. Spikes are then detected and sorted, after which examination of their statistical properties takes place. These include Single Unit Analysis (SUA) tools, Multi Unit Analysis (MUA) tools, and Ensemble Analysis (EA) tools that include functional and effective connectivity estimation as detailed below.
2.1.1 Data format
Most commercial data acquisition systems store the data according to a vendor-specific data structure. Proper data conversion is therefore an important step to facilitate the use of NeuroQuest. In the absence of a standard data format, data conversion is a challenge. There has been a consensus on unifying structures of neural data, and NeuroShare (neuroshare.org) is one such example. This data structure is designed to efficiently store neural data, and several commercial and academic software packages support this format. Therefore, NeuroQuest includes a conversion routine for Neuroshare support. In case the data is not available in the NeuroShare format, a generic MATLAB data file can be used as the primary input data format for NeuroQuest. The software works with a .MAT file containing a specific data structure (see Appendix A) that can be easily created from ASCII data with a data conversion tool provided in NeuroQuest. NeuroQuest automatically enables the modules to be applied to the loaded data.
2.2. Processing modules
2.2.1 Pre-conditioning module
The pre-conditioning modules are designed to improve the Signal-to-Noise Ratio (SNR) and reduce artifacts. This is believed to increase the accuracy of subsequent processes such as spike detection and spike sorting. Three pre-conditioning modules are provided in NeuroQuest: Local Field Potential (LFP) Extraction, Artifact Removal, and Wavelet Denoising.
2.2.1.1 LFP Extraction
LFPs are low frequency signals in the range of 0.5 Hz – 300Hz and are hypothesized to represent the aggregate activity of synchronized populations of neurons (Buzsáki, 2006). Many studies suggest that LFP contain information about neural representations of behavioral correlates such as hippocampus place fields (Murthy and Fetz, 1992; Mizuseki et al., 2009). NeuroQuest extracts LFP data from the extracellular recording and displays them using a spectrogram (Bokil et al., 2009). A 5th order Butterworth filter is implemented in NeuroQuest for LFP extraction where the upper and lower band limits of the filter can be adjusted (Geiger and Sanchez-Sinencio, 1985).
2.2.1.2 Artifact Removal
An often observed noise component in extracellular recording are artifacts caused by a number of sources such as muscle movement in the scalp, jaws, neck, or body, as well as other types of electrical artifacts from animal movements (Musial et al., 2002). These artifacts are highly correlated across electrodes with occasionally similar spectral content to the desired neural spikes. This similarity may cause neural spikes to overlap in a feature space and makes it difficult to define clear cluster boundaries for spike sorting.
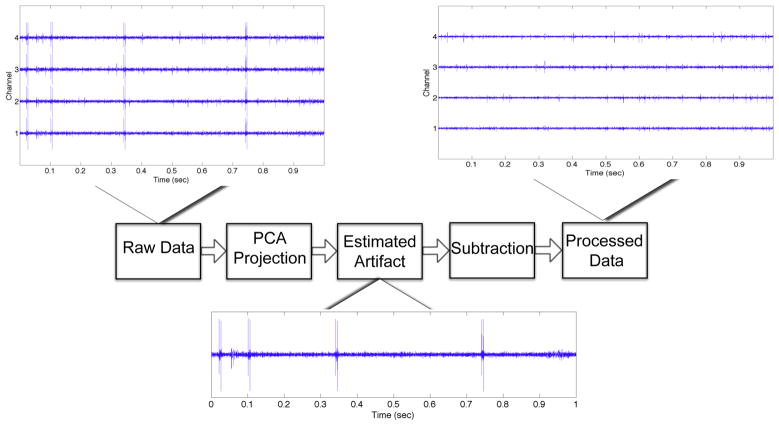
Artifacts can be identified using principal components analysis (PCA). Specifically, the data from all electrodes except for one are stacked together and projected onto the PCA domain using the first two or three principal components (PCs) that best represent the variance across the data (Musial et al., 2002). The selected electrode data is then projected onto the PCA domain. This projection is subtracted from the data yielding a cleaned signal. The entire process is performed sequentially for each electrode. The overall process of artifact removal is illustrated in Figure 2. It is noteworthy that this method should not be used for data with substantial correlation on neighboring electrodes such as stereotrodes or tetrodes, since any neural spikes appearing on multiple adjacent electrodes could be classified as artifacts in such case. More details of the method are found in (Musial et al., 2002).
Figure 2.
Steps of the artifact removal process
2.2.1.3 Denoising
Noise in extracellular recordings is predominantly neural, and therefore is temporally correlated. As a result, it often occupies the same frequency band as the signals of interest. Linear filtering techniques to enhance SNR in such cases have limited capability. Other non-neural noise components (for e.g. thermal and electrical) are typically suppressed by the amplification and bandpass filtering in the recording hardware and do not require further software-based processing.
Reducing colored noise in extracellular recordings can be achieved in a number of ways. Prewhitening - a linear transformation that de-correlates the signal - is one approach to enhance SNR (Pouzat et al, 2002). A similar effect can be achieved using the Discrete Wavelet Transform (DWT) as it substantially sparsifies the signal (Mallat, 1999; Oweiss and Anderson, 2002a; Oweiss, 2006; Oweiss and Aghagolzadeh, 2010), eventually concentrating the signal energy in very few coefficients with large amplitude while small amplitude noise coefficients are widely spread. This feature permits suppressing the noise by thresholding small coefficients. The performance of DWT denoising depends on the choice of the threshold and six different thresholding methods are provided: Heursure, Rigrsure, VisuShrink, SureShrink, BayesShrink, and Minimaxi. Details of these methods are found in (Mallat, 1999; Chang et al., 2002; Donoho, 2002).
2.2.2 Spike Detection module
Detection theory plays a crucial role in processing neural signals, largely because of the highly stochastic nature of these signals and the direct impact that the spike detection step has on subsequent analysis steps (Oweiss, 2010). Since it is widely accepted that spike arrival times carry all the information about information processing in the nervous system - not the actual spike waveform shape, detecting the arrival time of spikes is a fundamental step in the analysis of neural recordings. Some of the major challenges in spike detection are the presence of high levels of time-varying noise statistics as well as the similarity between the spike waveforms and the background noise. Many spike detection algorithms are available to overcome these challenges (Kim and Kim, 2002; Oweiss and Anderson, 2002a; 2002c; Oweiss and Anderson, 2002d; Obeid and Wolf, 2004; Nenadic and Burdick, 2005; Franke et al., 2009; Shahid et al., 2010).
NeuroQuest provides three time-domain spike detection methods based on single amplitude, absolute amplitude, and energy-based statistics. Single amplitude detection identifies any signal that crosses the threshold (only positive or negative) as a spike, while absolute amplitude detection applies both positive and negative thresholds simultaneously (Lewicki, 1998). Energy-based spike detectors compare the local power of the signal to a threshold estimated from the noise power. Manual detection is also provided to allow the user to select a threshold value from the selected data segments. Three different data segments, the lowest SNR, the smallest noise variance, and a large data segment (30 sec of data is selected empirically) can be selected for the threshold value estimation.
The energy-based spike detection method is more robust to noise than the amplitude threshold methods. To gain some insight on why this is the case, let’s express the detection problem as a binary hypothesis testing problem:
where y is the observations of length N, s denotes the L-dimensional spike to be detected, and n is an additive noise term. When the spike s is unknown, Bayesian analysis yields the generalized likelihood ratio test (GLRT) or the incoherent energy detector (see (Oweiss and Anderson, 2002d; Oweiss, 2010) for details) expressed as
| (1) |
Under the assumption of a zero mean Gaussian noise, the GLRT in (1) can be simplified as
| (2) |
where Σ is the noise covariance and η is a threshold value chosen by the user to achieve a constant false positive rate. One way to interpret this detector is to spectrally factor the kernel matrix Σ using singular value decomposition (SVD) to yield the form Σ=UDUT, where U is an eigenvector matrix and D is a diagonal matrix of eigenvalues arranged in decreasing order of magnitude. The detector in this case takes the form
| (3) |
Since Σ is a square positive definite covariance matrix, the eigenvector matrix inverse U−1 is simply its transpose UT. The transformation D−1/2U−1 is lower triangular and has the effect of whitening the observation vector with respect to the noise. The test statistic in this case becomes a blind energy detector, where the observations are whitened, squared, and then compared to a threshold. The whitening step de-correlates the observations and therefore improves the overall performance (Oweiss and Aghagolzadeh, 2010).
