Abstract
Genome structural variation (SV) is a major source of genetic diversity in mammals and a hallmark of cancer. While SV is typically defined by its canonical forms – duplication, deletion, insertion, inversion and translocation – recent breakpoint mapping studies have revealed a surprising number of “complex” variants that evade simple classification. Complex SVs are defined by clustered breakpoints that arose through a single mutation but cannot be explained by one simple end-joining or recombination event. Some complex variants exhibit profoundly complicated rearrangements between distinct loci from multiple chromosomes, while others involve more subtle alterations at a single locus. These diverse and unpredictable features present a challenge for SV mapping experiments. Here, we review current knowledge of complex SV in mammals, and outline techniques for identifying and characterizing complex variants using next-generation DNA sequencing.
The genomic landscape of structural variation
Structural variation (SV) is defined as differences in the copy number, orientation or location of relatively large genomic segments (typically >100 bp). The canonical forms include deletions, tandem duplications, insertions, inversions and translocations. Large-scale microscopically-visible genomic rearrangements have long been recognized for their role in evolution and disease, but the remarkable prevalence of submicroscopic SVs only became apparent this past decade with the development of high-resolution methods such as array-comparative genomic hybridization (array-CGH) and next20 generation DNA sequencing. Current data [1, 2] suggests that two humans differ by 5,000–10,000 inherited SVs and that both inherited and de novo SVs contribute to a number of normal and disease phenotypes [3]. Similar levels are apparent in other mammalian species including chimpanzee [4], mouse [5, 6], rat [7], dog [8, 9] and cattle [10].
Virtually all tumor genomes harbor somatically-acquired SV, but the landscape is extremely diverse. Some tumors have tens or hundreds [11–16], while others have very few [12, 15, 17, 18], and the abundance of different SV classes varies considerably within and among tumor types [12, 15, 19]. A subset of cancer-associated SV appears to be functional and under strong selection, such as amplification of oncogenes, deletion of tumor suppressors and translocations that produce fusion genes, but many appear benign. Tumor genome instability may be caused by mutations in DNA maintenance machinery, widespread telomere erosion and/or unstable chromosome architectures acquired during tumorigenesis (e.g., dicentrics) [20].
In this context the recent discovery of many complex variants that defy simple classification into the typical SV classes has led to a re-examination of the mechanisms and impact of structural variation. Here, we review recent findings regarding complex variation and discuss techniques for their identification and characterization. We limit our discussion to mammals, mainly human, but we note that these same issues are relevant to other species.
Complex structural variation defined
Structural variants are defined by their breakpoints, which are the novel sequence junctions generated by structural mutation (Figure 1a–c). Structural variants arise through four general mechanisms (reviewed in [21]): 1) ligation of double strand DNA breaks (DSBs) through non-homologous end-joining (NHEJ) or microhomology-mediated end-joining (MMEJ); 2) exchange between sequences sharing significant stretches of homology, as can occur either by non-allelic homologous recombination (NAHR) during DSB repair or meiosis, or by single-strand annealing (SSA) at DSBs; 3) DNA replication errors such as strand-slippage or template switching; and 4) transposition of mobile elements.
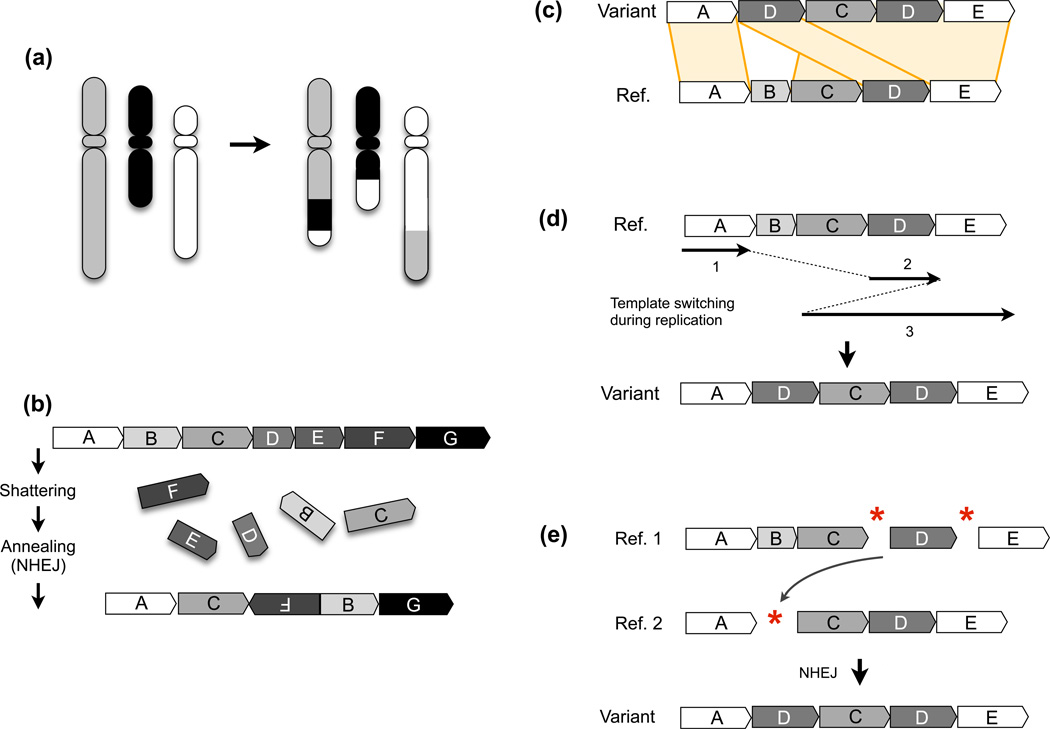
Figure 1.
High level view of complex rearrangements. (a) A depiction of a complex chromosomal rearrangement exhibiting 4 breakpoints from three differently-shaded chromosomes. (b) A complex rearrangement formed by chromothripsis, where a large section of a chromosome (segments B–F) is shattered and imprecisely stitched back together, leading to multiple intra-chromosomal rearrangements and loss of “D” and “E” segments. Chromosomal segments are indicated as blocks with different letters and shading, and the orientation by an arrow in each block. This diagram is loosely based on a figure in [94]. (c) A complex genomic rearrangement. The variant structure is shown at top and the reference genome (Ref.) below. Alignments are shown as orange blocks connecting the two sequences. This variant involves a deletion of “B” and a duplication of “D”, with “C” unaffected. (d) A model for how the variant structure in part (c) could arise through template switching during replication (e.g., FoSTeS/MMBIR), where continuously replicated segments are indicated as solid lines and template switches as dotted lines. (e) A model for how the variant structure in (c) could arise through NHEJ-mediated repair of three double strand DNA breaks. DNA breakages are indicated by red asterisks. In this example, a single DNA break destroys segment “B”, as can occur due to resection. The two copies of the reference locus (“Ref. 1” and “Ref. 2”) are present either as homologous chromosomes or sister chromatids.
