Abstract
BKPyV and JCPyV are closely related, ubiquitious human pathogens that cause disease in immunocompromised patients. The DNA sequence of the regulatory regions distinguishes two forms of these viruses, designated archetype and rearranged. Although cell culture systems exist for rearranged BKPyV and JCPyV, currently there is no robust cell culture system to study the archetype viruses. Large T antigen (TAg) is a virally encoded protein required to initiate viral DNA synthesis. Because archetype virus produces undetectable levels of TAg, we hypothesized that TAg overexpression would stimulate archetype virus replication. Efficient propagation of the archetype forms of BKPyV and JCPyV was observed in 293TT cells, human embryonic kidney cells overexpressing SV40 TAg. Importantly, the archetypal structure of the regulatory region was maintained during viral growth. Significant replication was not observed for Merkel cell, KI, or WU polyomaviruses. 293TT cells provide a means of propagating archetype BKPyV and JCPyV for detailed study.
Keywords: BKPyV, JCPyV, archetype, 293TT cells
Introduction
BK polyomavirus (BKPyV) and JC polyomavirus (JCPyV) are closely related human viruses originally isolated by two independent groups from patient samples in 1971 (Gardner et al., 1971; Padgett et al., 1971). BKPyV, JCPyV, and the related simian virus 40 (SV40) all share close to 70% whole genome sequence identity and differ mainly in their regulatory or transcriptional control regions (Imperiale, 2001). Both BKPyV and JCPyV are thought to be contracted in childhood or early adulthood. For BKPyV, the adult population seroprevalence is reached by the age of 10 and is estimated to be between 65% and 90% (Kean et al., 2009; Knowles, 2001). For JCPyV, the seroprevalence is lower than that of BKPyV, roughly 20% by the age of ten, and increasing to up to about 50% by the age of 70 (Kean et al., 2009). After what is thought to be a primary asymptomatic infection, these viruses disseminate to and persist in the urinary tract. In immunosuppressed patients, the viruses can reactivate and cause serious disease in specific sites. BKPyV can reactivate in bone marrow transplant patients to cause hemorrhagic cystitis in the bladder, and in kidney transplant patients to cause polyomavirus-associated nephropathy in the kidney (Ahsan and Shah, 2006). JCPyV can cause the fatal demyelinating disease, progressive multifocal leukoencephalopathy (PML), in which viral growth is restricted to glial cells of the brain, particularly in AIDS patients (Dorries et al., 1979).
BKPyV and JCPyV are small, non-enveloped, icosahedral viruses with circular double-stranded DNA genomes (Imperiale and Major, 2007). The genomes are divided into three genetic regions: the early coding region, the late coding region, and the regulatory region, which is also called the non-coding control region (NCCR) in BKPyV. The regulatory region contains the origin of DNA replication and the promoters for the divergently transcribed early and late coding regions. The early coding region encodes the non-structural T antigen proteins that are responsible for inducing a cellular environment conducive to replication and initiating viral DNA synthesis. The late coding region encodes the structural capsid proteins VP1, VP2, and VP3, and agnoprotein.
Two forms of the viral genomes of BKPyV and JCPyV exist, which are designated archetype and rearranged based on the DNA sequence of the regulatory region. The archetypal regulatory region is divided into defined blocks of sequence that are duplicated or deleted in the rearranged variants. In BKPyV, the archetype virus NCCR is divided into five blocks of sequence designated O, for the origin of replication, P, Q, R, and S (Moens and Van Ghelue, 2005). The JCPyV archetype regulatory region is similarly divided into blocks of sequence designated a, b, c, d, e, and f, with the origin of replication located between block a and the early genes (Jensen and Major, 2001). The archetype form of both viruses is thought to be the transmissible form that is capable of establishing a persistent infection in the host. For BKPyV, the archetype form can be periodically isolated in the urine of healthy individuals as a result of transient reactivation events (Doerries, 2006; Knowles, 2001). In contrast, rearranged variants are most often isolated from the serum of patients with BKPyV disease (Gosert et al., 2008). Similarly, JCPyV archetype virus isolation is often limited to the urine or urinary tract sites such as the kidney in healthy individuals (Loeber and Dorries, 1988; Yogo et al., 1990). The rearranged form of JCPyV can be isolated from the brain and lymphocytes in addition to the kidneys and urine in people with and without PML (Loeber and Dorries, 1988; Tornatore et al., 1992; White et al., 1992; Yogo et al., 1990). Rearranged variants of BKPyV and JCPyV are able to grow readily in the current cell culture models of renal proximal tubule epithelial (RPTE) cells and human fetal glial cells, respectively (Low et al., 2004; Padgett et al., 1977). To date, cell culture systems to study the archetype form of BKPyV and JCPyV have been extremely limited. However, it is important to be able to grow the archetype form of these polyomaviruses to understand the natural history of infection, develop means to prevent transmission and reactivation, and to aid in the search for a naturally permissive cell type for archetype virus in the host.
There is only one report of archetype BKPyV propagation in cell culture, but that propagation was not efficient. Archetype virus was shown to have limited replication capacity in a human endothelial cell line (Hanssen Rinaldo et al., 2005). Infectious progeny were not detected until 35 days post transfection (dpt) and only 4% of cells were infected upon secondary infection of the endothelial cells regardless of the multiplicity of infection of the inoculum. Additionally, the authors showed that while the archetype virus NCCR configuration was maintained, there were a significant number of rearranged variants identified in the progeny. Attempts to culture archetype JCPyV have been somewhat more successful. Archetype JCPyV has weak DNA replication ability in POJ cells, which are transformed human fetal glial cells that express JCPyV large T antigen (TAg) (Daniel et al., 1996). Archetype JCPyV DNA also replicates in COS-7 cells, monkey cells that constitutively express SV40 TAg, but infectious progeny are not produced (Hara et al., 1998). Additionally, it has recently been shown that archetype JCPyV propagates in COS-7 cells stably expressing HIV-1 Tat after 32 days in culture (Nukuzuma et al., 2009). HIV-1 Tat acts in coordination with the cellular protein Purα to stimulate viral DNA replication (Daniel et al., 2001). Importantly, these systems did not attain large scale propagation of archetype viruses. Therefore, in this study we sought to develop an improved culture system for archetype BKPyV and JCPyV.