Two additional improvements to the energy-based time domain spike detection method are provided in NeuroQuest. The first method deals with the case when the noise is colored and possibly correlated with the spike (e.g. neural noise). In such case, transform domain techniques that yield the most compact representation of the signal are the most desirable because of the transient nature of spikes, which in turn enables superior identification of the information bearing properties of the waveforms (Oweiss and Aghagolzadeh, 2010). NeuroQuest performs multilevel stationary wavelet packet decomposition (SWT) of the recorded data and uses the GLRT based on the transformed data for detection (Kwon and Oweiss, 2011). Different wavelet bases are provided in NeuroQuest for the user to select from which helps to maximize the compactness of wavelet coefficients.
The second method uses the additional information provided when spikes are recorded using an array of electrodes. The sufficient statistic in (2) in such case is computed from a snapshot of the entire array to minimize the effect of the spatially correlated noise component, particularly when the array is closely spaced. More details on this method are provided in (Oweiss, 2002).
2.2.3 Spike sorting module
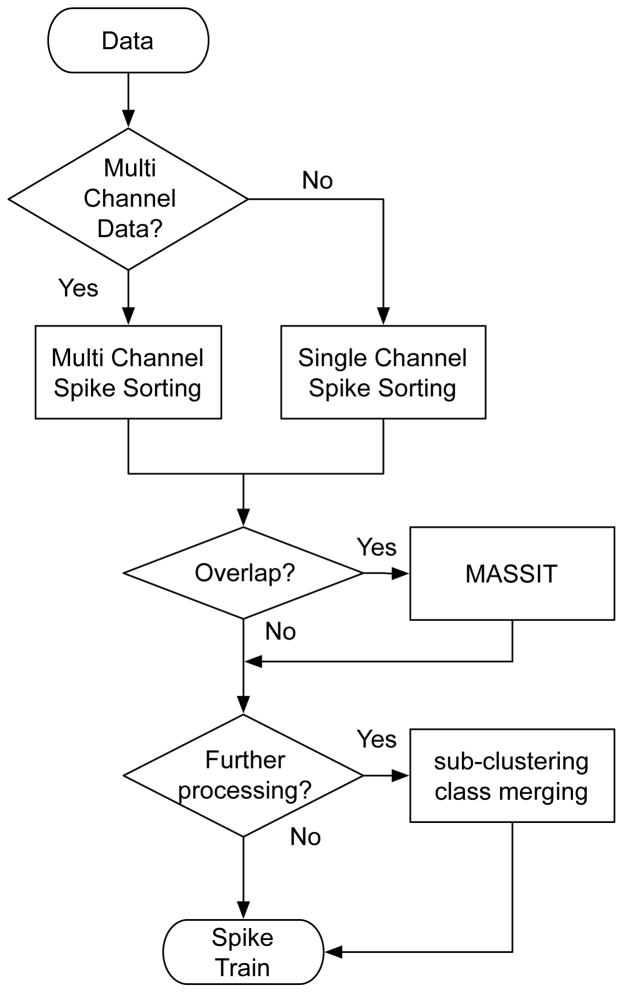
When a given electrode records the activity of multiple neurons, spike sorting is needed to identify the spikes belonging to each neuron. Spike sorting methods are categorized into two approaches: 1) The pattern recognition approach, which operates on the individual spikes extracted after the spike detection step and assumes that spikes from each neuron can be discriminated based on differences in their waveform shapes; 2) The Blind Source Separation (BSS) approach which operates on the raw data without the need to perform spike detection a priori (Oweiss, 2010). The spike sorting algorithm implemented in NeuroQuest combines these approaches as illustrated in Figure 3.
Figure 3. Flowchart of the spike sorting algorithms implemented in NeuroQuest.
Two spike sorting algorithms are available: single-channel and multi-channel. The Multi-channel mode uses spatial information about the distribution of the spike events across channels for sorting in addition to the temporal and spectral information used in the single-channel mode. MASSIT: Multiresolution Analysis of Signal Subspace Invariance Technique identifies the number of signal sources in overlapping spikes observed when two or more neurons fire nearly simultaneously (Oweiss and Anderson, 2002c; Oweiss, 2010).
Two types of spike sorting techniques are included in NeuroQuest: single- and multi-channel sorting. In the multi-channel mode, the design of the recording electrode device must be specified to choose the proper spike sorting method. If spacing between electrodes is small such as stereotrodes or tetrodes (McNaughton, 1983), there is a high probability to record the same action potential by an adjacent group of electrodes. Recordings from these electrodes allow additional information to be used for more accurate spike sorting (Harris et al., 2000; Oweiss and Anderson, 2002b; Oweiss, 2010). The multi-channel sorting mode takes into account the correlation among multiple electrodes and uses this information to resolve any potential ambiguity in the source of the spikes. On the other hand, single-channel sorting does not consider any correlation between the recordings from the adjacent electrodes and processes individual electrode channels independently.
If two or more adjacent spike events overlap, the event is considered to be a complex spike. In that case, the Multiresolution Analysis of Signal Subspace Invariance Technique (MASSIT) can be used to resolve the spike overlap (Oweiss and Anderson, 2007). This is based on an augmented representation of the observation space to simultaneously incorporate the spectral, temporal and spatial information of the spike waveform. The algorithm estimates the number of sources in the complex spike and uses singular value decomposition (SVD) to separate the individual eigenmodes of the data matrix. Multiple cases are considered and summarized in (Oweiss and Aghagolzadeh, 2010).
2.2.3.1 Spike Extraction and Alignment
In the pattern recognition approach to spike sorting, proper extraction and alignment of spikes is crucial for spike sorting accuracy. Spike alignment based on either positive or negative peak is commonly used. Alignment based on the energy of a spike, however, yields better results, particularly in low SNR (Franke et al., 2009). Energy-based spike alignment is performed as follows:
Detected spikes are extracted around the peak detection point.
The mean of all spikes is calculated.
Spikes are translated in time to minimize the Euclidean distance between each spike and the mean.
The process is repeated until convergence or a maximum number of iterations is reached
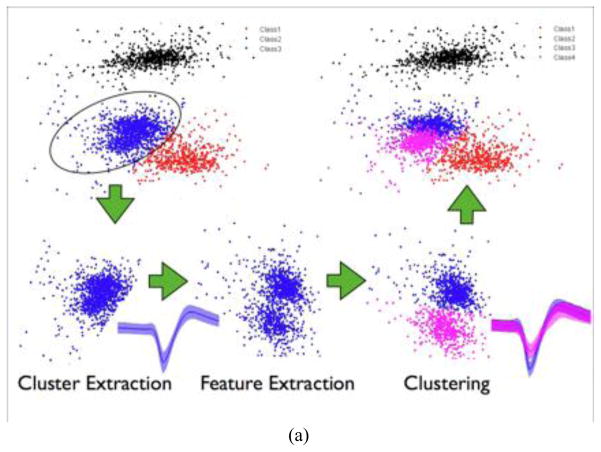
2.2.3.2 Feature Extraction
The second step in spike sorting is the feature extraction. Features that yield the largest separation between spike classes while maintaining the most compact spike representation in the feature space are the most desirable. NeuroQuest provides two time-domain feature extraction methods: peak-to-peak and temporal PCA. In addition, NeuroQuest provides a DWT-based feature extraction, which has been shown recently to outperform the PCA-based feature extraction with properly selected wavelet base (Oweiss and Anderson, 2002c; Oweiss and Anderson, 2007; Aghagolzadeh & Oweiss 2009; Geng et al., 2010; Takekawa et al., 2010). Different combinations of features or wavelet bases can be adjusted to achieve the best cluster separation in the selected feature space.