Breakpoints are usually identified by comparing the structure of an experimental genome to that of the reference genome, and breakpoint positions are reported based on the coordinate system of the reference (Figure 2). This can cause some confusion since the number of sites may be different depending on which genome one is referring to. For example, a deletion produces a single junction in the experimental genome, but this junction is defined by two coordinates in the reference; the term “breakpoint” has in various studies been used to describe either, or both points of view. The Variant Call Format (VCF) definition resolves this ambiguity by using the terms “novel adjacency” and “breakend” to refer to sites in the experimental and reference genomes, respectively [22]. For simplicity, we define breakpoints based upon their number and position in the experimental genome. This facilitates technical discussion since it is the genome for which experimental data is generated and interpreted, and usually the genome that harbors the derived SV allele produced by recent mutation. However, we note that the reference genome harbors a finite number of derived alleles that can only be reliably discerned by comparison to related species [2, 23].
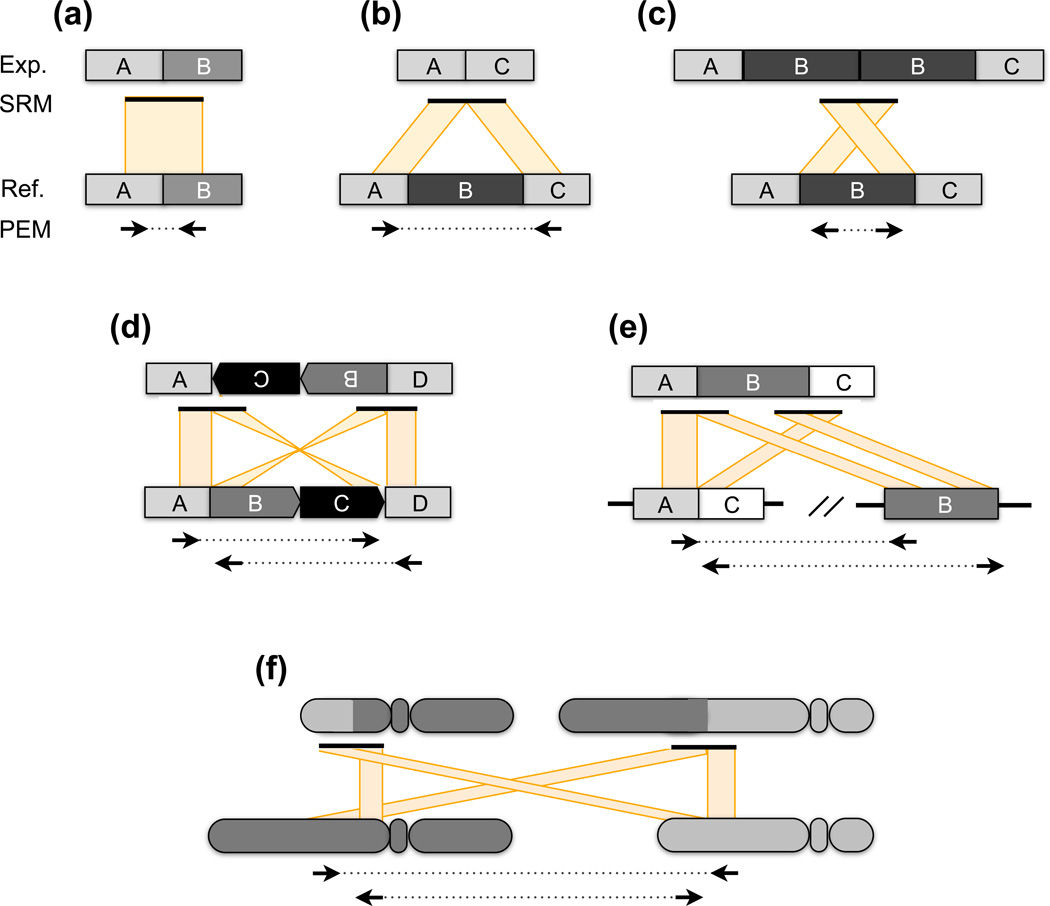
Figure 2.
Detecting canonical SV breakpoints through sequencing. When DNA sequences are collected from an experimental (Exp.) genome and aligned to a reference (Ref.) genome, each structural variant class generates a distinct alignment pattern. The patterns observed for paired-end mapping (PEM) and split-read mapping (SRM) are illustrated when both genomes have identical structure (a), and cases where the experimental genome contains a deletion (b), a tandem duplication (c), an inversion (d), a transposon insertion (e) and a reciprocal translocation (f). PEM relies upon readpairs whose unsequenced portion (dotted lines) spans a SV breakpoint. When aligned to the reference genome, the alignment distance and orientation of such readpairs indicates the type of rearrangement that has occurred. Reads that map to the plus strand are shown as right-facing arrows, those that map to the negative strand as leftward-facing arrows. All examples depict Illumina paired-end sequence data, where in the absence of SV the normal concordant orientation is plus for the leftmost read and minus for the rightmost read. Note that the expected orientation is different for Illumina mate-pair libraries and for other sequencing platforms such as SOLiD. In the case of a deletion (b), the readpairs ends will align much farther apart than expected for the DNA library. In contrast to PEM, SRM depends on contiguous sequences that contain an SV breakpoint. Consequently, the sequences before and after the breakpoint will align to disjoint regions of the reference genome. In contrast to PEM, breakpoints are identified at single-base resolution.
By definition, complex structural variants are composed of multiple breakpoints whose origin cannot be explained by a single end-joining or DNA exchange event. Complex SVs vary considerably in their architecture. The most extreme forms exhibit multiple rearrangements between distinct loci and/or different chromosomes, sometimes involving complex patterns of copy number alteration at or near rearrangement breakpoints [16, 24, 25]. Many are comprised of multiple deletions, duplications and/or rearrangements at a single locus [6, 26–29]. The most subtle forms contain one or more small65 scale insertions, deletions or rearrangements at the breakpoint of a larger SV [6, 30–33]. As one might expect, the most extreme forms of complex SV are generally associated with cancer or sporadic disorders, and the majority of complex SVs identified in healthy individuals are ostensibly benign.
By definition, complex SVs arise through a single mutational event. A central caveat is that this fact can be difficult to establish. Any complex variant structure can, in theory, also be produced by independent temporally-distinct mutations (Figure 1c–e), and repeated mutation is known to occur at localized regions within dicentric chromosomes subjected to breakage-fusion-bridge cycles [34], or at unstable loci such as fragile sites [35], recombination hotspots [36], palindromes [37], and “core duplicons” [38]. Artificially complex breakpoint patterns can also be produced by one simple mutation at an otherwise complex locus in the reference genome, such as those formed by repeated segmental duplication during evolution [39]. Thus, it can be difficult to accurately distinguish between simple and complex forms of structural mutation.
While the methods for detecting complex breakpoint patterns formed by sequential versus complex mutation are essentially the same, their origins and consequences are different. We mainly focus on variants formed by complex mutation and attempt to distinguish between these classes when possible.
Complex SV in the germline
The observation of complex structural mutation is not new. Using standard cytogenetic methods such as G-banded karyotyping and fluorescent in situ hybridization (FISH), numerous complex chromosome rearrangements (CCRs) have been identified in patients suffering from sporadic disorders or infertility (reviewed in [24, 40]). CCRs involve at least 3 breakpoints from 2 or more chromosomes (Figure 1a), and are estimated to comprise ~3% of spontaneous rearrangements detected in prenatal diagnoses [41]. Some events are remarkably complex: ~26% of 251 well-characterized CCRs have more than 5 breakpoints [24, 40], and two contain 15–17 breakpoints [42, 43]. Similarly complex intra-chromosomal rearrangements have also been reported [44]. Interestingly, when rearrangements are fine-mapped, many apparently simple rearrangements are found to be complex, the number of detected breakpoints tends to increase and additional copy number mutations and local rearrangements are often found near breakpoints [43, 45, 46] (prior work reviewed in [24, 40]). The fact that most CCRs are identified as spontaneous events strongly argues that they arise through a single complex mutation rather than multiple independent simple mutations.