We have previously shown that the BKPyV NCCR is the major genetic region that determines the ability of rearranged variants and not archetype virus to replicate viral DNA in RPTE cells (Broekema et al., 2010). We created a system that allowed us to exchange the major genetic regions between archetype and rearranged forms and showed that the rearranged NCCR present in an archetype virus background is sufficient for viral replication. It is also known that, in addition to the contribution of the NCCR, the early coding region product, TAg, is required in trans for viral replication by binding to the viral origin of replication to initiate DNA synthesis (Fanning and Zhao, 2009). Previous research has shown that SV40 TAg is capable of binding the origins of both JCPyV and BKPyV and initiating replication of their viral DNAs in vitro as well as in vivo (Daniel et al., 1996; Mahon et al., 2009; Sock et al., 1993). Additionally, BKPyV TAg and JCPyV TAg have some capacity to function in BKPyV, JCPyV, and SV40 DNA replication.
In this study we show that archetype BKPyV produces undetectable levels of TAg in cell culture models that support viral replication of rearranged variants, whose TAg production is robust. This knowledge, combined with data from previous studies, led us to ask if TAg overexpression could stimulate archetype BKPyV and JCPyV replication. We looked at the contribution of TAg transient overexpression by cotransfecting a BKPyV TAg cDNA with the viral genome and found that this could stimulate viral DNA replication and capsid protein production in archetype virus, which suggests progeny virus was produced. Therefore, we reasoned that constitutive high TAg levels may support archetype virus propagation. We chose to assess the contribution of TAg overexpression to viral replication in 293TT cells, human embryonic kidney cells constitutively expressing SV40 TAg (Buck et al., 2004), and successfully developed a means to efficiently propagate both BKPyV and JCPyV archetype virus for future study. Additionally, we determined whether SV40 TAg overexpression could stimulate the replication of other unculturable human polyomaviruses: Merkel cell polyomavirus (MCPyV), KI polyomavirus (KIPyV), and WU polyomavirus (WUPyV). 293TT cells were found to only propagate viruses closely related to SV40 whereas MCPyV, KIPyV, and WUPyV all failed to replicate efficiently in these cells.
Results
Archetype BKPyV produces undetectable level of TAg in RPTE cells
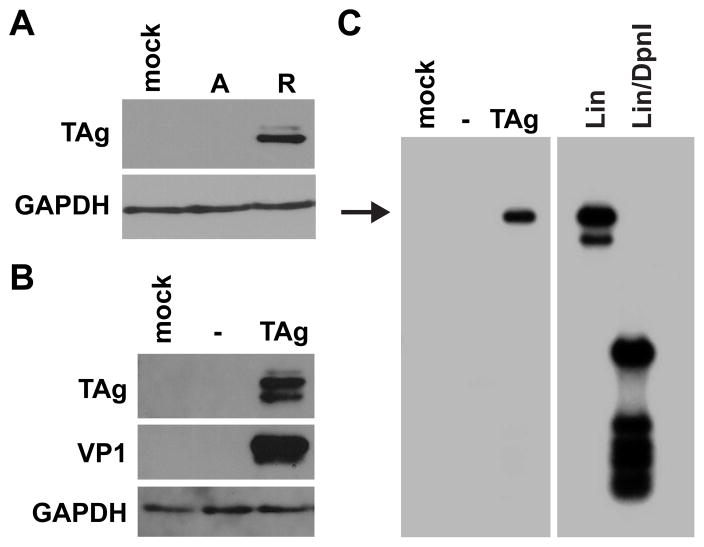
Rearranged forms of BKPyV are able to replicate in the natural host cell culture model RPTE cells, whereas archetype virus is unable to efficiently replicate in culture (Broekema et al., 2010; Hara et al., 1986; Watanabe and Yoshiike, 1985). TAg is the viral protein responsible for initiating viral DNA synthesis as a result of binding to the origin of replication to recruit DNA polymerase (Fanning and Zhao, 2009). Therefore, TAg protein production was first assessed in cells transfected with rearranged or archetype viral genomes to determine if the TAg level was a factor limiting archetype virus replication in RPTE cells. Recombinant archetype (Dik) and rearranged (Dunlop) BKPyV genomes were excised from the vector backbone, recircularized, and transfected into RPTE cells. We assayed for TAg production by collecting total cell proteins 4 dpt and immunoblotting for TAg protein (Fig. 1A). TAg was only detectable when the rearranged genome, and not the archetype genome, was transfected into RPTE cells. Since TAg is necessary for viral DNA replication, we hypothesized that limited TAg expression from the archetype NCCR production was likely one key factor limiting the propagation of archetype virus in RPTE cells and that TAg overexpression, therefore, may be able to rescue archetype virus replication. Transient TAg overexpression in RPTE cells, by cotransfecting a BKPyV TAg cDNA with the archetype genome, did result in a detectable level of capsid protein production at 4 dpt (Fig. 1B). Viral DNA replication was assessed in the same experiment by a DpnI resistance assay, which distinguishes methylated input plasmid DNA from unmethylated DNA that has been replicated in eukaryotic cells (Pipas et al., 1983). At 4 dpt, low molecular weight DNA was isolated, linearized, digested with DpnI, and assayed by Southern blotting. Replication was not detectable in control cells transfected with empty vector and the archetype genome, consistent with previous results (Broekema et al.). However, viral DNA replication was detectable when the archetype genome was cotransfected with BKPyV TAg cDNA. This demonstrates that TAg overexpression can stimulate archetype virus replication and capsid protein production.
Fig 1.
TAg expression limits archetype BKPyV replication in RPTE cells. A. RPTE cells were transfected with recircularized archetype (A) or rearranged (R) viral genomes. Total cell protein was harvested 4 dpt and 25 ug of protein was analyzed by Western blotting for the expression of TAg and GAPDH. B. RPTE cells were transfected with recircularized archetype genome and empty vector (−) or BKPyV TAg cDNA. Total cell protein was harvested 4 dpt and 30 μg was analyzed by Western blotting for the expression of TAg, VP1, and GAPDH. C. RPTE cells were transfected with recircularized archetype genome and empty vector (−) or BKPyV TAg cDNA, and low molecular weight DNA was harvested 4dpt. Samples were linearized, digested with DpnI, and analyzed by Southern blotting. Arrow indicates the replicated viral genome. Input DNA is not visible on this exposure. Mock, mock transfection. Lin, linearized BKPyV plasmid control. Lin/DpnI, linearized and DpnI-digested BKPyV plasmid control. The blots shown are representative of three independent experiments.