2.2.3.3 Clustering
Estimating the number of clusters in a given ensemble recording is an important – yet one of the most challenging – problem, often requiring substantial user supervision. Because automation of the analysis is a highly desirable feature in any software, it is important to provide users with some degree of automation without compromising accuracy – a well-established tradeoff in multivariate data analysis. NeuroQuest therefore was designed to give users the ability to choose the level of manual versus automated analysis as a function of desired accuracy. For clustering, automated methods for determining the number of clusters yield an accurate estimate provided enough separability between different clusters exists (Duda et al., 2001). NeuroQuest employs the modified Expectation-Maximization (EM) algorithm to estimate the number of clusters (Figueiredo and Jain, 2002). This specific algorithm can be applied to any type of parametric mixture models. The estimated number of clusters can then be used for clustering the extracted features using four clustering methods provided in NeuroQuest: Fuzzy c-means, EM, k-means, linkage, and manual cluster cutting (Bezdek and Ehrlich, 1984; Duda et al., 2001).
2.2.3.4 Sub-Clustering
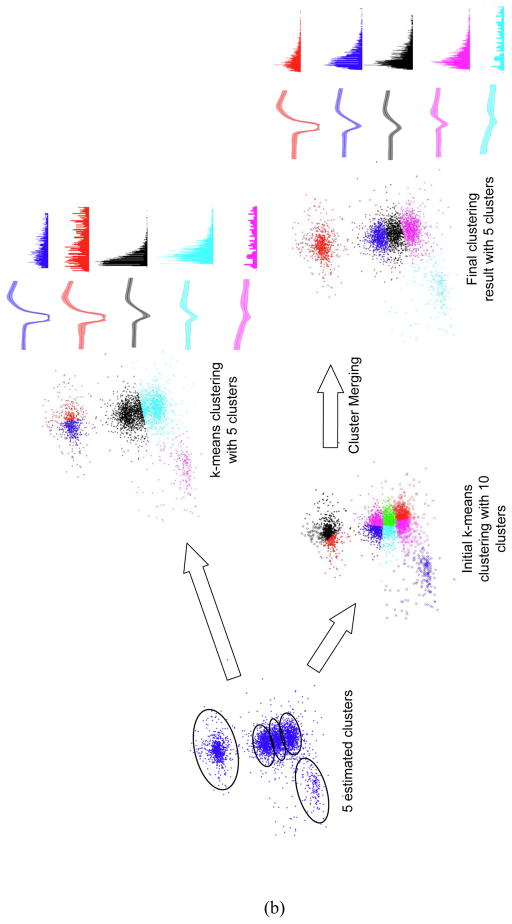
NeuroQuest provides two graphical tools to evaluate and enhance the spike sorting result. Once an initial run of the spike sorting algorithm has been performed, spike clusters can be further split into smaller clusters using the sub-clustering tool. Sub-clustering allows selecting one of the labeled clusters, and projecting it onto a different basis in the original feature space or a new feature space. If there is potential for further splitting of the labeled cluster in the new feature space, one (or more) new cluster is identified. Details of sub-clustering are illustrated in Figure 4.a. The sub-clustering algorithm is performed as follows
Figure 4. Sub-Clustering and cluster merging.
(a) Example of a sub-clustering process (b) Example of cluster merging process
Classified clusters in feature space, spike templates, and ISIH plots are displayed. Color indicates the corresponding clusters and their spike templates and ISIH. Spike template displays an average of a cluster with a centerline and variance of a cluster with a shaded area.
An initially-formed cluster is selected.
The corresponding spikes are projected onto the selected feature space.
The modified EM algorithm estimates the numbers of clusters in that feature space.
The features are clustered into a number of sub-clusters corresponding to the number estimated in the previous step.
The initial results are updated with the new clustering results.
The process is repeated until all the initial clusters are processed.
The process also can be done manually by selecting individual clusters and providing an estimated number of sub-clusters.
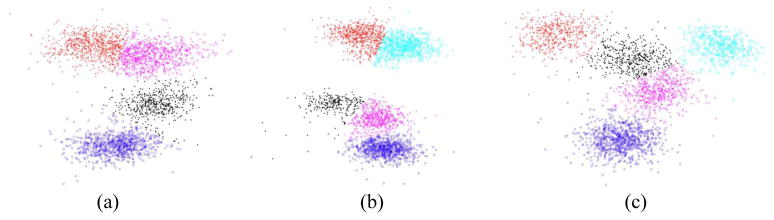
The Inter-spike Interval Histogram (ISIH) metric is used to validate the sorting result to determine whether individual clusters from the first run should be further split or selectively merged together. The ISIH is a distribution of the observed times that elapsed between successive spikes collected in ‘bins’ of fixed width. A significant number of inter-spike intervals within the refractory period would indicate that the sorting results are not perfectly indicative of single units and that sub-clustering should be used to further refine the results. The Class merging tool prevents overfitting the data by combining multiple labeled clusters into a single one without re-clustering the entire data. It can also be used as a manual clustering tool by initially clustering the data into many clusters and then combining them based on the user’s judgment. This technique is particularly useful when several clusters are overlapping in the feature space. Figure 4.b illustrates a 2D feature space that has a total of clusters: two clear clusters, located in the bottom left and the top of the feature space, respectively, and three close clusters, located at the center. Applying the k-means algorithm with 5 clusters gave an undesired clustering result by cutting a well separated cluster into two clusters as shown in Figure 4.b in which the top well separated clusters are split into two clusters, while the three center clusters are clustered as two. Manual clustering of the center cluster was done by first clustering the data into ten clusters then merging them into five clusters. As demonstrated by the ISIH and the uniformity of the spike variance around the mean across time of each cluster in Figure 4.b, the manual clustering using cluster merging yields more accurate spike sorting result.
2.3. Analysis modules
The spike train of a given neuron represents its output in response to the dynamics of its intrinsic properties, such as the membrane conductance, the rebound and refractory effects, and the interactions with other presynaptic neurons. After spike sorting, the spike train is represented as a series of binary zeros and ones, where the presence of a spike is indicated by a ‘1’ and ‘0’ otherwise, provided the sampling rate is sufficiently small. NeuroQuest provides a number of spike train analysis tools categorized into three groups: single unit analysis (SUA), multi unit analysis (MUA), and ensemble analysis modules.
2.3.1 Single Unit Analysis (SUA) module
One of the simplest ways to study the pattern of activity in a spiking neuron is to construct an ISIH. This is useful not only for evaluation of spike sorting accuracy, but also for estimating intrinsic properties of the neuron. The ISIH of an excitatory pyramidal neuron exhibits negative exponential distribution with histogram offset due to the refractory period. One-way to model neuronal firing characteristics is using an inhomogeneous Poisson distribution where the distribution of the inter-spike interval becomes exponentially distributed. A curve fitting tool provided in NeuroQuest validates the closeness of a given spike train to a Poisson distributed by fitting the distribution of the ISIH with an exponential density (Brown et al., 2004).
The Post-stimulus Time Histogram (PSTH) is a histogram of the times at which the neuron fires (Perkel et al., 1967). It is used to visualize the rate and timing of neuronal spike discharges in response to an external stimuli or events, or during the preparation for an upcoming movement (Ellaway, 1978). The SUA GUI displays the raster plot of multiple spike trains and the stimulus, if present, in addition to the number of parameter selections for ISIH and PSTH, such as bin size and pre- and post-stimulus duration for the PSTH display.
2.3.2 Multi Unit Analysis (MUA) module
NeuroQuest provides two well-known multi-unit analysis tools: Cross-correlogram (CC) and Joint Peri-stimulus Time Histogram (JPSTH). The CC is a function that correlates the firing pattern of a target neuron to that of a reference one. Therefore, it provides some information about the statistical dependence between the neuron pair (Perkel et al., 1967). The JPSTH quantifies a similar relationship conditioned on the presence of a stimulus. This allows the JPSTH to be used in studying neuronal functional connectivity over short time scales in the order of a few milliseconds (Perkel et al., 1967). Similar to the SUA GUI, the MUA GUI also provides a number of parameter selections for both JPSTH and CC such as bin size, window size for the CC, and pre- and post-stimulus duration for the JPSTH display.