More recently, array-CGH has revealed a number of smaller-scale submicroscopic complex genomic rearrangements associated with sporadic disease. These mutations are generally “non-recurrent” in that they exhibit novel breakpoints, as opposed to recurrent mutations formed by NAHR. The most detailed studies characterized a series of non-recurrent pathogenic de novo SVs at the PLP1 [26] and MECP2 genes [28], and at a 3Mb locus associated with Potocki-Lupski and Smith-Magenis syndromes [27]. Remarkably, the authors found complex structures in 41% of 61 non-recurrent mutations. Taking into account previous (reviewed in [24]) and subsequent [47–50] reports of complex SV at disease-associated loci, these data indicate that complex mutations account for a significant fraction of de novo SVs. Reported patterns include adjacent copy number alterations separated by unaltered intervening sequence, deletions or duplications embedded within larger duplications, and triplications.
These observations, as well as previous data from bacterial studies [51], led to two related models for the generation of complex SVs: fork stalling and template switching (FoSTeS) [26], and microhomology-mediated break-induced replication (MMBIR) [29]. In these models, a stalled or broken replication fork undergoes template switching events utilizing microhomology (e.g., 2–5 bp) between the 3’ end of the newly synthesized strand and non-allelic loci (Figure 1d). Complex SVs are produced when multiple switches occur at a single broken/stalled fork. Importantly, template switches may occur between distant loci spanning entire chromosome arms [52], presumably due to proximity in the nucleus, which implies that they may also be involved in many complex chromosomal rearrangements. Fine-scale mapping of CCRs using modern sequencing technologies [46] will help resolve this question.
One might predict that many inherited germline SVs, most of which are likely benign, might also exhibit these features. An early study found that 5 of 24 deletion breakpoints showed small-scale insertions or rearrangements, or multiple deletions separated by non-deleted sequence [30]. Another clue came from sequencing breakpoints in synteny between the human and gibbon genomes [31]. Of 24 rearrangement breakpoints, 11 contained insertions ranging from 9 bp to 20 kb, and some insertions were mosaic structures composed of common repeats and segmental duplications originating from nearby genomic regions.
Three recent genome-wide DNA sequencing-based studies have assessed the prevalence of complex SVs by characterizing inherited SV breakpoints at single base resolution. The first [6] examined 1171 breakpoints in the mouse genome and found ~16% of variants to be complex. Of these, 84% were composed of multiple breakpoints in close proximity (<1 kb), often with intertwined breakpoint patterns caused by one or more adjacent deletion/duplication events plus local rearrangement, and the remainder contained small breakpoint insertions or rearrangements. Common patterns included duplications separated by small non-duplicated segments, deletions adjacent to larger duplications, and deletions with an internal sequence transposed to edge of the breakpoint, often in inverted orientation (Figure 3). Two subsequent studies in human focused mainly on breakpoint insertions. One [32] used DNA capture technology to sequence 324 breakpoints predicted by array-CGH [53], and found that 5.2% contained breakpoint insertions, most of which were derived from nearby loci and inserted in inverted orientation. Another [33] sequenced 1054 SV breakpoints identified by fosmid paired-end mapping [54], and found that 5.5% contained insertions of DNA larger than 20bp, and 73% of the breakpoint insertions were derived from a locus less than 250 kb away. Thus, three studies, using distinct methods and definitions of variant complexity, have converged on a fairly similar estimate for inherited complex SV: 5–16%. Given the technical difficulties associated with high-throughput mapping, assembly and interpretation of breakpoint sequences, as well as the apparently higher incidence of de novo complex variants (discussed above) we suspect that the true number is somewhat higher.
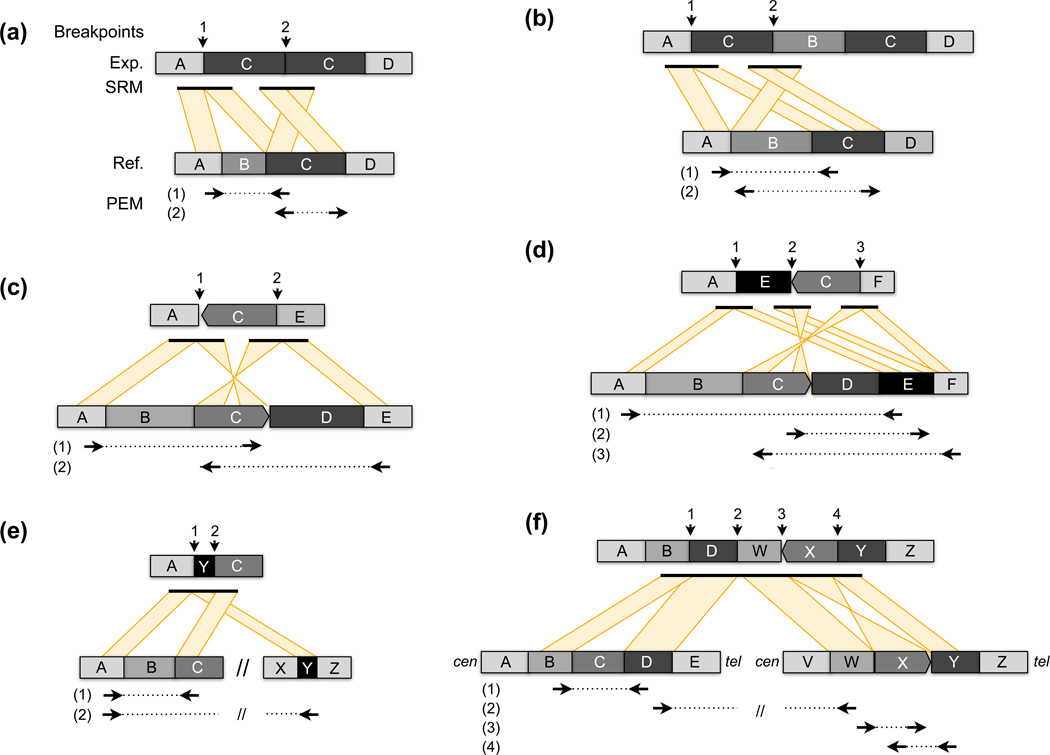
Figure 3.
Some common complex SV architectures. In each example the structure of the experimentally-sequenced genome (Exp.) is shown above the reference genome (Ref.), with genomic segments represented as shaded blocks with letters. Distinct loci in the reference genome, either from a nearby or distant location, are separated by “//”. From top, the positions of “breakpoints” in the experimental genome are shown with arrows. “SRM” shows the alignment patterns generated by split-read mapping of long-reads or assembled contigs that span the breakpoint(s). Shown beneath the reference genome are the discordant alignment patterns generated by paired-end mapping (PEM) using Illumina paired-end sequencing, following the conventions outlined in Figure 2. (a) A deletion of segment “B” and tandem duplication of “C”. (b) A duplication of “C” with an intervening unaffected segment (“B”). (c) A deletion of “B” and “D”, with an inversion of the intervening “C” segment. (d) A highly complex variant involving deletion of “B” and “D”, inversion of “C”, and rearrangement of “E” between “A” and “C”. (e) A deletion of “B” where a segment from a different locus “Y”, either nearby or from an entirely different chromosome, has inserted directly into the deletion breakpoint. (f) A large-scale rearrangement between distant loci that is associated with additional local alterations. In this example there is a rearrangement between “D” and “W”, segment “C” has been deleted, and segment “X” has been inverted.