Archetype BKPyV and JCPyV replicate efficiently and produce capsid protein in 293TT cells
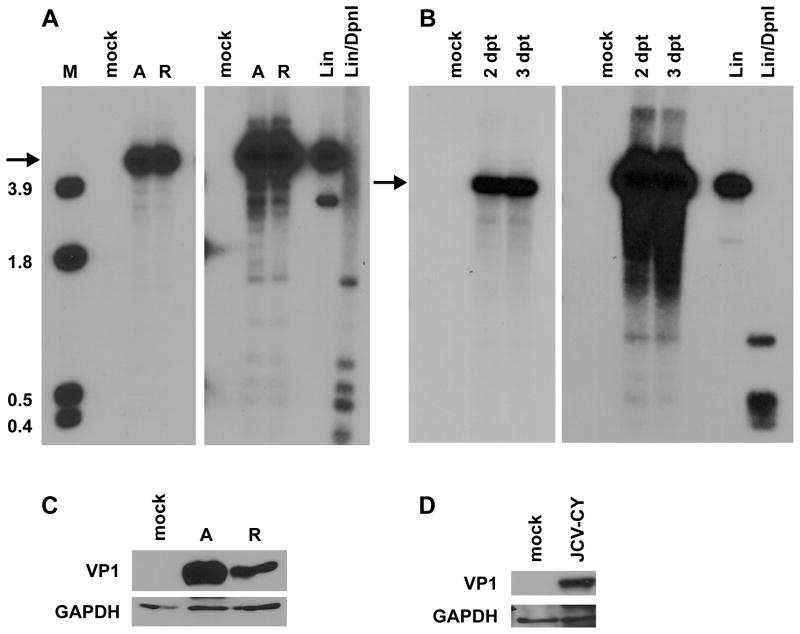
To determine whether sustained higher levels of TAg overexpression could promote robust archetype BKPyV replication, we performed viral replication assays in 293TT cells, human embryonic kidney cells that constitutively express high levels of SV40 TAg (Buck et al., 2004). 293TT cell contain an SV40 TAg expression plasmid in addition to an integrated SV40 genome which produces low levels of TAg due to a splicing bias that favors small t antigen. We reasoned that SV40 is a closely related virus and it has been shown that SV40 TAg can drive DNA replication from the origins of BKPyV and JCPyV (Daniel et al., 1996; Lynch and Frisque, 1990; Mahon et al., 2009). Recombinant archetype and rearranged BKPyV genomes were excised from the vector backbone, recircularized, and transfected into 293TT cells. At 4 dpt, low molecular weight DNA was isolated, linearized, digested with DpnI, and assayed by Southern blotting (Fig. 2A). Day 4 was chosen because rearranged variant replication can be easily detected at this time point (Broekema et al., 2010). Replication was quantified by normalizing the replicated genome band to a DpnI-digested input band, which controls for transfection efficiency. The quantified value of archetype virus DNA replication was set to 100. In two of four independent experiments, archetype virus replicated better than the rearranged virus while in the other two experiments there were almost equal levels of replication between the two viruses. When the data was averaged for all experiments, rearranged variant replication was 83.8% that of archetype virus with a standard deviation of 29.8%. This was not statistically different from that of archetype virus (Student’s t test p value of 0.3). Therefore, both archetype and rearranged BKPyV replicated well in 293TT cells. This is in contrast to RPTE cells, in which rearranged BKPyV replicates ~75 fold better than archetype (Broekema et al., 2010). Archetype JCPyV replication was also assessed at 2 and 3 dpt by DpnI resistance assay (Fig. 2B). Archetype JCPyV efficiently replicated in 293TT cells, and somewhat faster than BKPyV. We also measured capsid protein production in 293TT cells for archetype BKPyV and JCPyV by collecting total cell proteins 4 dpt and Western blotting for VP1 protein. Consistent with the replication assay results, archetype BKPyV and JCPyV produced capsid protein in 293TT cells (Fig. 2C and D). Additionally, archetype BKPyV appeared to produce more VP1 than rearranged. Encouraged by the results with these archetype viruses, we assessed replication ability and capsid protein production of three other unculturable human polyomaviruses: MCPyV, KIPyV, and WUPyV. These viruses were not capable of significant replication as measured by a DpnI resistance assay (data not shown). Furthermore, we obtained capsid protein antibody for KIPyV and WUPyV and these viruses did not produce detectable levels of capsid protein as assayed by immunoblotting (data not shown). Capsid production was not tested for MCPyV due to lack of an available antibody. These results show that archetype BKPyV and JCPyV but not MCPyV, KIPyV, and WUPyV are able to replicate their DNA and express late proteins in 293TT cells.
Fig 2.
Archetype BKPyV and JCPyV replicate efficiently in 293TT cells. A. 293TT cells were transfected with recircularized archetype (A) or rearranged (R) BKPyV genomes, and low molecular weight DNA was harvested at 4 dpt. Samples were linearized, digested with DpnI, and analyzed by Southern blotting. The right panel shows a darker exposure where the input DpnI digested bands and digested plasmid controls are visible. Arrow indicates the replicated viral genome. M, HindIII digest of pGEM-TU (size of bands in kb). Lin, linearized BKPyV plasmid control. Lin/DpnI, linearized and DpnI-digested BKPyV plasmid control. B. Low molecular weight DNA was harvested 2 and 3 dpt from archetype JCPyV transfection and analyzed by Southern blotting. Light and dark exposures are shown as in A. C, D. Analysis of VP1 expression. C. Total cell protein was harvested from 293TT cells 4 dpt and 90 μg of protein was analyzed by Western blotting for BKPyV VP1 and GAPDH expression. Mock, mock transfection. D. Total cell protein was harvested 4 dpt and 60 μg of protein was analyzed by Western blotting for JCPyV VP1 and GAPDH expression. The blots shown are representative of three independent experiments.