2.3.3 Ensemble Analysis modules
NeuroQuest provides a number of ensemble level analysis tools that are superior to pairwise measures such as the CC and the JPSTH. The tools are based on the rich literature of graphical models - an elegant blend of probability and graph theories to capture complex dependencies between random variables. They have been widely used in mathematics and machine learning applications such as bioinformatics, communication theory, statistical physics, signal and image processing, and information retrieval (Wainwright and Jordan, 2008; Koller and Friedman, 2009). The ability of these models to capture multimodal, nonlinear and non-Gaussian characteristics of joint population density makes them well suited for analyzing neuronal interactions. Surprisingly, it’s not until recently that they have been introduced to analyze spike trains (Eldawlatly et al., 2009; Eldawlatly and Oweiss, 2010a; Eldawlatly and Oweiss, 2010b)
Briefly, a graphical model of a neuronal population uses a vertex (or a node) to represent a neuron’s state during a specific time interval and an edge to connect the neuron to other neurons (vertices) in the observed population. Inferring graphs with undirected edges would be equivalent to discovering the functional connectivity between the neurons, i.e., the statistical dependency between their spike trains. Graphs with directed edges, on the other hand, would indicate their effective connectivity, i.e., the causal influence exercised between the neurons (Friston, 1994; Bullmore and Sporns, 2009). Graphical representation of neuronal interactions is intuitive because it can be easily visualized and allows the use of many established graph metrics to quantify certain features of the inferred graph that can be compared to imaging and anatomical neural data.
NeuroQuest offers two algorithms that fall under probabilistic graphical models: Multiscale clustering and Dynamic Bayesian Network (DBN). We detail each algorithm below.
2.3.3.1 Multiscale Clustering
This algorithm identifies functional connectivity between spike trains (Eldawlatly et al., 2009). First, the spike trains are projected onto a scale space using Haar wavelet transform (Mallat, 1999). This projection step enables studying neuronal interactions at different time scales. A similarity matrix is then formed at each time scale where each entry in that matrix corresponds to the Pearson correlation coefficient between the projections of a given pair of neurons. Other similarity metrics (for e.g., mutual information) can also be used to fill this matrix. The computed similarity matrices across time scales are then fused together using Singular Value Decomposition (SVD) to obtain a single similarity matrix. Neurons are then represented as nodes in a graph connected with edges whose weights correspond to the entries of the fused similarity matrix. We then use a probabilistic spectral clustering algorithm to cluster the neurons in the obtained graph by solving a minimum graph cut optimization problem (Jin et al., 2006; Eldawlatly et al., 2009).
2.3.3.2 Dynamic Bayesian Networks (DBN)
This algorithm infers the effective connectivity between neurons from the spike trains using a Bayesian framework (Eldawlatly et al., 2010). In the case of time varying stochastic processes with potentially many possible causes such as spike trains, Bayesian analysis provides a powerful framework because it treats all model quantities as random variables. In this approach, connections are randomly proposed one at a time. A score is then computed to assess how well the proposed network fits the data. The network space is searched to identify the network with the highest score. One notable difference between DBN and other measures of effective connectivity (such as cross-correlogram and directed coherence (Jarvis and Mitra, 2001)), DBN does not rely on pair-wise relationships. Rather, it takes into account the activity of the entire observed ensemble when searching for relationships between the neurons. This enables DBN to identify only direct, and not indirect, relationships among observed neurons. We have recently demonstrated the ability of DBN to infer stable, stimulus-specific networks in the medial prefrontal cortex as well as the somatosensory cortex of rats (Eldawlatly and Oweiss, 2011; Eldawlatly and Oweiss, 2010a; Eldawlatly and Oweiss, 2010b). NeuroQuest provides a MATLAB interface with the Bayesian Network Inference with Java Objects (BANJO) toolbox developed in Java (Smith et al., 2006).
Together, the two algorithms constitute a two-stage framework for statistical inference in large-scale neural population data. The multiscale clustering algorithm is used first to reduce the dimensionality of the problem to a smaller number of neuronal clusters. The DBN algorithm is then applied to identify the connectivity within each cluster (Eldawlatly and Oweiss, 2010b).
3. Results
We evaluated the performance of the software by comparing: 1) the data handling capability; 2) the spike detection performance; 3) the effectiveness and flexibility in extracting features for advanced analysis; and 4) the spike sorting performance. Data handing capability is a major feature to consider when evaluating software usability. Spike detection helps demonstrate the robustness of the software under low SNR scenarios. Comparison of different feature extraction methods helps demonstrate that the plurality of feature extraction methods available in NeuroQuest is necessary to deal with variable data quality under different experimental conditions. Spike sorting helps assess the reliability and accuracy of NeuroQuest relative to other software packages when analyzing the same data sets. We demonstrate the performance of NeuroQuest compared to Osort, KlustaKwik, and wave_clus. We analyzed four simulated datasets provided by Osort, wave_clus and one experimental dataset from our archive. Each software was used as is, with only a slight adjustment to the detection threshold values to fix the false detection rates and assess the corresponding true detection rates.
3.1 Data handling capability
Osort supports Neuralynx data format (*.Ncs, both analog and digital cheetah versions) and a text file, and the data extraction routine in the software can be customized. KlustaKwik is not a stand-alone spike sorting software, and has been used primarily as a clustering method. The precompiled Windows version of KlustaKwik used in this study only supported a text file. Wave_clus supports cheetah data format (*.csc), ASCII text file and *.mat. NeuroQuest supports Neuroshare, TDT data tank, and *.mat with data conversion routines. Since most major data acquisition system vendors support the Neuroshare standard, experimental data can be analyzed with NeuroQuest with minimal data conversion effort. NeuroQuest, in addition, is the only software capable of using information provided by multichannel data with different array geometries such as stereotrodes, tetrodes and polytrodes, while other software packages are limited to a single channel at any given time.
3.2 Spike detection comparison
Spike detection performance of three software packages, Osort, wave_clus, and NeuroQuest, was compared. Three simulated datasets (labeled sim1, sim2, and sim3) with four different SNRs obtained from OSort were used (Rutishauser et al., 2006). Simulated datasets were generated by using a database of 150 spike waveform templates taken from well-separated neurons recorded in previous experiments. The background noise was generated by randomly selecting and scaling spike waveforms from the database before adding them to the noise traces. Sim1 dataset contained three neurons, each simulated by a renewal Poisson process with a refractory period of 3 ms and a mean firing rate of 5, 7 and 4 Hz, respectively. The different noise levels (1, 2, 3, and 4) were processed and evaluated independently. In sim1 dataset, they corresponded to an average SNR of 1.2, 2.2, 3.4, and 6.7 dB, respectively. Sim2 dataset contained three neurons and was generated in a similar manner to sim1 with an average SNR of 1.3, 1.7, 2.6, and 5.2 dB, respectively. Spikes in sim2 dataset were hard to detect compared to sim1 dataset, mainly because the spike waveforms in sim2 dataset have similar peak-to-peak values. Sim3 dataset contained five neurons and was the most challenging to process due to the similarity among the spike waveforms. The five neurons in sim3 had an average SNR of 1.2, 1.6, 2.3, and 4.7 dB, respectively (Rutishauser et al., 2006).
We used the wavelet-based spike detection method in NeuroQuest. This particular algorithm enables integrating information about spike transitions represented across scales – often referred to as a wavelet footprint (Dragotti and Vetterli, 2003). A wavelet footprint is defined as a scale space vector obtained by gathering all the wavelet coefficients around the discontinuities of a signal, which usually carry information about piecewise polynomial signals. Spike detection implemented in OSort is an energy-based spike detector that compares the local power of the signal with a threshold estimated from the noise power (Kim and Kim, 2002). Wave_clus provides spike detection based on amplitude thresholding with 4 × standard deviation of the background noise (Donoho and Johnstone, 1994). The spike detection results obtained by NeuroQuest were compared to the results obtained by Osort and wave_clus.
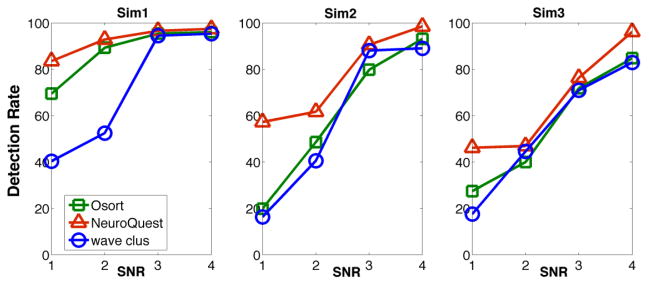
Figure 6 illustrates the performance, where the detection rate is defined as the total number of correctly detected spikes divided by the total number of actual spikes. As Figure 6 shows, NeuroQuest provides more robust detection under various noise levels at a similar false detection rate. Details of the spike detection comparison between NeuroQuest, OSort, and wave_clus results are summarized in Table 1.