Complex SV in tumor genomes
The architecture of a somatic genome is less constrained than that of a germline genome, which must complete meiosis and development to survive, and tumors evolve under diverse selective pressures and mutational forces. As a result, the types and numbers of de novo SVs in different tumors varies widely, and diverse karyotypic configurations have been observed. Many tumors show complex patterns of gene amplification [55], presumably due to repeated mutation and strong selection. In some breast tumors, “firestorms” of amplification and deletion have been observed [56] on chromosome arms, likely resulting from breakage-fusion-bridge. These complex patterns have historically been explained by a gradual accumulation of mutations during tumorigenesis [20].
The field has been upended by the discovery of extraordinarily complex intra- and inter-chromosomal rearrangements in certain tumor genomes. In the initial finding, sequencing of a single chronic lymphocytic leukemia (CLL) genome revealed 42 somatically-acquired SV breakpoints in several clusters on the long arm of chromosome 4 (4q) [25]. These included deletions, intra-chromosomal rearrangements, and inter-chromosomal rearrangements to a single site each on chromosomes 1, 12 and 15. Remarkably, only one additional somatic SV was discovered in the rest of the genome. The 4q region exhibited numerous hemizygous deletions (1 copy) separated from each other by unaltered segments (2 copies), and the boundaries of deleted segments corresponded to intra- and inter-chromosomal rearrangement breakpoints. This pattern differs markedly from previously described tumors, but is not rare; the authors mined SNP array data and found similar patterns in 18 of 746 (2.4%) diverse cancers/cell lines, 4 of which were confirmed by whole-genome sequencing, and in 5 of 20 (25%) unselected bone cancers also analyzed by genome sequencing.
The authors presented three lines of evidence that these unprecedented rearrangements are generated through a single catastrophic event [25]. First, simulations revealed that breakpoints are clustered in a highly nonrandom manner. Second, the copy number profiles associated with complex events only exhibit two states – either losses or gains but not both – interdigitated with unaltered segments, whereas sequential mutation should produce many states. Third, within breakpoint clusters harboring intertwined deletions and rearrangements, losses derive from the same parental chromosome and heterozygosity is preserved at unaltered segments, which greatly constrains the order of events under a model of sequential mutation. The authors refer to this mutational process as chromothripsis, and propose that a chromosome is shattered in a one-off event, perhaps by ionizing radiation or one dramatic cycle of breakage-fusion-bridge, and stitched back together again in imprecise fashion (Figure 1b). Interestingly, a recent study [43] reported an inherited complex rearrangement with a similar structure, which indicates that chromothripsis-like mechanisms also operate in the germline.
More recently, a single complex rearrangement was identified in 3 of 7 prostate cancer genomes analyzed by whole-genome sequencing [16]. One involved 4 loci on a single chromosome, another involved 4 loci on two chromosomes, and the third involved 9 loci on 4 chromosomes. Strikingly, two involved a novel “closed chain” breakpoint pattern, such that each locus was connected to two other distinct loci. While the precise structure of “closed chain” rearrangements is unclear (Figure 4), there are two key differences between them and those attributed to chromothripsis: 1) there is no obvious clustering of breakpoints on a single chromosome; and 2) the breakpoint regions do not exhibit copy number mutations. It is an open question whether these rearrangements are caused by chromothripsis or a distinct mechanism such as FoSTeS/MMBIR. Perhaps indicating the latter, the data shown for one rearrangement are more consistent with 3 small insertions into a single locus rather than a series of translocations.
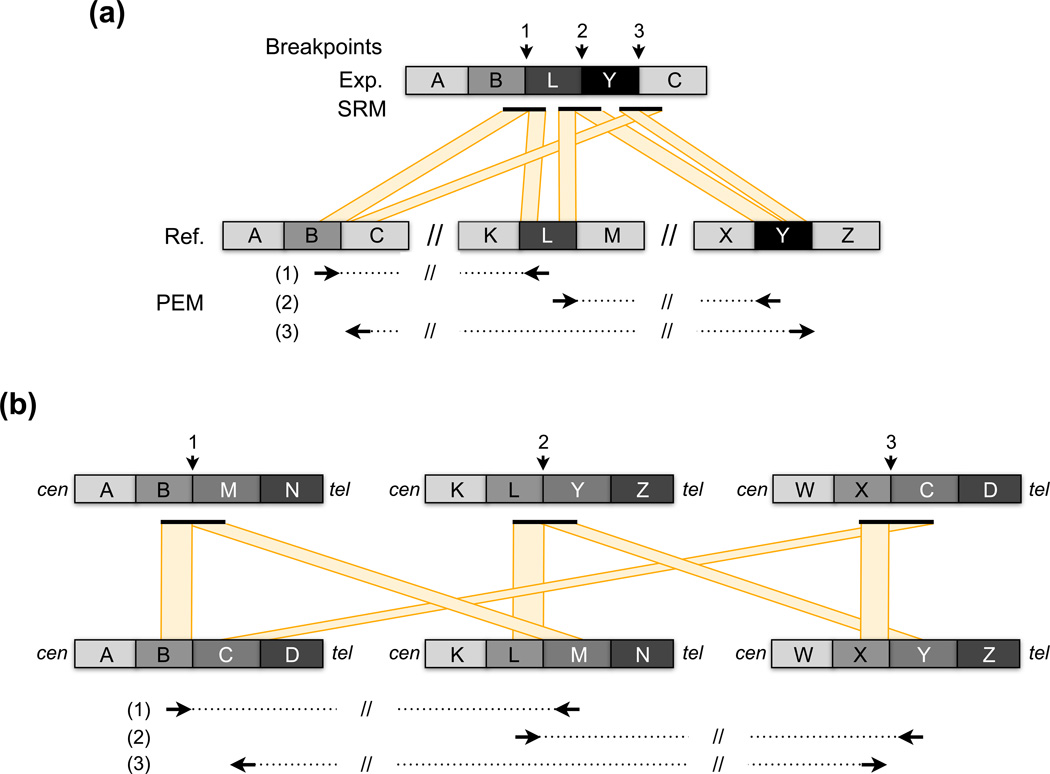
Figure 4.
Two potential explanations for “closed chain” rearrangement patterns. The breakpoint calls and the experimental (“Exp.”) and reference (“Ref.”) genomes are shown exactly as in Figure 3. (a) DNA segments from two donor loci “L” and “Y” each insert the recipient locus between “B” and “C” at adjacent positions This generates a closed chain of PEM calls whereby each locus connects to exactly two other loci. (b) A series of translocations involving three chromosomes can also generate a closed chain rearrangement pattern. The direction of the centromere and telomere are indicated by “cen” and “tel”, respectively. Note that these two forms of closed chain rearrangement can be distinguished because insertion donor loci have a distinct pattern of two outward facing PEM calls (see “L” and “Y” in part (a)), whereas the PEM calls at translocation breakpoints are inward facing (e.g., see breakpoint between “L” and “M” in part (b)).