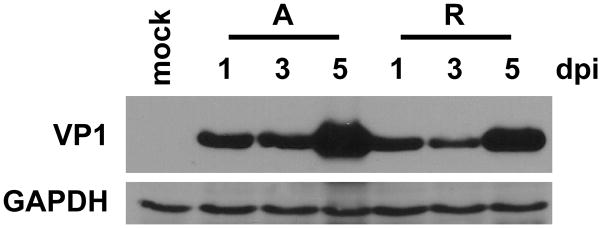
Infectious progeny production of archetype BKPyV and JCPyV
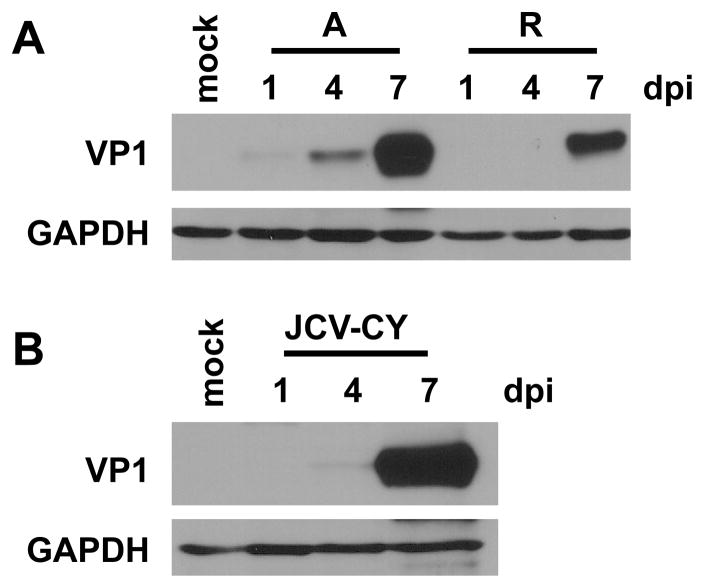
Production of VP1 by archetype virus suggests that viral progeny are also produced in 293TT cells. To test whether archetype virus was in fact producing infectious progeny virus we harvested whole cell lysates 7 dpt. We then infected fresh 293TT cells with 1:2 dilutions of the transfection lysate in serum-free medium and assessed capsid protein production over time as a measure of infection. We could not use BKPyV or JCPyV TAg protein expression as a readout of infection because of the abundant SV40 TAg expression in 293TT cells, which is detected by the SV40 TAg pAb416 antibody. Therefore, VP1 capsid protein was assessed at 1, 4, and 7 days post infection (dpi). Archetype BKPyV lysates showed increased capsid protein production over time in 293TT cells. Consistent with the transfection DNA replication and capsid protein results, archetype virus appeared to have somewhat accelerated kinetics of infection when compared to the rearranged variant (Fig. 3A). Infection on fresh 293TT cells with 7 dpt lysates of archetype JCPyV also resulted in an increased VP1 protein over time (Fig. 3B). These results show that archetype BKPyV and JCPyV are able to produce infectious progeny in 293TT cells. Lysates harvested from cells transfected with the KIPyV and WUPyV genomes were unable to produce infectious progeny as measured by passage on fresh 293TT cells (data not shown). This result is consistent with the inability of these viruses to replicate DNA in 293TT cells.
Fig 3.
Archetype BKPyV and JCPyV produce infectious progeny in 293TT cells. Cell lysates were harvested from 293TT cells 7 dpt. The lysates were then used to infect fresh 293TT cells. A. Total cell protein was harvested on 1, 4, and 7 dpi with BKPyV lysate, and 50 μg of protein was analyzed by Western blotting for BKPyV VP1 and GAPDH expression. A, archetype. R, rearranged. B. Total cell protein was harvested on 1, 4, and 7 dpi with JCPyV lysate, and 50 μg of protein was analyzed for JCPyV VP1 and GAPDH expression. The blots shown are representative of three independent experiments.
Previously, it has been shown that rearrangements in the NCCR of archetype BKPyV can occur when it is passaged in cell culture (Rubinstein et al., 1991). Therefore, the integrity of the regulatory region of the virus in the lysates used for infection was confirmed by PCR amplifying the regulatory region from one transfection lysate, cloning the PCR products, and sequencing. Three clones from the rearranged BKPyV lysate were sequenced and no mutations were found. Nine clones from the archetype BKPyV lysate also had no rearrangements or point mutations. Ten clones were sequenced from the JCPyV regulatory region, none of which contained any rearrangements. Three of the ten, however, did contain base changes, at G108A, G235A, and C253T, where 1 is the first nucleotide of the regulatory region. Single nucleotide changes are not rearrangements and therefore all archetype BKPyV and JCPyV clones sequenced maintained an archetype regulatory region structure.
Purification of archetype BKPyV from 293TT cells and virion morphology
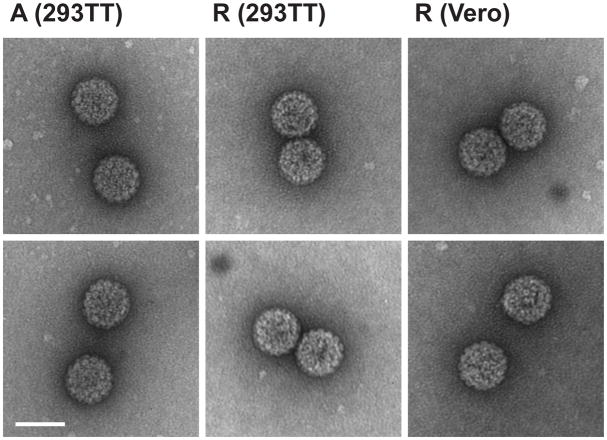
We next examined the physical properties of the BKPyV produced in 293TT cells. Archetype and rearranged BKPyV from 293TT cells were purified on a CsCl gradient. The refractive index of the mature virus band for both forms was measured and the density was calculated to be 1.34g/cm3, the expected polyomavirus density. Purified virus morphology was then examined by transmission electron microscopy (TEM) (Fig. 4). As a comparison, infectious rearranged (TU) BKPyV viral particles from Vero cells, which support rearranged variant replication, are shown. The average sizes of the archetype virions were 44.9 +/− 2.0 nm (n=85), rearranged virions were 46.6 +/− 2.5 nm (n=34), and particles purified from Vero cells were 46.4 +/− 1.4 nm (n=36). The number of virus particles observed on the grids was roughly equivalent between the viruses purified from 293TT and Vero cells, indicating equivalent progeny production.
Fig 4.
Morphology of BKPyV produced in 293TT cells. Virus was purified from cells by CsCl gradient centrifugation. Mature virus particles were applied to grids as described in the text. Viral particles were visualized by negative staining at a magnification of 180,000X. Archetype virions from 293TT cells are shown in the left column, rearranged virions from the same cells in the middle column, and rearranged virions purified from Vero cells in the right column. Scale bar = 50 nm. The particles shown are representative of three independent purifications.
To examine the integrity of the NCCR of the purified virus, the NCCR was PCR amplified and cloned, and ten clones were sequenced from each virus. No rearrangements were identified in the archetype virus NCCR, but one clone from the rearranged variant had a deletion of the first two P blocks of the Dunlop NCCR. Single nucleotide changes were identified in each form, in 3 of 10 clones from the rearranged variant and 1 of 10 clones from the archetype virus. The rearranged variant base substitutions were T28C, A46G, and T322C, where 1 is the first nucleotide of the O block. The archetype virus substitution was C141T, which maps to the BKPyV TAg binding site III. However, this substitution does not make the site more similar to that of SV40 TAg binding site III, which may be expected if selection for use of SV40 TAg were occuring.