Figure 6. Comparison of spike detection performance in NeuroQuest, wave_clus, and OSort.
Sim1 dataset contains three neurons with different noise levels (1, 2, 3, and 4) that is an average SNR of 1.2, 2.2, 3.4, and 6.7 dB, respectively. Sim2 dataset contains three neurons with an average SNR of 1.3, 1.7, 2.6 and 5.2 dB, respectively. Sim3 dataset contains five neurons, some of which have very similar waveforms, with an average SNR of 1.2, 1.6, 2.3, and 4.7 dB, respectively.
Table 1. Spike detection results wave_clus, Osort, and NeuroQuest with three simulation datasets (Sim1, Sim2, and Sim3).
TP indicates true positive and FP indicates false positive of spike detection.
| SNR | 4 | 3 | 2 | 1 | |
|---|---|---|---|---|---|
| Sim1 (1576) | Wave clus | TP:1502 (95.30%) FP: 0 |
TP:1489 (94.48%) FP: 0 |
TP:827 (52.47%) FP: 2 |
TP:636 (40.36%) FP: 5 |
| Osort | TP:1513 (96%) FP: 0 |
TP:1503 (95.37%) FP: 2 |
TP:1407 (89.28%) FP: 46 |
TP:1094 (69.42%) FP: 163 |
|
| Neuro Quest | TP:1535 (97.40%) FP: 0 |
TP:1523 (96.57 %) FP: 0 |
TP:1463 (92.83%) FP: 13 |
TP:1316 (83.50%) FP: 75 |
|
| Sim2 (1568) | Wave clus | TP:1397 (89.01%) FP: 0 |
TP:1381 (88.07%) FP: 2 |
TP:636 (40.56%) FP: 3 |
TP:256 (16.33%) FP: 6 |
| Osort | TP:1460 (93.11%) FP: 0 |
TP:1251 (79.78%) FP: 14 |
TP:762 (48.60%) FP: 202 |
TP:312 (19.90%) FP: 134 |
|
| Neuro Quest | TP:1544 (98.47%) FP: 0 |
TP:1420 (90.56%) FP: 39 |
TP:967 (61.67%) FP: 85 |
TP:857 (57.27%) FP: 225 |
|
| Sim3 (2986) | Wave clus | TP:2475 (82.89%) FP: 0 |
TP:2097 (70.94%) FP: 4 |
TP:1330 (44.54%) FP: 10 |
TP:524 (17.55%) FP: 14 |
| Osort | TP:2531 (84.76%) FP: 7 |
TP:2145 (71.84%) FP: 317 |
TP:1193 (39.95%) FP: 270 |
TP:820 (27.46%) FP: 308 |
|
| Neuro Quest | TP:2870 (96.12%) FP: 6 |
TP:2278 (76.29%) FP: 60 |
TP:1400 (46.89%) FP: 321 |
TP:1378 (46.15%) FP: 348 |
3.3 Effectiveness of feature extraction
We evaluated the effectiveness of feature selection and spike alignment. Three different feature sets were extracted: 1) peak-to-peak, 2) temporal PCA, and 3) wavelet footprint. The effectiveness of each method was assessed based on clustering separability. Specifically, the separability of spike clusters {Ci| i = 1, 2, …, P} was defined as
| (4) |
where SB is the between-clusters scatter and SW is the within-clusters scatter, defined respectively as (Tan et al., 2006)
| (5) |
| (6) |
where |Cj| equals the number of spikes belonging to class Cj and ||.|| represents the Euclidean norm.
For a more strenuous comparison between the different feature extraction methods, 12 simulated data sets with different noise levels and spike waveforms were randomly generated from a database of 131 spike waveform templates that were detected, extracted and averaged from spontaneous activity recorded in the primary motor cortex of an anesthetized rat. The Institutional Animal Care and Use Committee at Michigan State University following National Institutes of Health (NIH) guidelines approved all procedures. Details of the experimental procedures to obtain these recordings are described in (Oweiss, 2006). Noise obtained from spontaneous activity was added to the spike templates selected from the database using a renewal Poisson process with a refractory period of 3 ms and a fixed firing rate of 5Hz.
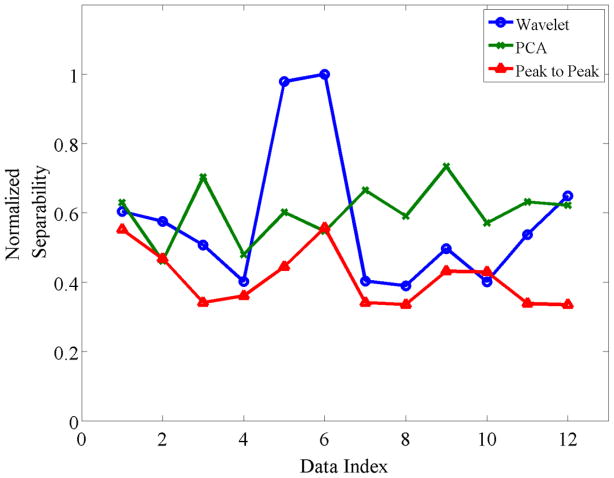
Overall, the temporal PCA feature selection exhibited good separability, while the wavelet footprint feature showed higher separability in datasets 2, 5 and 6 as illustrated in Figure 7. These specific datasets contained spike waveforms with large similarities that could not be captured by the first few PCs. Peak-to-peak features yielded poor performance in most cases, but showed equal separability to the PCA in datasets 1, 2, and 6, and to the wavelet footprint method in datasets 1, 4, and 10. These results indicate that there is no single feature extraction method that is superior to the others across all datasets.
Figure 7. Separability comparison between three feature extraction methods.
A total of 12 datasets generated using a renewal Poisson process with a refractory period of 3 ms and a fixed firing rate 5Hz were examined. Each dataset has different noise levels and spike waveforms that were randomly selected from a database of 131 average spike waveforms obtained from spontaneous activity recorded in the primary motor cortex of an anesthetized rat.
To demonstrate how spike alignment can affect the spike sorting results, we processed three simulated data (sim1, sim2, and sim3 with SNR3) obtained from Osort using three different spike alignment methods: 1) negative peak; 2) positive peak; and 3) energy-based. The results are summarized in Table 2. Figure 8 shows an example PCA feature space of the sim3 dataset with different alignment methods. These results highlight the importance of proper spike alignment for accurate spike sorting.
Table 2. Spike sorting results with three spike alignment methods.
( ) indicates the number of classified units.
| Negative | Positive | Absolute | |
|---|---|---|---|
| sim1 (3) | TP : 99.8% (3) | TP : 97.8% (3) | TP : 99.8% (3) |
| sim2 (3) | TP : 98% (3) | TP : 97.3% (3) | TP : 100% (3) |
| sim3 (5) | TP : 85% (4) | TP : 95% (5) | TP : 92% (5) |
Figure 8. Sample Clustering result of detected spikes in a temporal PCA feature space with different alignment methods.
(a) Negative Peak Alignment (b) Absolute Peak Alignment. (c) Positive Peak Alignment
3.4 Spike sorting comparison
Four simulated datasets and one experimental dataset were used to evaluate the spike sorting performance of NeuroQuest. The simulated data used in spike detection comparison were reused with some modifications. The three datasets (sim1, sim2, and sim3) were combined into one dataset (3 channel data), and the dataset was duplicated into three datasets with three different noise levels (SNR2, SNR3, and SNR4). Each dataset (3 channels) contained 11 total units (3 units in channel 1 and channel 2, and 5 units in channel 3). Another simulated dataset obtained from wave_clus (labeled difficult1_010.mat) was also analyzed for comparison. The data was generated in a similar manner as the Osort data: three distinct spikes with a Poisson distribution of interspike intervals with mean ring rate of 20 Hz. This particular dataset was considered difficult to sort because three spike classes share the same peak amplitude with very similar widths and shapes. Relative noise level of the data was 10% of average spike peak values. The experimental dataset analyzed was recorded in the dorsal cochlear nucleus of an anesthetized Guinea pig where five units were clearly isolated and sorted by an expert.
Spikes were detected using Osort and spike event times were given to NeuroQuest and wave_clus where the first five principal components of the extracted spikes obtained by NeuroQuest were used as an input to KlustaKwik. The spike sorting results are shown in Table 3. NeuroQuest yielded consistent spike sorting performance in all datasets that was also superior to other software packages.