Identification and interpretation of complex variation
Advances in DNA sequencing technologies have enabled the exploration of genome structure with exquisite detail. Unlike conventional cytogenetic methods or array-CGH, sequencing permits genome-wide characterization of breakpoints from all classes of SV with high precision. The general algorithmic approaches and available tools for detecting SV breakpoints from DNA sequence data have been reviewed elsewhere [62, 63]. In essence, the identification and interpretation of complex SV involves 3 steps: 1) genome-wide breakpoint detection using one or more of the techniques discussed in Box 1; 2) screening for clusters or interconnected chains of breakpoints that comprise a single complex variant; and 3) reconstructing the architecture of the variant locus to infer the causal mechanism and potential functional impact.
Box 1. Mapping SV breakpoints with modern sequencing technologies.
Depth of coverage (DOC)
When DNA sequences are aligned to the reference genome, copy number variations (CNVs) are evident as significant increases or decreases in the depth of aligned sequence [57]. Inferring CNV via DOC analysis is conceptually similar to array-CGH and typically yields similar or moderately better resolution (1–15kb). DOC is inadequate for mapping fine-scale locus complexity, but permits initial identification of complex CNVs and helps determine whether complex breakpoint patterns involve copy number mutations. DOC can also detect NAHR-derived CNVs whose breakpoints lie within large repeats (which can confound the methods below).
Paired-end mapping (PEM)
PEM strategies identify SV breakpoints by examining the alignments of relatively short sequences from the ends of larger DNA molecules [58]. Sequencing libraries are created with fragments of known length (generally 200–500 bp for paired-end libraries and 1–10 kb for mate-pair libraries). Paired-end sequences (readpairs) that are “concordant” with the reference genome align with the expected distance and orientation, whereas readpairs spanning an SV breakpoint will produce “discordant” alignments with an unexpected alignment distance and/or orientation. Each SV class produces a distinct mapping signature (Figure 2). However, current fragment sizes limit sensitivity, discordant mapping patterns can be difficult to interpret at complex SVs (Figure 4), and PEM cannot map breakpoints to single-base resolution.
Split-read mapping (SRM)
SRM identifies sequences that actually contain a breakpoint [2, 59] (Figure 2). The alignments for such sequences are “split” because DNA segments flanking the breakpoint align to disjoint locations in the reference genome. SRM inherently maps breakpoints to single base resolution and thus provides mechanistic insight. Owing to the repetitive structure of mammalian genomes, genome-wide SRM requires reads longer than ~200 bp. Long-read (> 500 bp) SRM is a particularly powerful approach for studying complex SV because multiple breakpoints can potentially be captured by a single read, greatly aiding in variant locus reconstruction (Figure 3e,f).
Assembly
Local de novo sequence assembly [60] can be used to reconstruct a variant locus. Once an assembled sequence is aligned to the reference genome, breakpoint(s) are discerned following the same principles as SRM. Unlike SRM, assembly typically generates substantially larger “contigs” that are more amenable to characterizing complex SV. While currently infeasible for most laboratories, whole-genome assemblies promise the most comprehensive description of SV, as large portions of entire chromosomes can be aligned to precisely identify both canonical and complex rearrangements [1, 61].
Screening for complex SV
Once raw breakpoints have been mapped the primary goal is to distinguish clusters of breakpoints delineating complex variants from nearby, yet potentially simple SV breakpoints caused by independent mutations. The development of robust tools for identifying complex events is a difficult and unsolved problem because at present there are no defined rules for constraining the expected breakpoint patterns. It is not clear whether such rules exist. Nevertheless, discerning complex mutations can be relatively straightforward when analyzing human families or minimally mutated cancer genomes, since spontaneous events can be readily distinguished from inherited variants by analyzing related samples. However, detecting complex variants in a “sea” of simple variants, as in studies of inherited SV or highly rearranged cancer genomes, is problematic because breakpoints may lie in close proximity due to chance alone. This may not be a concern for functional studies but is crucial for inferring mechanism. There is no simple solution to this conundrum, and thus most studies have focused on the most obvious examples of complex SV.
Simple and flexible approaches are therefore preferable. Screens must begin by accounting for simple multi-breakpoint variants such as inversions, retrotranspositions and reciprocal translocations (Figure 2e,f). Merging these breakpoint calls is conceptually simple, but we are not aware of any available software that does so comprehensively. Breakpoint clusters can then be identified by simple sliding window schemes that compare local breakpoint density to a null model. Ideally, this screening method should take into account the non-uniform distribution of simple SV in normal and tumor genomes, as well as commonly observed complex variant architectures. It may be possible to use homology profiles to tease apart nearby or overlapping clusters that arose through distinct mechanisms, but since breakpoints formed by template switching and end-joining can display similar levels of microhomology, in practice this will be difficult. Complex SVs that do not involve obvious breakpoint clusters at a single locus can be identified by computationally searching for chains of interconnected breakpoints that share at least one locus in common. Tools in the BEDTools software suite [64] can be adapted for this purpose [6]. By integrating results from clustering and chaining approaches, most classes of complex SV can be discerned. We stress, however, that these higher-order clustering steps can produce falsely complex SVs at repetitive or poorly-assembled loci in the reference genome that generate abundant breakpoint calls, as often occurs at or near centromeres, telomeres, simple tandem repeats, and regions laden with segmental duplications. Thus, subsequent annotation and characterization steps are crucial.
The above methods may fail to detect complex SVs that possess neither clustered nor chained breakpoints, but rather are composed of nested or overlapping variant calls that affect a common genomic interval. This pattern is trivial to detect, but is also commonly produced by sequential mutation and should be interpreted with caution. These methods may also miss cryptic complex variants that contain small-scale insertions or rearrangements at the breakpoint itself. For these it is necessary to carefully inspect breakpoints at single-base resolution and to align the breakpoint sequence to the reference genome. Sensitive alignment is crucial because small breakpoint alterations can masquerade as non-templated addition of nucleotides during NHEJ, merely due the inability of aligners to find significant matches.
Interpreting complex variants
A key question for any complex variant is: what exactly does it look like? Integration of breakpoints identified by PEM, SRM and/or local assembly (Box 1), combined with DOC analysis to distinguish between balanced rearrangements and copy number mutations, is theoretically sufficient to infer the architecture of most variants (Figure 3 and Figure 4). However, this remains a major challenge for two reasons. First, neither reconstructing nor visualizing complex variant structures are trivial problems and there is a notable dearth of suitable computational tools. Thus, to our knowledge, all DNA sequencing-based studies to date have relied heavily on manual curation and human expertise to interpret complex breakpoint patterns. This laborious approach has proven effective and resulted in detailed architectural information for over 250 complex SVs [6, 16, 25, 32, 33, 43], but is unsustainable given the scale of current genome sequencing projects. Second, the accuracy of interpretation depends entirely on the accuracy of the underlying breakpoint calls, and current breakpoint mapping strategies suffer from either high false positive or high false negative rates, and sometimes both. It is therefore likely that complex SVs are more prevalent, and more architecturally diverse, than currently recognized owing to under-ascertainment and misinterpretation.