Purified virus was treated with DNase to digest any unencapsidated viral DNA and progeny virions were quantified by real-time PCR. In a representative experiment, archetype virus purified from 293TT cells had 2.84 × 1011 genomes/ml and rearranged virus had 1.65 × 1011 genomes/ml. Infectivity of the purified virus was confirmed by infecting 293TT cells with 500 genomes/cell and measuring VP1 expression at 1, 3, and 5 dpi (Fig. 5). Both purified archetype and rearranged viruses were able to infect 293TT cells and showed an increase in capsid protein over the five day time course. We noted no difference in viral kinetics when cells were infected with equal genome numbers. Next we estimated the amount of infectious virus present by infecting 293TT cells and immunostaining for VP1. As a control to validate the assay, rearranged virus purified from Vero cells was quantified by both VP1 immunostaining in 293TT cells and TAg immunostaining in RPTE cells, a standard infectious unit (IU) assay; it was calculated to have an equal titer in both assays. In the representative experiment of virus whose genomes were quantified above, the viral titers for archetype and rearranged virus were measured to be 2.14 × 108 and 2.40 × 107 IU/ml, respectively. In comparison, the yield from rearranged virus grown in Vero cells in three fold more cells is, on average, 108 IU/ml. Archetype and rearranged virus purified from 293TT cells therefore have a ratio of 1352 and 6875 genomes/IU, respectively. This result was reproducible. Taken together, these results show that archetype BKPyV purified from 293TT cells is infectious and that the yields of infectious virus are similar to those obtained with rearranged variants in Vero cells.
Fig 5.
Purified archetype BKPyV is infectious in 293TT cells. 293TT cells were infected with 500 genomes/cell of purified BKPyV and total cell protein was harvested 1, 3, and 5 dpi. 50 μg of protein was analyzed by Western blotting for BKPyV TAg and GAPDH expression. A, archetype. R, rearranged. The blots shown are representative of three independent experiments.
Discussion
BKPyV and JCPyV cause severe clinical disease in immunosuppressed and immunocompromised patients, respectively. It is thought that the archetype form of these viruses is acquired in childhood or early adulthood and establishes a persistent infection in the host. Thus far, cell culture systems to propagate archetype polyomaviruses have been very limited in their availability and robustness. Prior to this study we noted that archetype BKPyV replicated very poorly in RPTE cells, and we showed here that when RPTE cells were transfected, the archetype virus genome produced undetectable levels of TAg protein. Since TAg is required to initiate viral DNA synthesis, we hypothesized that overexpression of TAg would stimulate archetype virus replication. We show here that transient BKPyV TAg overexpression with archetype BKPyV transfection is able to rescue replication as assayed by capsid protein production. Additionally we show that 293TT cells, which greatly overexpress SV40 TAg, are capable of propagating both archetype BKPyV and JCPyV. These viruses were able to replicate their DNA, express VP1, and produce infectious progeny that were capable of reinfecting 293TT cells. This represents an improved culture model system for archetype BKPyV and JCPyV.
In addition to testing replication of archetype BKPyV and JCPyV, we also assessed three other unculturable polyomaviruses: MCPyV, KIPyV, and WUPyV. Each of these viruses showed only minimal replication in 293TT cells as assessed by the DpnI resistance assay, akin to what is seen for archetype BKPyV in RPTE cells (Broekema et al., 2010). Additionally, KIPyV and WUPyV were unable to express detectable levels of capsid protein. The TAg coding sequences of MCPyV, KIPyV, and WUPyV have only a 47.6%, 64.4%, and 62.8% similarity to SV40 TAg, respectively, whereas BKPyV has 85.9% and JCPyV has 84.1% similarity. Our results suggest that SV40 TAg is likely not similar enough to bind to and initiate replication from the MCPyV, KIPyV, and WUPyV origins of replication.
It has been shown previously that SV40 TAg is capable of initiating replication from both the archetype and rearranged JCPyV and BKPyV origins (Daniel et al., 1996; Mahon et al., 2009; Sock et al., 1993). BKPyV TAg and JCPyV TAg also have some capacity to initiate replication of BKPyV, JCPyV, and SV40 DNA, although replication of SV40 DNA initiated by JCPyV TAg is limited. Our results show robust replication of JCPyV at early timepoints post transfection of 293TT. This is consistent with previous results showing replication of JCPyV DNA initiated by SV40 TAg is better than that of BKPyV DNA (Mahon et al., 2009). Archetype JCPyV DNA can replicate but not efficiently produce virus in POJ cells and COS-7 cells overexpressing JCPyV and SV40 TAg, respectively (Daniel et al., 1996; Hara et al., 1998; Nukuzuma et al., 2009). The difference in progeny production of archetype JCPyV from COS-7 cells compared to 293TT cells likely has to do with the very high levels of SV40 TAg produced in 293TT cells. Furthermore, archetype BKPyV grown in 293TT cells is able to produce as many IU/ml as rearranged virus grown in Vero cells, indicating efficient propagation. In contrast, previous research in a human endothelial cell line, archetype BKPyV infectious progeny production was poor and not detected until 35 dpt (Hanssen Rinaldo et al., 2005).
Previous attempts to propagate archetype BKPyV in human embryonic kidney cells resulted in selection for rearrangements (Rubinstein et al., 1991). It also appears that significant rearrangement of the NCCR was selected for upon propagation in endothelial cells (Hanssen Rinaldo et al., 2005). Therefore, we confirmed the integrity of the regulatory regions of the viruses produced in 293TT cells. Single nucleotide changes were observed in a minority of the clones sequenced. These may be bona fide mutations or possibly PCR errors, although we did use high fidelity Taq polymerase. However, single base changes have been shown to be common in archetype BKPyV and are not likely to affect replication (Yogo et al., 2008). There was only one major rearrangement identified in one clone from the BKPyV rearranged NCCR, which was a deletion of two of three P blocks. However, no rearrangements were observed in the archetype virus regulatory region of BKPyV and JCPyV. These results suggest that there is no selective pressure in 293TT cells to induce rearrangements which might facilitate replication. The TEM images of the purified BKPyV confirm the identity of the purified virus as authentic polyomavirus.
Archetype JCPyV DNA replicates in COS-7, simian kidney cells expressing SV40 TAg, and COS-7 cells expressing HIV-1 Tat (Hara et al., 1998; Nukuzuma et al., 2009). However, these systems show weak progeny production, taking up to 32 days to produce JCPyV as detected by hemagglutination assay. Our system has the advantage of faster progeny production, since we detect significant titers, measured by infecting fresh 293TT cells with lysate harvested 7 dpt. 293TT cells are a human cell line and thus can be more effectively used to study unique host-virus interactions that may be present, such as interactions between cellular transcription factors and archetype virus promoters. However, since archetype virus replication in 293TT cells is driven by SV40 TAg, these cells cannot be used as a natural host cell to study the life cycle of polyomaviruses. Regardless, these cells will allow for further detailed comparative study of archetype polyomaviruses by providing a means for their propagation in the laboratory.