Table 3. Spike sorting results of four software packages, Klusta-Kwik, wave_clus, Osort, and NeuroQuest, with five datasets, four simulated data and one real data.
( ) indicates the number of classified units and TP represents true positive. For NeuroQuest, details of spike sorting methods, selected feature and clustering method, are listed. The number of clusters was decided by the modified EM algorithm (Figueiredo and Jain, 2002).
| SNR4 (11 units) | SNR3 (11 units) | SNR2 (11 units) | Wave_clus data (3 units) | real data (5 units) | |
|---|---|---|---|---|---|
| Klusta Kwik | (11 units) TP: 99.96% | (11 units) TP: 95.32% | (9 units) TP : 86.82% | (1 units) TP : 42.51% | (3 units) TP : 67.43% |
| Wave_clus | (11 units) TP : 99.96% | (10 units) TP : 92.50% | (8 units) TP : 86.48% | (3 units) TP : 98.48% | (4 units) TP : 84.72% |
| Osort | (11 units) TP : 99.84% | (11 units) TP : 93.13% | (9 units) TP : 85.83% | (1 units) TP : 46.72% | (2 units) TP : 41.75% |
| Neuro Quest | (11 units) Temporal PCA Fuzzy c-mean TP : 99.92% |
(11 units) Temporal PCA Fuzzy c-mean TP : 98.84% |
(10 units) Temporal PCA Fuzzy c-mean Class merging Sub-clustering TP : 91.43% |
(2 units) wavelet footprint Fuzzy c-mean TP : 73.24% |
(5 units) Temporal PCA Fuzzy c-mean TP : 98.85% |
4. Discussion
NeuroQuest was developed to address one fundamental need in the neuroscience community: the analysis of massive amounts of neural data with highly sophisticated algorithms while keeping simplicity and automation as primary features for wide user adoption. As summarized in Table 4, most of the current academic software packages fall into either spike sorting or spike train analysis software categories. Since both are required to perform a comprehensive data analysis, multiple software packages are typically needed to analyze a given dataset. Table 5 lists the features available in several known software packages. Among the spike sorting packages, only NeuroMAX and OSort offer limited spike train analysis tools (Rutishauser et al., 2006). Among these tools, FIND is the only software equipped with spike detection and a rather elementary spike sorting tool (Meier et al., 2008).
Table 4.
Existing software packages
| Category | Spike Sorting | Spike Train Analysis |
|---|---|---|
| Software | MClust, OSort, Klusters, KlustaKwik, Chronux, NeuroMAX, , Wave_clus | MatOFF, Spike Train Analysis Toolkit, FIND |
Table 5.
Features of the software packages
| Chornux | KlustaKwik | MClust | NeuroMAX | FIND | MatOFF | STAToolkit | Wave_Clus | OSort | NeuroQuest | |
|---|---|---|---|---|---|---|---|---|---|---|
| GUI | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Pre-Conditioning | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✔ | ✔ |
| Spike Detection | ✕ | ✕ | ✕ | ✔ | ✔ | ✕ | ✕ | ✔ | ✔ | ✔ |
| Spike Sorting | ✔ | ✔ | ✔ | ✔ | ✕ | ✕ | ✕ | ✔ | ✔ | ✔ |
| Single Unit Analysis | ✕ | ✕ | ✕ | ✔ | ✔ | ✔ | ✔ | ✕ | ✔ | ✔ |
| Multi Unit Analysis | ✕ | ✕ | ✕ | ✕ | ✔ | ✔ | ✔ | ✕ | ✕ | ✔ |
| Connectivity Estimation | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✔ |
The comprehensive analysis capability in NeuroQuest reduces the burden of converting the data between different software packages and speeds up the analysis. The simple GUI in NeuroQuest reduces the complexity in parameter selection and shortens the learning time. The graphical tools provided help the user during manual parameter selection by instantaneously illustrating the effect of parameter changes on the analysis results. The extensive visualization of this process helps to improve the accuracy of the analysis results. Since individual processing modules are designed as independent GUIs in NeuroQuest, algorithms developed by users can be easily integrated into NeuroQuest for performance comparison using a template GUI provided.
Although NeuroQuest runs under MATLAB environment, we circumvented possible memory usage issues by dividing large datasets into smaller segments. This segmentation helps to speed up the processing and resolves the ‘out of memory’ problem. The segmentation process also helps to improve the accuracy of the analysis when signals are nonstationary by estimating signal parameters independently for each segment.
Extracellular recordings using microelectrode arrays with different geometries require specially designed algorithms that most software packages do not offer, which hinders efficient use of spatial information that can improve sorting results, particularly when resolving complex overlapping spikes. NeuroQuest is the only software - to our knowledge - that is comprehensive in offering spike detection and sorting algorithms that can analyze data recorded with closely spaced microelectrode arrays.
The current version has some limitations that we continue to overcome. For example, the GUIs were designed using the (GUIDE) tools in MATLAB. Although the GUIDE tools provide basic elements for building a simple GUI structure, they are not efficient to allow more sophisticated user interfaces. Careful optimization and more efficient implementation in a programming language such as C++ and a better GUI programming language such as C# will provide substantial improvements in speed and more interactive user interfaces. Another limitation is that the current version does not allow processing in real time. Online spike detection and spike sorting are sometimes desirable, particularly in BMI applications where real-time neural decoding is needed. Future versions of NeuroQuest will be equipped with real-time algorithm implementation that is adapted to most of the available commercial data acquisition systems. This will provide a platform to test custom-built analysis tools and enable replicating experimental data analysis across labs to facilitate objective comparison between scientific findings. We believe NeuroQuest is a good first step towards standardizing the analysis of neural recordings in a variety of experimental designs.
The current beta version of the software can be downloaded from (www.neuroquest.org) with and end-user licensing agreement (EULA). In the EULA, we required user feedback primarily to help tailor the software design at this initial development stage to users needs and further provide adequate support as needed.
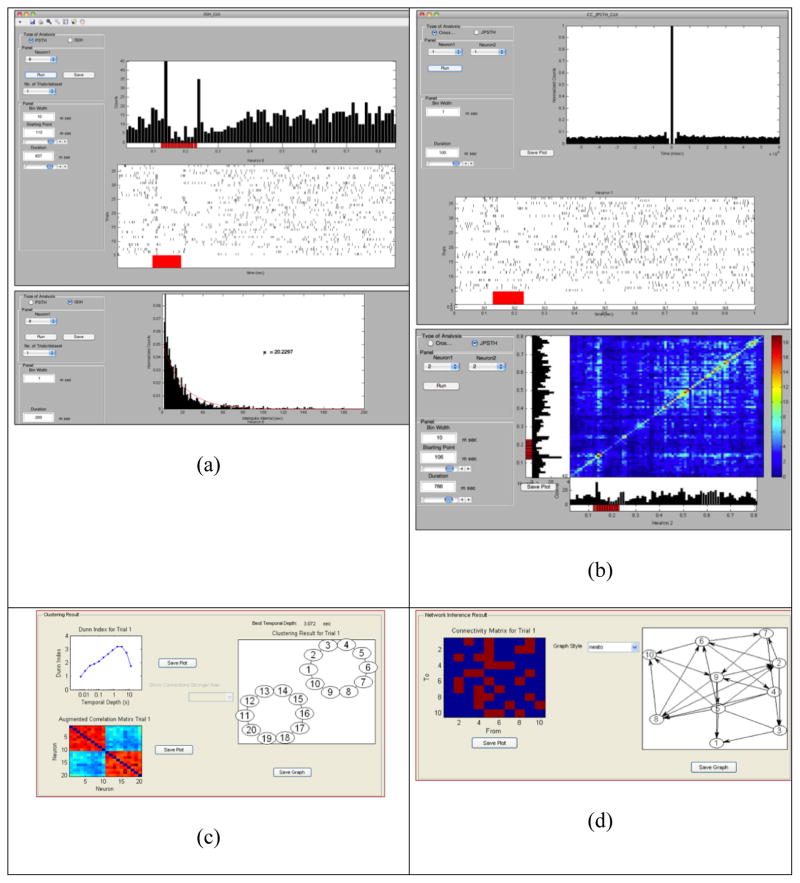
Figure 5. Sample screen shot of the spike train analysis tools.