Manual variant reconstruction is greatly aided by data visualization software (Figure 5). The UCSC Genome Browser [65], Integrative Genomics Viewer (IGV) [66] and Savant [67] excel at displaying raw sequence data aligned to the reference genome and can also display annotation tracks, but are only practical for visualizing small genomic regions (< 100 kb). A current advantage of IGV is the ability to visualize two distinct loci in “split-screen” mode, but Savant offers superior visualization of readpair connectivity. At the other end of the spectrum, visualization tools like CIRCOS [68] or GREMLIN [69] provide aesthetically-pleasing rearrangement depictions, but are mainly useful for summarizing results, not interpreting data. A major limitation of the above tools is that they display data solely with respect to the reference genome, which does not allow one to easily infer variant architecture.
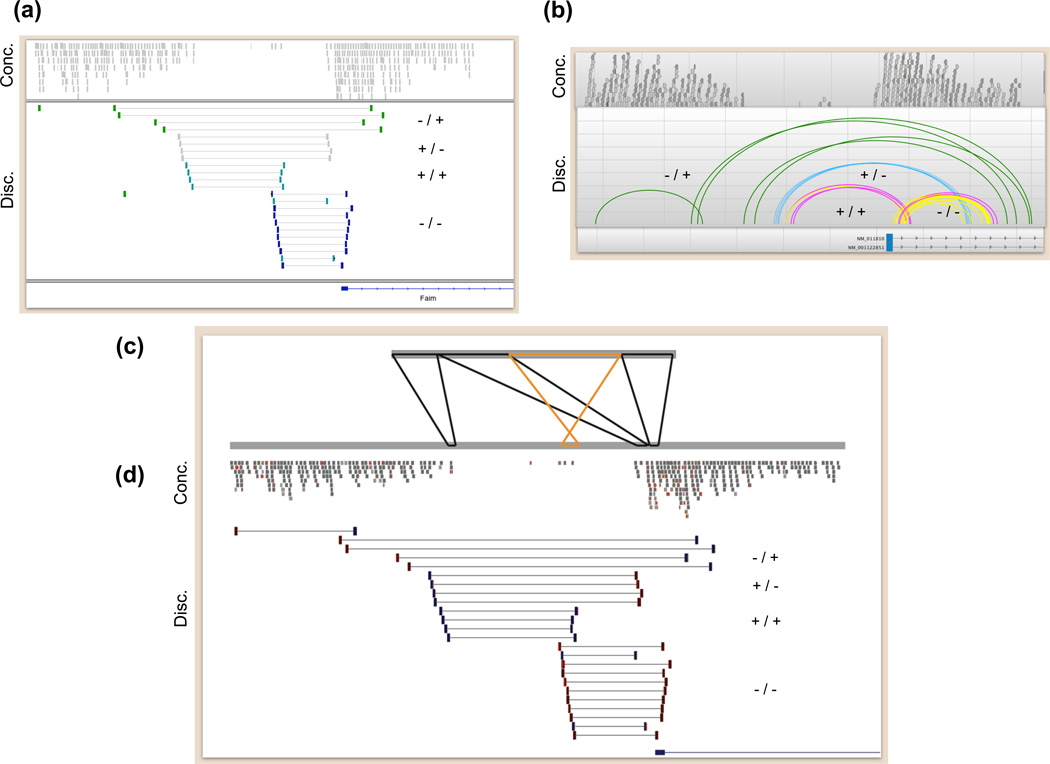
Figure 5.
Visualizing complex loci. Snapshots of aligned paired-end sequence data from a complex locus (chr9: 98,880,333–98,889,602; NCBI37/mm9) in the DBA/2J mouse strain are depicted with (a) the Integrated Genomics Viewer (IGV), (b) SAVANT and (d) the UCSC Genome Browser. In (c) alignment of a partial de novo assembly of this locus is displayed with PARASIGHT. In each panel, concordant readpairs (Conc.) are displayed above the discordant (Disc.) readpairs that collectively indicate the complex rearrangement at this locus. The orientation of the “left” and “right” ends of each type of discordant readpairs are displayed next to the alignments: “−/+” for tandem duplication, “+/−” for deletion, “−/−” and “+/+” for distinct inversion breakpoints. In panel (c), black alignments between the assembled locus and the reference genome are in the same orientation, whereas orange alignments are in opposite orientation. Note that this assembled contig does not represent the entire locus, as the outermost readpairs suggesting a tandem duplication (−/+) were not incorporated into the assembly. However, the assembled sequence suggests that, at a minimum, this complex locus involves two adjacent deletions of 2.5 kb and 0.9 kb, which are separated by an intervening 300-bp segment that was not deleted, but rather inverted.
Rapid interpretation requires a direct comparison of the structure of assembled breakpoint sequences, or entire variant loci, to the structure of the reference genome. In some cases a simple dotplot may suffice. The PARASIGHT software (J. Bailey et al., unpublished: http://eichlerlab.gs.washington.edu/jeff/parasight) is ideally suited to this task because it shows pairwise alignments in an informative format that preserves the structure of both variant and reference sequences (Figure 5c), and can display annotation tracks. For example, an automated PARASIGHT pipeline enabled visualization and interpretation of several thousand assembled breakpoints in several days [6]. Unfortunately, while PARASIGHT is extremely flexible, it is difficult to use and often requires substantial customization for informative viewing. Other tools such as MIROPEATS [70] and BARAVI (R. Ophoff et al., unpublished: http://www.genetics.ucla.edu/labs/ophoff/BARAVI/) support pairwise alignment and visualization but cannot display tracks. The paucity of user-friendly breakpoint visualization software presents a major bottleneck for interpreting complex variants and underscores the need for improved tools.
Manual curation is the most accurate approach for variant reconstruction, but as the study of complex SV expands to thousands of genomes it is neither practical nor reproducible. In theory it should be possible to develop software that infers variant architecture from breakpoint predictions and DOC profiles, but we are unaware of any that explicitly attempts to do so. Moreover, we suspect that automated reconstruction of complex SVs would require impeccable input data. For example, sophisticated algorithms have proven necessary merely to integrate breakpoint calls and DOC profiles for simple deletions [2, 71]. As sequencing methods continue to improve, automated approaches will eventually be feasible through increased read lengths, emerging technologies such as “strobe” sequencing [72] and, ultimately, routine generation of high-quality diploid genome assemblies.
If a complex SV can be assembled into a single contig, variant reconstruction becomes a tractable problem of describing the relative structure of two DNA sequences. The first step is to align the variant sequence to the reference genome. A complication is that portions of the variant “query” sequence containing repeats will align to multiple loci. This problem is trivial for variants that involve a single well-defined locus, but for rearrangements that involve repetitive regions or multiple loci resolving these ambiguities can be difficult. This is also a significant problem for the initial detection of complex SVs from long-reads or draft assemblies. Most suitable aligners report all significant alignments including irrelevant “sub-alignments” contained within larger aligned sections of the query [73–75], which necessitates subsequent selection of the “best” minimal set for locus reconstruction. The BWA-SW aligner uses a greedy heuristic strategy to discard sub-alignments that are subsumed by larger alignments [76]; we have found that this, or similar, heuristic strategies are adequate for moderately complex variants composed mainly of unique sequence. Otherwise, it is preferable to pursue a more optimal alignment selection strategy.