In conclusion, 293TT cells are able to support growth of the closely related archetype BKPyV and JCPyV. Together, these findings strongly suggest that TAg production is a limiting factor for archetype polyomavirus propagation in currently-available cell culture models. Based on the results of this study, we speculate that cell culture systems overexpressing TAg from other unculturable polyomaviruses may be able to support propagation of those viruses. The findings in this paper expand our understanding of archetype virus and are an early step toward characterizing the differences between archetype and rearranged polyomaviruses. This study and knowledge gained from future studies may lead to new approaches to prevent transmission or reactivation of archetype virus in healthy individuals and immunocompromised hosts.
Materials and Methods
Cell culture
Primary human RPTE cells were maintained for up to six passages in renal epithelial growth medium as previously described (Abend et al., 2007). 293TT cells were obtained from Chris Buck at the National Cancer Institute (Buck et al., 2004) and were maintained under 400 μg/ml hygromycin selection in DMEM with 10% fetal bovine serum and 100 U/ml penicillin, 100 μg/ml streptomycin.
Plasmids
The pBR322-Dunlop plasmid (ATCC #45025, pBKV) contains a full length Dunlop (rearranged NCCR) genome (Seif et al., 1979) and the pBR322-Dik plasmid contains a full length Dik (archetype NCCR) genome (Nishimoto et al., 2006). The Dik and Dunlop genomes each were subcloned from these plasmids into pGEM7 at the BamHI site to facilitate plasmid growth. The pJC-CY plasmid containing archetype JCPyV genome cloned into pUC19 at the BamHI (Yogo et al., 1990) was received from Richard Frisque at Pennsylvania State University. The pBlu-WU and pBlu-KI plasmids were received from David Wang at Washington University. We obtained pMCV-R17a (Schowalter et al., 2010) from Addgene (plasmid 24729), where it had been deposited by Chris Buck. The BKPyV TAg cDNA plasmid was engineered for other purposes with a silent mutation at nucleotide 4406 (Harris et al., 1998). For the present study, the cDNA was subcloned into pcDNA3.1(+)-hygro (Invitrogen) using the BamHI and EcoRI sites.
Transfection
The viral genome was excised from the vector backbone by BamHI or EcoRI digestion (New England Biolabs, Ipswich, MA) and then recirularized at a concentration of 10 ng/μL using T4 DNA ligase (New England Biolabs). RPTE or 293TT cells were seeded into 12 well plates and transfected with 0.5 μg DNA at 70–80% confluency using TransIT LT-1 transfection reagent (Mirus Bio, Madison, WI) according to the manufacturer’s instructions, with a DNA to transfection reagent ratio of 1:6. For the TAg cotransfection experiments, 0.5 μg of total DNA was transfected at a ratio of 1:9 of TAg to viral genome. Transfection complexes were removed 18 hr post-transfection by washing once with PBS and adding fresh media. DNA was extracted from cell lysates by phenol-chloroform extraction, and the integrity of the regulatory regions of BKPyV and JCPyV was confirmed in transfection lysates 7 dpt by cloning PCR products of the BKPyV NCCR and JCPyV regulatory region (RR) into pGEM using primers BKPyV NCCR Forward (5′ TTTGTCAGGGTGAAATTCCTT 3′) and BKPyVV NCCR Reverse (5′ TCACCTCTACAAA ATTCCAGCA 3′) and JCPyV RR Forward (5′ GAATGTTTCCCCATGCAGAT 3′) and JCPyV RR Reverse (5′ CTGTGCAAAAGTCCAGCAAA 3′). The PCR reaction consisted of 0.5 μM each primer, 1 mM dNTPs, 2 mM MgSO4, and 1 U Platimun Tag High Fidelity polymerase (Invitrogen) in 1X buffer provided by the manufacturer. The PCR program consisted of an initial 5 min denaturation at 95°C followed by 30 cycles each of denaturation at 95°C for 45 sec, annealing at 55°C for 1 min, and elongation at 68°C for 1 min. The PCR product was purified using the QIAquick PCR Purification Kit (Qiagen), ligated into pGEM-T Easy vector (Promega), and clones were sequenced.
Southern Blotting
DpnI resistance assays were performed as previously described (Broekema et al., 2010) using low molecular weight DNA harvested at the times stated in the text. For BKPyV, the probe was a 3239 bp PvuII fragment excised from pGEM7-TU (Abend et al., 2007) which spans the early region and the NCCR. The size marker was pGEM7-TU digested with HindIII. For JCPyV, the probe was the whole 5120 bp JCPyV-CY genome excised from pJC-CY by BamHI digestion.
Western Blotting
E1A lysis buffer was used to harvest total cell proteins, which were analyzed as previously described (Jiang et al., 2009) with the following modifications. The membrane was blocked for 30 min at room temperature (RT) in 2% nonfat dried milk in PBS with 0.1% Tween 20 (PBS-T), then incubated with primary antibody in 2% nonfat dried milk in PBS-T for 2 hr at RT or overnight at 4°C. The following antibodies and concentrations were used: SV40 pAb416 (Harlow et al., 1981) for TAg at 1:3,000; P5G6 (received from D. Galloway) for BKPyV VP1 at 1:10,000; pAb597 (Atwood et al., 1995) (received from R. Frisque) for JCPyV VP1 at 1:5,000; WUPyV VP1 and KIPyV VP1 antibodies at 1:1,000 (Nguyen et al., 2009) (received from D. Wang); and Ab9484 (Abcam) for GAPDH at 1:10,000. The membrane was then washed 3 × 10 min with PBS-T and incubated with horseradish peroxidase-conjugated sheep anti-mouse antibody (Amersham) at 1:5,000 in 2% nonfat dried milk at room temperature for 2 hr. The membrane was washed 3 × 10 min and developed using HyGLO (Denville Scientific Inc.) or Luminol (Millipore).
Virus Purification
Virus purification was performed by density centrifugation on CsCl gradients as previously described (Jiang et al., 2009). The mature virus bands were collected by gravity flow from the side of the centrifuge tube using an 18-gauge needle. The collected virus was dialyzed against buffer A (10mM HEPES [pH7.9], 1mM CaCl2, 1mM MgCl2, 5 mM KCl) overnight at 4°C. The density of each band was determined by measuring the refractive index. The integrity of the NCCR was confirmed by cloning PCR products of the BKPyV NCCR using primers BKPyV NCCR Forward and BKPyVV NCCR Reverse as described above and sequencing 10 clones for each. Purified virus was DNase-treated to digest unencapsidated viral DNA and progeny genomes were quantified by real-time PCR as previously described (Jiang et al., 2011) using primers TAg-Forward (5′ TGTGATTGGGATTCAGTGCT 3′) and TAg-Reverse (5′ AAGGAAAGGCTGGATTCTGA 3′).