(a) Single Unit Analysis Tools (b) Multi Unit Analysis Tools (c) Functional connectivity estimation GUI (d) Effective connectivity estimation GUI
Highlights.
NeuroQuest is a MATLAB-based toolbox for analyzing extracellular neural data
NeuroQuest provides an efficient and time saving unified processing
Spike detection and sorting in NeuroQuest outperform other software packages
Acknowledgments
This work was supported by NINDS grant number NS054148.
Appendix A
File format
NeuroQuest works with MAT files that contain the following data structure.
data: A cell array where each cell contains 1 sec of raw data. Each cell contains a matrix with each row representing a single channel and each column representing one sample.
BinWidth: Inverse of the data sampling rate.
chanInfo: Channel labels in a row vector. (e.g. Four channel data:[1 2 3 4])
plotOption: A structure array containing the following flags: raw, denoise, detection, stimulus, LFP, spiketrain, and Trials. Each flag should be set to ‘1’ if the file contains that type of data or ‘0’ if it does not.
fileDescription: a cell array that contains a short description, a string, of the data.
The processed results in each step are added to the above data structure. For example the spike detection process produces spike position data and updates plotOption and fileDescription. If spikes have already been detected by another software package, the spike position data can be included in the data file using the following data structure.
denoised_data: A cell array structured identically to the data array containing the raw data that has undergone noise reduction. This can be calculated using the PreConditioning tool.
spikeposition: A cell array where each cell contains the time stamps of detected spikes in 1 sec data. This can be calculated using the spike detection tool.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beck JM, Ma WJ, Kiani R, Hanks T, Churchland A, Roitman J, Shadlen MN, Latham P, Pouget A. Probabilistic population codes for Bayesian decision making. Neuron. 2008;60:1142–1152. doi: 10.1016/j.neuron.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezdek JC, Ehrlich R. FCM: The fuzzy c-means clustering algorithm. Computers & Geosciences. 1984;10:191–203. [Google Scholar]
- Bokil H, Andrews P, Maniar H, Pesaran B, Kulkarni J, Loader C, Mitra P. Chronux: a platform for analyzing neural signals. BMC Neuroscience. 2009;10:S3. doi: 10.1016/j.jneumeth.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E, Kass R, Mitra P. Multiple neural spike train data analysis: state-of-the-art and future challenges. Nature Neuroscience. 2004;7:456–461. doi: 10.1038/nn1228. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nature Reviews Neuroscience. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Buzsáki G. Rhythms of the Brain. Oxford University Press; USA: 2006. [Google Scholar]
- Chang S, Yu B, Vetterli M. Adaptive wavelet thresholding for image denoising and compression. Image Processing, IEEE Transactions on. 2002;9:1532–1546. doi: 10.1109/83.862633. [DOI] [PubMed] [Google Scholar]
- DeWeese MR, Wehr M, Zador AM. Binary spiking in auditory cortex. Journal of Neuroscience. 2003;23:7940. doi: 10.1523/JNEUROSCI.23-21-07940.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoho D. De-noising by soft-thresholding. Information Theory, IEEE Transactions on. 2002;41:613–627. [Google Scholar]
- Donoho D, Johnstone J. Ideal spatial adaptation by wavelet shrinkage. Biometrika. 1994;81(3):425–455. [Google Scholar]
- Dragotti P, Vetterli M. Wavelet footprints: Theory, algorithms, and applications. Signal Processing, IEEE Transactions on. 2003;51:1306–1323. [Google Scholar]
- Duda R, Hart P, Stork D. Pattern classification. Citeseer: 2001. [Google Scholar]
- Eldawlatly S, Jin R, Oweiss K. Identifying functional connectivity in large-scale neural ensemble recordings: a multiscale data mining approach. Neural computation. 2009;21:450–477. doi: 10.1162/neco.2008.09-07-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldawlatly S, Zhou Y, Jin R, Oweiss K. On the use of dynamic bayesian networks in reconstructing functional neuronal networks from spike train ensembles. Neural computation. 2010;22:158–189. doi: 10.1162/neco.2009.11-08-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldawlatly S, Oweiss K. Causal neuronal networks provide functional signatures of stimulus encoding. Proc. of 32nd IEEE Eng. in Medicine and Biology; 2010a. [DOI] [PubMed] [Google Scholar]
- Eldawlatly S, Oweiss K. Statistical Signal Processing for Neuroscience and Neurotechnology. Academic Press, Elesevier; 2010b. Graphical Models of Functional and Effective Neuronal Connectivity; pp. 129–174. [Google Scholar]
- Eldawlatly S, Oweiss K. Millisecond-Timescale Local Network Coding in the Rat Primary Somatosensory Cortex. PLos ONE. 2011;6(6) doi: 10.1371/journal.pone.0021649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellaway PH. Cumulative sum technique and its application to the analysis of peristimulus time histograms. Electroencephalography and Clinical Neurophysiology. 1978;45:302–304. doi: 10.1016/0013-4694(78)90017-2. [DOI] [PubMed] [Google Scholar]
- Fee MS, Mitra PP, Kleinfeld D. Automatic sorting of multiple unit neuronal signals in the presence of anisotropic and non-Gaussian variability. Journal of neuroscience methods. 1996;69:175–188. doi: 10.1016/S0165-0270(96)00050-7. [DOI] [PubMed] [Google Scholar]
- Figueiredo M, Jain A. Unsupervised learning of finite mixture models. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 2002;24:381–396. doi: 10.1109/TPAMI.2004.1273970. [DOI] [PubMed] [Google Scholar]
- Franke F, Natora M, Boucsein C, Munk MHJ, Obermayer K. An online spike detection and spike classification algorithm capable of instantaneous resolution of overlapping spikes. Journal of Computational Neuroscience. 2009:1–22. doi: 10.1007/s10827-009-0163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ. Functional and effective connectivity in neuroimaging: a synthesis. Human Brain Mapping. 1994;2:56–78. [Google Scholar]
- Fujisawa S, Amarasingham A, Harrison MT, Buzsáki G. Behavior-dependent short-term assembly dynamics in the medial prefrontal cortex. Nature neuroscience. 2008;11:823–833. doi: 10.1038/nn.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger R, Sanchez-Sinencio E. Active filter design using operational transconductance amplifiers: a tutorial. IEEE Circuits and Devices Magazine. 1985;1:20–32. [Google Scholar]
- Geng X, Hu G, Tian X. Neural spike sorting using mathematical morphology, multiwavelets transform and hierarchical clustering. Neurocomputing. 2010;73:707–715. [Google Scholar]
- Harris KD, Csicsvari J, Hirase H, Dragoi G, Buzsáki G. Organization of cell assemblies in the hippocampus. Nature. 2003;424:552–556. doi: 10.1038/nature01834. [DOI] [PubMed] [Google Scholar]
- Harris KD, Henze DA, Csicsvari J, Hirase H, Buzsáki G. Accuracy of tetrode spike separation as determined by simultaneous intracellular and extracellular measurements. Journal of Neurophysiology. 2000;84:401–414. doi: 10.1152/jn.2000.84.1.401. [DOI] [PubMed] [Google Scholar]
- Hazan L, Zugaro M, Buzsáki G. Klusters, NeuroScope, NDManager: a free software suite for neurophysiological data processing and visualization. Journal of neuroscience methods. 2006;155:207–216. doi: 10.1016/j.jneumeth.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Hermle T, Schwarz C, Bogdan M. Employing ICA and SOM for spike sorting of multielectrode recordings from CNS. Journal of Physiology-Paris. 2004;98:349–356. doi: 10.1016/j.jphysparis.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Hulata E, Segev R, Ben-Jacob E. A method for spike sorting and detection based on wavelet packets and Shannon’s mutual information. Journal of Neuroscience Methods. 2002;17:1–12. doi: 10.1016/s0165-0270(02)00032-8. [DOI] [PubMed] [Google Scholar]