Once alignments are defined, reconstructing variant architecture is a semantic problem of describing the relationship between alignment blocks based upon their relative positions and orientations in the variant and reference sequences. The VCF 4.1 specification offers a sensible solution for this practical problem [22].
Mechanistically minded studies might seek to reconstruct the mutational events that generated each complex variant. Similar problems has been studied in the context of ancestral genome reconstruction using breakpoint graphs [77–80], and for inferring the mutational history of segmental duplications using modified A-Bruijn graphs [81] or DAWGs [82]. Genome-scale models are subjected to various simplifying assumptions to prevent intractable computational complexity, but for any given complex variant optimal solutions are possible. An unsolved problem is how to define optimal solutions that take into account current models of mutation.
Concluding remarks
Studies of complex SV have provided new insights into the processes that generate genome variation, and this has clear implications for conventional models of species and cancer evolution that generally assume progressive, step-wise mutations. In both contexts, complex mutations represent a form of punctuated genome evolution. Resulting variants may have more subtle, unpredictable and multi-faceted phenotypic impacts than simple variants. For example, complex mutations can rearrange exons to create novel proteins, shuffle promoters, enhancers and/or repressors into a novel regulatory configuration, or simultaneously disrupt multiple genes and pathways. In the context of a developing tumor, simultaneous formation of multiple fusion genes, amplified oncogenes or deleted tumor suppressors may lead to rapid expansion of a clone with very different characteristics than neighboring cells.
A major unresolved question in the field is how complex variants arise. The two general models for complex SV formation – template switching during DNA replication (FoSTeS/MMBIR) [26, 29] and chromosome shattering (chromothripsis) [25] – each have eminently sensible features, but it is worth remembering that neither has been directly implicated. This begs the question of whether these mechanisms indeed account, either alone or through collusion, for the architecturally diverse rearrangements that have been observed. Or is another as-yet undescribed mechanism at work? At present, there is not sufficient data to answer these questions. However, we speculate that most complex variants arise through a common mechanism. The rearrangements thus far attributed to chromothripsis differ from those explained by FoSTeS/MMBIR mainly in their greater size and complexity; the patterns are ostensibly similar. We further note that a recent study of germline rearrangements [83] has proposed that FoSTeS/MMBIR may explain complex breakpoint clusters that resemble those attributed to chromothripsis [25, 43, 84]. These clusters contain 3 copy number states, including duplications and triplications, and small breakpoint insertions derived from nearby loci. These features are much easier to explain by replication than by chromosome shattering. On the other hand, shattering is a more simple explanation for the staggeringly complex variants that exhibit frequent oscillation between 2 copy number states (deleted and unaltered), as observed in tumor genomes. We expect future breakpoint sequencing studies to yield additional clues, but we are not confident that the true mechanism(s) can be resolved by sequencing alone, since neither variant architectures nor breakpoint homology profiles appear sufficient to distinguish the two models. Direct experimental studies may be necessary to yield clarity.
The likelihood that complex mutations primarily arise through processes that are active in somatic cells, and not concentrated in meiosis, also implies that many other simple mutations do as well, and thus each individual may be a mosaic composition of cells with different genome structures. Indeed, evidence of somatic variation is growing [85–91], and this may potentially account for certain phenotypes that emerge during development and aging. The potential link to replication also implies that environmental conditions or trans-acting mutations that affect replication fidelity can modulate mutation rates. It has been proposed that replication stress may lead to flurries of structural mutation [21, 29], and there is direct evidence for this in E. coli [51] and cultured human cells [92, 93]. Further work is necessary to prove this theory, but the potential existence of genetic and environmental modulators of complex mutation is intriguing.
In most cases the functional consequences of complex SVs are unclear, and their true contribution to natural variation remains an open question. Whether these variants turn out to be a curious sideshow of mutational complexity or a driving force of functional innovation can only be answered by ongoing and future whole-genome sequencing of well-phenotyped samples. Rapidly improving DNA sequencing technologies will aid this effort, but perhaps the greater challenge lies in bioinformatic interpretation. At present, there is a notable paucity of high-throughput methods for complex SV identification, visualization, reconstruction or interpretation. We expect this challenge to be met in coming years, and we look forward to a more complete understanding of the mechanisms and functional ramifications of complex structural variation.
Acknowledgments
Our work has been sponsored by the National Institutes of Health (DP2OD006493-01 to IMH; 1F32HG005197-01 to ARQ), the Burroughs Wellcome Fund (IMH) and the March of Dimes (IMH). We thank R.A. Clark for implementing our SV visualization pipeline.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pang AW, et al. Towards a comprehensive structural variation map of an individual human genome. Genome biology. 2010;11:R52. doi: 10.1186/gb-2010-11-5-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mills RE, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang F, et al. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perry GH, et al. Hotspots for copy number variation in chimpanzees and humans. Proc Natl Acad Sci U S A. 2006;103:8006–8011. doi: 10.1073/pnas.0602318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graubert TA, et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinlan AR, et al. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res. 2010;20:623–635. doi: 10.1101/gr.102970.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guryev V, et al. Distribution and functional impact of DNA copy number variation in the, rat. Nat Genet. 2008;40:538–545. doi: 10.1038/ng.141. [DOI] [PubMed] [Google Scholar]
- 8.Chen WK, et al. Mapping DNA structural variation in dogs. Genome Res. 2009;19:500–509. doi: 10.1101/gr.083741.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholas TJ, et al. The genomic architecture of segmental duplications and associated copy number variants in dogs. Genome Res. 2009;19:491–499. doi: 10.1101/gr.084715.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu GE, et al. Analysis of copy number variations among diverse cattle breeds. Genome Research. 2010;20:693–703. doi: 10.1101/gr.105403.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephens PJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welch JS, et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA : the journal of the American Medical Association. 2011;305:1577–1584. doi: 10.1001/jama.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Hastings PJ, et al. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danecek P, et al. The Variant Call Format and VCFtools. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam HY, et al. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nature biotechnology. 2010;28:47–55. doi: 10.1038/nbt.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F, et al. Complex human chromosomal and genomic rearrangements. Trends in genetics : TIG. 2009;25:298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JA, et al. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 27.Zhang F, et al. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carvalho CM, et al. Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching. Hum Mol Genet. 2009;18:2188–2203. doi: 10.1093/hmg/ddp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hastings PJ, et al. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000327. e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perry GH, et al. The fine-scale and complex architecture of human copy-number variation. Am J Hum Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Girirajan S, et al. Sequencing human-gibbon breakpoints of synteny reveals mosaic new insertions at rearrangement sites. Genome Res. 2009;19:178–190. doi: 10.1101/gr.086041.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conrad DF, et al. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet. 2010;42:385–391. doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kidd JM, et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–847. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 36.Myers S, et al. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 37.Inagaki H, et al. Chromosomal instability mediated by non-B DNA: cruciform conformation and not DNA sequence is responsible for recurrent translocation in humans. Genome Res. 2009;19:191–198. doi: 10.1101/gr.079244.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marques-Bonet T, Eichler EE. The evolution of human segmental duplications and the core duplicon hypothesis. Cold Spring Harbor symposia on quantitative biology. 2009;74:355–362. doi: 10.1101/sqb.2009.74.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey JA, et al. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pellestor F, et al. Complex chromosomal rearrangements: origin and meiotic behavior. Human reproduction update. 2011;17:476–494. doi: 10.1093/humupd/dmr010. [DOI] [PubMed] [Google Scholar]