Infection
293TT cells were infected at 70–80% confluency with 500 genomes/cell of purified archetype or rearranged virus, or crude viral lysate, in low serum media at 4°C. After 1 hr the infection media was replaced with fresh warmed maintenance media. Viral titer was determined by an infectious unit assay. Fifty percent confluent 293TT cells were infected at 4°C with 10 fold dilutions of purified virus. Infection was allowed to proceed for 3 dpi at 37°C. The cells were then fixed for 5 min in 95% ethanol/5% acetic acid and stained with the monoclonal anti-VP1 antibody P5G6 (received from D. Galloway) at 1:500 followed by a fluorescein-conjugated anti-mouse antibody at 1:100. Viral titer was determined by averaging five fields of view in triplicate infections.
Transmission Electron Microscopy
Dialyzed virus was diluted 1:10 in buffer A and applied to glow-discharged 300-mesh copper grids (Electron Microscopy Sciences) for 5 min at RT. Grids were then washed in buffer A, fixed with glutaraldehyde, and stained with uranyl acetate as previously described (Christensen et al., 2008).
Sequence alignment
TAg coding sequences were aligned by pairwise sequence alignment (http://www.ebi.ac.uk/Tools/psa/).
Acknowledgments
We thank members of the Imperiale laboratory for comments and discussion and Amanda Pattridge and Dorothy Sorenson for technical assistance with TEM. We thank Richard Frisque for the pJC-CY plasmid and VP1 antibody, David Wang for the pBlu-KI and pBlu-WU plasmids and VP1 antibodies, Peter Howley for the pBR322-Dunlop plasmid, and John Lednicky for the pBR322-Dik plasmid. This work was supported by NIH grant AI060584 awarded to M.J.I and in part by NIH grant CA046592 awarded to the University of Michigan Cancer Center. N.M.B. was supported in part by the NIH National Research Service Award T32-GM07544 from the National Institute of General Medicine Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Nicole M. Broekema, Email: broekenm@umich.edu.
Michael J. Imperiale, Email: imperial@umich.edu.
References
- Abend JR, Low JA, Imperiale MJ. Inhibitory effect of gamma interferon on BK virus gene expression and replication. J Virol. 2007;81:272–279. doi: 10.1128/JVI.01571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahsan N, Shah KV. Polyomaviruses and human diseases. Adv Exp Med Biol. 2006;577:1–18. doi: 10.1007/0-387-32957-9_1. [DOI] [PubMed] [Google Scholar]
- Atwood WJ, Wang L, Durham LC, Amemiya K, Traub RG, Major EO. Evaluation of the role of cytokine activation in the multiplication of JC virus (JCV) in human fetal glial cells. J Neurovirol. 1995;1:40–49. doi: 10.3109/13550289509111009. [DOI] [PubMed] [Google Scholar]
- Broekema NM, Abend JR, Bennett SM, Butel JS, Vanchiere JA, Imperiale MJ. A system for the analysis of BKV non-coding control regions: application to clinical isolates from an HIV/AIDS patient. Virology. 407:368–373. doi: 10.1016/j.virol.2010.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekema NM, Abend JR, Bennett SM, Butel JS, Vanchiere JA, Imperiale MJ. A system for the analysis of BKV non-coding control regions: application to clinical isolates from an HIV/AIDS patient. Virology. 2010;407:368–373. doi: 10.1016/j.virol.2010.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck CB, Pastrana DV, Lowy DR, Schiller JT. Efficient intracellular assembly of papillomaviral vectors. J Virol. 2004;78:751–757. doi: 10.1128/JVI.78.2.751-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JB, Byrd SA, Walker AK, Strahler JR, Andrews PC, Imperiale MJ. Presence of the adenovirus IVa2 protein at a single vertex of the mature virion. J Virol. 2008;82:9086–9093. doi: 10.1128/JVI.01024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel AM, Swenson JJ, Mayreddy RP, Khalili K, Frisque RJ. Sequences within the early and late promoters of archetype JC virus restrict viral DNA replication and infectivity. Virology. 1996;216:90–101. doi: 10.1006/viro.1996.0037. [DOI] [PubMed] [Google Scholar]
- Daniel DC, Wortman MJ, Schiller RJ, Liu H, Gan L, Mellen JS, Chang CF, Gallia GL, Rappaport J, Khalili K, Johnson EM. Coordinate effects of human immunodeficiency virus type 1 protein Tat and cellular protein Puralpha on DNA replication initiated at the JC virus origin. J Gen Virol. 2001;82:1543–1553. doi: 10.1099/0022-1317-82-7-1543. [DOI] [PubMed] [Google Scholar]
- Doerries K. Human polyomavirus JC and BK persistent infection. Adv Exp Med Biol. 2006;577:102–116. doi: 10.1007/0-387-32957-9_8. [DOI] [PubMed] [Google Scholar]
- Dorries K, Johnson RT, ter Meulen V. Detection of polyoma virus DNA in PML-brain tissue by (in situ) hybridization. J Gen Virol. 1979;42:49–57. doi: 10.1099/0022-1317-42-1-49. [DOI] [PubMed] [Google Scholar]
- Fanning E, Zhao K. SV40 DNA replication: from the A gene to a nanomachine. Virology. 2009;384:352–359. doi: 10.1016/j.virol.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971;1:1253–1257. doi: 10.1016/s0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- Gosert R, Rinaldo CH, Funk GA, Egli A, Ramos E, Drachenberg CB, Hirsch HH. Polyomavirus BK with rearranged noncoding control region emerge in vivo in renal transplant patients and increase viral replication and cytopathology. J Exp Med. 2008;205:841–852. doi: 10.1084/jem.20072097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen Rinaldo C, Hansen H, Traavik T. Human endothelial cells allow passage of an archetypal BK virus (BKV) strain--a tool for cultivation and functional studies of natural BKV strains. Arch Virol. 2005;150:1449–1458. doi: 10.1007/s00705-005-0511-3. [DOI] [PubMed] [Google Scholar]
- Hara K, Oya Y, Kinoshita H, Taguchi F, Yogo Y. Sequence reiteration required for the efficient growth of BK virus. J Gen Virol. 1986;67 ( Pt 11):2555–2559. doi: 10.1099/0022-1317-67-11-2555. [DOI] [PubMed] [Google Scholar]
- Hara K, Sugimoto C, Kitamura T, Aoki N, Taguchi F, Yogo Y. Archetype JC virus efficiently replicates in COS-7 cells, simian cells constitutively expressing simian virus 40 T antigen. J Virol. 1998;72:5335–5342. doi: 10.1128/jvi.72.7.5335-5342.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Crawford LV, Pim DC, Williamson NM. Monoclonal antibodies specific for simian virus 40 tumor antigens. J Virol. 1981;39:861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KF, Christensen JB, Radany EH, Imperiale MJ. Novel mechanisms of E2F induction by BK virus large-T antigen: requirement of both the pRb-binding and the J domains. Mol Cell Biol. 1998;18:1746–1756. doi: 10.1128/mcb.18.3.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperiale MJ. The Human Polyomaviruses: an Overview. In: Khalili KS, GL, editors. Human Polyomaviruses: Molecular and Clinical Perspectives. Wiley-Liss; New York: 2001. pp. 53–71. [Google Scholar]