- Jarvis MR, Mitra PP. Sampling properties of the spectrum and coherency of sequences of action potentials. Neural Computation. 2001;13:717–749. doi: 10.1162/089976601300014312. [DOI] [PubMed] [Google Scholar]
- Jin R, Ding C, Kang F. A probabilistic approach for optimizing spectral clustering. Advances in neural information processing systems. 2006;18:571. [Google Scholar]
- Kim K, Kim S. Neural spike sorting under nearly 0-dB signal-to-noise ratio using nonlinear energy operator and artificial neural-network classifier. Biomedical Engineering, IEEE Transactions on. 2002;47:1406–1411. doi: 10.1109/10.871415. [DOI] [PubMed] [Google Scholar]
- Koller D, Friedman N. Probabilistic Graphical Models: Principles and Techniques. The MIT Press; 2009. [Google Scholar]
- Kwon K, Eldawlatly S, Oweiss K. NeuroQuest: A comprehensive tool for large scale neural data processing and analysis. 4th International IEEE/EMBS Conference on Neural Engineering; 2009. pp. 622–625. [Google Scholar]
- Kwon K, Oweiss K. NeuroQuest: Detection and Extraction of Wavelet Footprint in Extracellular Recording. IEEE International Conference on Acoustics, Speech, and Signal Processing; 2011. [Google Scholar]
- Lewicki MS. A review of methods for spike sorting: the detection and classification of neural action potentials. Network: Computation in Neural Systems. 1998;9:53. [PubMed] [Google Scholar]
- Mallat S. A wavelet tour of signal processing. Academic Pr; 2009. [Google Scholar]
- McNaughton S. Compensatory plant growth as a response to herbivory. Oikos. 1983;40:329–336. [Google Scholar]
- Meier R, Egert U, Aertsen A, Nawrot M. FIND--A unified framework for neural data analysis. Neural Networks. 2008;21:1085–1093. doi: 10.1016/j.neunet.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Sirota A, Pastalkova E, Buzsáki G. Theta oscillations provide temporal windows for local circuit computation in the entorhinal-hippocampal loop. Neuron. 2009;64:267–280. doi: 10.1016/j.neuron.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy VN, Fetz EE. Coherent 25-to 35-Hz oscillations in the sensorimotor cortex of awake behaving monkeys. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:5670. doi: 10.1073/pnas.89.12.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musial P, Baker S, Gerstein G, King E, Keating J. Signal-to-noise ratio improvement in multiple electrode recording. Journal of neuroscience methods. 2002;115:29–43. doi: 10.1016/s0165-0270(01)00516-7. [DOI] [PubMed] [Google Scholar]
- Nenadic Z, Burdick JW. Spike detection using the continuous wavelet transform. IEEE Transactions on Biomedical Engineering. 2005;52:74–87. doi: 10.1109/TBME.2004.839800. [DOI] [PubMed] [Google Scholar]
- Obeid I, Wolf PD. Evaluation of spike-detection algorithms fora brain-machine interface application. IEEE Transactions on Biomedical Engineering. 2004;51:905–911. doi: 10.1109/TBME.2004.826683. [DOI] [PubMed] [Google Scholar]
- Oweiss K. Statistical Signal Processing for Neuroscience and Neurotechnology. East Lansing: Elsevier; 2010. [Google Scholar]
- Oweiss K, Anderson DJ. A new approach to array denoising. IEEE Asilomar Conference on Signals, Systems and Computers; 2002a. pp. 1403–1407. [Google Scholar]
- Oweiss K, Anderson DJ. A new technique for blind source separation using subband subspace analysis in correlated multichannel signal environments. IEEE Acoustics, Speech, and Signal Processing. 2002c:2813–2816. [Google Scholar]
- Oweiss K, Anderson DJ. MASSIT-Multiresolution Analysis of Signal Subspace Invariance Technique: a novel algorithm for blind source separation. IEEE Asilomar Conference on Signals, Systems and Computers; 2002b. pp. 819–823. [Google Scholar]
- Oweiss K, Aghagolzadeh M. Detection and Classication of Extracellular Action Potential Recordings. Statistical Signal Processing for Neuroscience. 2010:15. [Google Scholar]
- Oweiss K, Anderson D. A multiresolution generalized maximum likelihood approach for the detection of unknown transient multichannel signals in colored noise with unknown covariance. IEEE Acoustics, Speech, and Signal Processing. 2002d [Google Scholar]
- Oweiss K. PhD dissertation. Univ. Michigan; Ann Arbor: 2002. Multiresolution analysis of multichannel neural recordings in the context of signal detection, estimation, classification and noise suppression. [Google Scholar]
- Oweiss K. A systems approach for data compression and latency reduction in cortically controlled brain machine interfaces. Biomedical Engineering, IEEE Transactions on. 2006;53:1364–1377. doi: 10.1109/TBME.2006.873749. [DOI] [PubMed] [Google Scholar]
- Oweiss K, Anderson D. Tracking signal subspace invariance for blind separation and classification of nonorthogonal sources in correlated noise. EURASIP journal on Applied Signal Processing. 2007:194. [Google Scholar]
- Panzeri S, Petersen RS, Schultz SR, Lebedev M, Diamond ME. The role of spike timing in the coding of stimulus location in rat somatosensory cortex. Neuron. 2001;29:769–777. doi: 10.1016/s0896-6273(01)00251-3. [DOI] [PubMed] [Google Scholar]
- Pavlov A, Makarov VA, Makarova I, Panetsos F. Sorting of neural spikes: When wavelet based methods outperform principal component analysis. Natural Computing. 2007;6:269–281. [Google Scholar]
- Perkel DH, Gerstein GL, Moore GP. Neuronal Spike Trains and Stochastic Point Processes:: I. The Single Spike Train Biophysical journal. 1967;7:391–418. doi: 10.1016/S0006-3495(67)86596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouzat C, Mazor O, Laurent G. Using noise signature to optimize spike-sorting and to assess neuronal classification quality. Journal of neuroscience methods. 2002;122:43–57. doi: 10.1016/s0165-0270(02)00276-5. [DOI] [PubMed] [Google Scholar]
- Quiroga Q, Nadasdy Z, Ben-Shaul Y. Unsupervised spike detection and sorting with wavelets and superparamagnetic clustering. Neural Comput. 2004;16:1661–87. doi: 10.1162/089976604774201631. [DOI] [PubMed] [Google Scholar]
- Reimer J, Hatsopoulos NG. Periodicity and Evoked Responses in Motor Cortex. Journal of Neuroscience. 2010;30:11506. doi: 10.1523/JNEUROSCI.5947-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser U, Schuman EM, Mamelak AN. Online detection and sorting of extracellularly recorded action potentials in human medial temporal lobe recordings, in vivo. Journal of neuroscience methods. 2006;154:204–224. doi: 10.1016/j.jneumeth.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Shahid S, Walker J, Smith L. A new spike detection algorithm for extracellular neural recordings. Biomedical Engineering, IEEE Transactions on. 2010;57:853–866. doi: 10.1109/TBME.2009.2026734. [DOI] [PubMed] [Google Scholar]
- Smith VA, Yu J, Smulders TV, Hartemink AJ, Jarvis ED. Computational inference of neural information flow networks. PLoS Comput Biol. 2006;2:e161. doi: 10.1371/journal.pcbi.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takekawa T, Isomura Y, Fukai T. Accurate spike sorting for multi unit recordings. European Journal of Neuroscience. 2010;31:263–272. doi: 10.1111/j.1460-9568.2009.07068.x. [DOI] [PubMed] [Google Scholar]
- Tan PN, Steinbach M, Kumar V. Introduction to data mining. Pearson Addison Wesley; Boston: 2006. [Google Scholar]
- Tiesinga P, Fellous JM, Sejnowski TJ. Regulation of spike timing in visual cortical circuits. Nature Reviews Neuroscience. 2008;9:97–107. doi: 10.1038/nrn2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velliste M, Perel S, Spalding MC, Whitford AS, Schwartz AB. Cortical control of a prosthetic arm for self-feeding. Nature. 2008;453:1098–1101. doi: 10.1038/nature06996. [DOI] [PubMed] [Google Scholar]
- Wainwright MJ, Jordan MI. Graphical models, exponential families, and variational inference. Foundations and Trends in Machine Learning. 2008;1:1–305. [Google Scholar]
- Wiest MC, Nicolelis MAL. Behavioral detection of tactile stimuli during 7–12 Hz cortical oscillations in awake rats. Nature neuroscience. 2003;6:913–914. doi: 10.1038/nn1107. [DOI] [PubMed] [Google Scholar]