- 41.Giardino D, et al. De novo balanced chromosome rearrangements in prenatal diagnosis. Prenatal diagnosis. 2009;29:257–265. doi: 10.1002/pd.2215. [DOI] [PubMed] [Google Scholar]
- 42.Tupler R, et al. A complex chromosome rearrangement with 10 breakpoints: tentative assignment of the locus for Williams syndrome to 4q33----q35.1. Journal of medical genetics. 1992;29:253–255. doi: 10.1136/jmg.29.4.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kloosterman WP, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Human molecular genetics. 2011;20:1916–1924. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- 44.Lindstrand A, et al. Molecular cytogenetic characterization of a constitutional, highly complex intrachromosomal rearrangement of chromosome 1, with 14 breakpoints and a 0.5 Mb submicroscopic deletion. American journal of medical genetics. Part A. 2008;146A:3217–3222. doi: 10.1002/ajmg.a.32570. [DOI] [PubMed] [Google Scholar]
- 45.Feenstra I, et al. Balanced into array: genome-wide array analysis in 54 patients with an apparently balanced de novo chromosome rearrangement and a meta-analysis. European journal of human genetics : EJHG. 2011 doi: 10.1038/ejhg.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Talkowski ME, et al. Next-generation sequencing strategies enable routine detection of balanced chromosome rearrangements for clinical diagnostics and genetic research. American Journal of Human Genetics. 2011;88:469–481. doi: 10.1016/j.ajhg.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang F, et al. Mechanisms for nonrecurrent genomic rearrangements associated with CMT1A or HNPP: rare CNVs as a cause for missing heritability. American Journal of Human Genetics. 2010;86:892–903. doi: 10.1016/j.ajhg.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu P, et al. Copy number gain at Xp22.31 includes complex duplication rearrangements and recurrent triplications. Human molecular genetics. 2011;20:1975–1988. doi: 10.1093/hmg/ddr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang F, et al. Identification of uncommon recurrent Potocki-Lupski syndrome-associated duplications and the distribution of rearrangement types and mechanisms in PTLS. American Journal of Human Genetics. 2010;86:462–470. doi: 10.1016/j.ajhg.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi BO, et al. Inheritance of Charcot-Marie-Tooth disease 1A with rare nonrecurrent genomic rearrangement. Neurogenetics. 2011;12:51–58. doi: 10.1007/s10048-010-0272-3. [DOI] [PubMed] [Google Scholar]
- 51.Slack A, et al. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS genetics. 2006;2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koumbaris G, et al. FoSTeS, MMBIR and NAHR at the human proximal Xp region and the mechanisms of human Xq isochromosome formation. Human molecular genetics. 2011;20:1925–1936. doi: 10.1093/hmg/ddr074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2009 doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korkola J, Gray JW. Breast cancer genomes--form and function. Current opinion in genetics & development. 2010;20:4–14. doi: 10.1016/j.gde.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hicks J, et al. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res. 2006;16:1465–1479. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chiang DY, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raphael BJ, et al. Reconstructing tumor genome architectures. Bioinformatics. 2003;19 Suppl 2:ii162–ii171. doi: 10.1093/bioinformatics/btg1074. [DOI] [PubMed] [Google Scholar]
- 59.Mills RE, et al. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–1190. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller JR, et al. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–327. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y, et al. Structural variation in two human genomes mapped at single-nucleotide resolution by whole genome de novo assembly. Nature biotechnology. 2011;29:723–730. doi: 10.1038/nbt.1904. [DOI] [PubMed] [Google Scholar]
- 62.Medvedev P, et al. Computational methods for discovering structural variation with next-generation sequencing. Nat Methods. 2009;6:S13–S20. doi: 10.1038/nmeth.1374. [DOI] [PubMed] [Google Scholar]
- 63.Alkan C, et al. Genome structural variation discovery and genotyping. Nature reviews. Genetics. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson JT, et al. Integrative genomics viewer. Nature biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fiume M, et al. Savant: genome browser for high-throughput sequencing data. Bioinformatics. 2010;26:1938–1944. doi: 10.1093/bioinformatics/btq332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Research. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O'Brien TM, et al. Gremlin: an interactive visualization model for analyzing genomic rearrangements. IEEE transactions on visualization and computer graphics. 2010;16:918–926. doi: 10.1109/TVCG.2010.163. [DOI] [PubMed] [Google Scholar]
- 70.Parsons JD. Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci. 1995;11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- 71.Handsaker RE, et al. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nature Genetics. 2011;43:269–276. doi: 10.1038/ng.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ritz A, et al. Structural variation analysis with strobe reads. Bioinformatics. 2010;26:1291–1298. doi: 10.1093/bioinformatics/btq153. [DOI] [PubMed] [Google Scholar]
- 73.Altschul SF, et al. Basic local alignment search tool. Journal of molecular biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 74.Ning Z, et al. SSAHA: a fast search method for large DNA databases. Genome Research. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pevzner P, Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc Natl Acad Sci U S A. 2003;100:7672–7677. doi: 10.1073/pnas.1330369100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bourque G, et al. Reconstructing the genomic architecture of ancestral mammals: lessons from human, mouse, and rat genomes. Genome Res. 2004;14:507–516. doi: 10.1101/gr.1975204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murphy WJ, et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- 80.Alekseyev MA, Pevzner PA. Breakpoint graphs and ancestral genome reconstructions. Genome Research. 2009;19:943–957. doi: 10.1101/gr.082784.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jiang Z, et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat Genet. 2007;39:1361–1368. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- 82.Kahn CL, Raphael BJ. A parsimony approach to analysis of human segmental duplications. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing. 2009:126–137. [PubMed] [Google Scholar]
- 83.Liu P, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Magrangeas F, et al. Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood. 2011;118:675–678. doi: 10.1182/blood-2011-03-344069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liang Q, et al. Extensive genomic copy number variation in embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:17453–17456. doi: 10.1073/pnas.0805638105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Piotrowski A, et al. Somatic mosaicism for copy number variation in differentiated human tissues. Hum Mutat. 2008;29:1118–1124. doi: 10.1002/humu.20815. [DOI] [PubMed] [Google Scholar]
- 87.Bruder CE, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lam KW, Jeffreys AJ. Processes of de novo duplication of human alpha-globin genes. Proc Natl Acad Sci U S A. 2007;104:10950–10955. doi: 10.1073/pnas.0703856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Flores M, et al. Recurrent DNA inversion rearrangements in the human genome. Proc Natl Acad Sci U S A. 2007;104:6099–6106. doi: 10.1073/pnas.0701631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 91.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arlt MF, et al. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. American Journal of Human Genetics. 2009;84:339–350. doi: 10.1016/j.ajhg.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arlt MF, et al. Comparison of constitutional and replication stress-induced genome structural variation by SNP array and mate-pair sequencing. Genetics. 2011;187:675–683. doi: 10.1534/genetics.110.124776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meyerson M, Pellman D. Cancer genomes evolve by pulverizing single chromosomes. Cell. 2011;144:9–10. doi: 10.1016/j.cell.2010.12.025. [DOI] [PubMed] [Google Scholar]