- Imperiale MJ, Major EO. Polyomaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Vol. 2. Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 2263–2298. 2 vols. [Google Scholar]
- Jensen PN, Major EO. A classification scheme for human polyomavirus JCV variants based on the nucleotide sequence of the noncoding regulatory region. J Neurovirol. 2001;7:280–287. doi: 10.1080/13550280152537102. [DOI] [PubMed] [Google Scholar]
- Jiang M, Abend JR, Tsai B, Imperiale MJ. Early events during BK virus entry and disassembly. J Virol. 2009;83:1350–1358. doi: 10.1128/JVI.02169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Entezami P, Gamez M, Stamminger T, Imperiale MJ. Functional reorganization of promyelocytic leukemia nuclear bodies during BK virus infection. MBio. 2011;2:e00281–00210. doi: 10.1128/mBio.00281-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean JM, Rao S, Wang M, Garcea RL. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009;5:e1000363. doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles WA. The epidemiology of BK virus and the occurence of antigenic and genomic subtypes. In: Khalili K, Stoner GL, editors. Human Polyomaviruses: Molecular and Clinical Perspectives. Wiley-Liss; New York: 2001. [Google Scholar]
- Loeber G, Dorries K. DNA rearrangements in organ-specific variants of polyomavirus JC strain GS. J Virol. 1988;62:1730–1735. doi: 10.1128/jvi.62.5.1730-1735.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low J, Humes HD, Szczypka M, Imperiale M. BKV and SV40 infection of human kidney tubular epithelial cells in vitro. Virology. 2004;323:182–188. doi: 10.1016/j.virol.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Lynch KJ, Frisque RJ. Identification of critical elements within the JC virus DNA replication origin. J Virol. 1990;64:5812–5822. doi: 10.1128/jvi.64.12.5812-5822.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon C, Liang B, Tikhanovich I, Abend JR, Imperiale MJ, Nasheuer HP, Folk WR. Restriction of human polyomavirus BK virus DNA replication in murine cells and extracts. J Virol. 2009;83:5708–5717. doi: 10.1128/JVI.00300-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens U, Van Ghelue M. Polymorphism in the genome of non-passaged human polyomavirus BK: implications for cell tropism and the pathological role of the virus. Virology. 2005;331:209–231. doi: 10.1016/j.virol.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Nguyen NL, Le BM, Wang D. Serologic evidence of frequent human infection with WU and KI polyomaviruses. Emerg Infect Dis. 2009;15:1199–1205. doi: 10.3201/eid1508.090270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto Y, Takasaka T, Hasegawa M, Zheng HY, Chen Q, Sugimoto C, Kitamura T, Yogo Y. Evolution of BK virus based on complete genome data. J Mol Evol. 2006;63:341–352. doi: 10.1007/s00239-005-0092-5. [DOI] [PubMed] [Google Scholar]
- Nukuzuma S, Kameoka M, Sugiura S, Nakamichi K, Nukuzuma C, Miyoshi I, Takegami T. Archetype JC virus efficiently propagates in kidney-derived cells stably expressing HIV-1 Tat. Microbiol Immunol. 2009;53:621–628. doi: 10.1111/j.1348-0421.2009.00166.x. [DOI] [PubMed] [Google Scholar]
- Padgett BL, Rogers CM, Walker DL. JC virus, a human polyomavirus associated with progressive multifocal leukoencephalopathy: additional biological characteristics and antigenic relationships. Infect Immun. 1977;15:656–662. doi: 10.1128/iai.15.2.656-662.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet. 1971;1:1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- Pipas JM, Peden KW, Nathans D. Mutational analysis of simian virus 40 T antigen: isolation and characterization of mutants with deletions in the T-antigen gene. Mol Cell Biol. 1983;3:203–213. doi: 10.1128/mcb.3.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein R, Schoonakker BC, Harley EH. Recurring theme of changes in the transcriptional control region of BK virus during adaptation to cell culture. J Virol. 1991;65:1600–1604. doi: 10.1128/jvi.65.3.1600-1604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seif I, Khoury G, Dhar R. The genome of human papovavirus BKV. Cell. 1979;18:963–977. doi: 10.1016/0092-8674(79)90209-5. [DOI] [PubMed] [Google Scholar]
- Sock E, Wegner M, Fortunato EA, Grummt F. Large T-antigen and sequences within the regulatory region of JC virus both contribute to the features of JC virus DNA replication. Virology. 1993;197:537–548. doi: 10.1006/viro.1993.1627. [DOI] [PubMed] [Google Scholar]
- Tornatore C, Berger JR, Houff SA, Curfman B, Meyers K, Winfield D, Major EO. Detection of JC virus DNA in peripheral lymphocytes from patients with and without progressive multifocal leukoencephalopathy. Ann Neurol. 1992;31:454–462. doi: 10.1002/ana.410310426. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Yoshiike K. Decreasing the number of 68-base-pair tandem repeats in the BK virus transcriptional control region reduces plaque size and enhances transforming capacity. J Virol. 1985;55:823–825. doi: 10.1128/jvi.55.3.823-825.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White FA, 3rd, Ishaq M, Stoner GL, Frisque RJ. JC virus DNA is present in many human brain samples from patients without progressive multifocal leukoencephalopathy. J Virol. 1992;66:5726–5734. doi: 10.1128/jvi.66.10.5726-5734.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogo Y, Kitamura T, Sugimoto C, Ueki T, Aso Y, Hara K, Taguchi F. Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J Virol. 1990;64:3139–3143. doi: 10.1128/jvi.64.6.3139-3143.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogo Y, Zhong S, Xu Y, Zhu M, Chao Y, Sugimoto C, Ikegaya H, Shibuya A, Kitamura T. Conserved archetypal configuration of the transcriptional control region during the course of BK polyomavirus evolution. J Gen Virol. 2008;89:1849–1856. doi: 10.1099/vir.0.2008/000836-0. [DOI] [PubMed] [Google Scholar]