Abstract
Huntington’s disease (HD) is a progressive, fatal neurodegenerative disease caused by an expanded polyglutamine repeats in the HD gene. HD is characterized by chorea, seizures, involuntary movements, dystonia, cognitive decline, intellectual impairment and emotional disturbances. Research into mutant huntingtin (Htt) and mitochondria has found that mutant Htt interacts with the mitochondrial protein dynamin-related protein 1 (Drp1), enhances GTPase Drp1 enzymatic activity, and causes excessive mitochondrial fragmentation and abnormal distribution, leading to defective axonal transport of mitochondria and selective synaptic degeneration. This article summarizes latest developments in HD research and focuses on the role of abnormal mitochondrial dynamics and defective axonal transport in HD neurons. This article also discusses the therapeutic strategies that decrease mitochondrial fragmentation and neuronal damage in HD.
Keywords: Mutant huntingtin, Abnormal mitochondrial dynamics, Defective axonal transport, RNA silencing, BACHD mice, Mitochondrial trafficking
1. Introduction
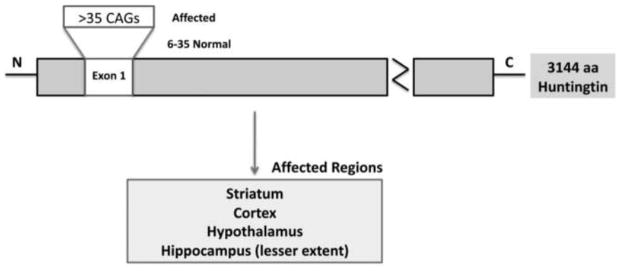
Huntington’s disease (HD) is characterized by chorea, seizures, involuntary movements, dystonia, cognitive decline, intellectual impairment and emotional disturbances [1–4]. HD has an autosomal dominant pattern of inheritance and an age-dependent penetrance. HD occurs in 4–10 per 100,000 persons, mainly of Caucasian origin, with mean age of onset at about 40 years. HD patients survive for about 15–20 years from disease onset and die mainly due to complications from the disease. It is caused by polyglutamine (polyQ) repeat expansion within the exon 1 of HD gene, that encodes an expanded polyQ stretch in the huntingtin (Htt) protein (Fig. 1). Wild-type (WT) and mutant Htt proteins are expressed ubiquitously in the peripheral and central nervous systems of patients with HD. In the postmortem brains of patients with HD, extensive medium spiny neuronal loss was observed in the striatum and also loss of pyramidal neurons was observed in the cerebral cortex and hippocampus. Neuronal loss has also been reported in the hypothalamus in postmortem brains from HD patients and from HD mouse models [1,5–8], and Htt protein aggregates have been found in pathological sites in the postmortem brains of patients with HD and of HD mouse models [9–18]. The causes of selective and premature death of medium spiny projection neurons are not completely understood. Currently, there are no drugs or agents available to treat or delay or prevent HD progression.
Figure 1.
Schematic illustration of expanded polyQ repeats within exon 1 of Huntington’s disease gene.
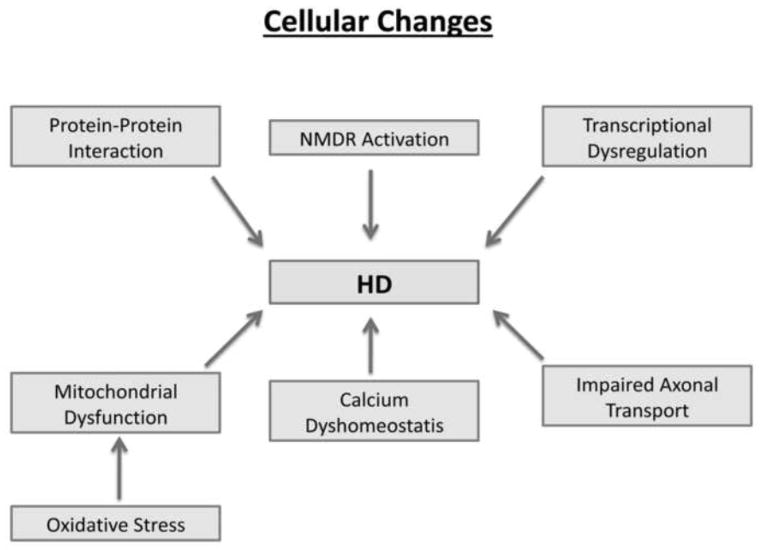
Over two decades of intense research using cell models, animal models, and postmortem HD brains have implicated multiple cellular changes in HD progression and pathogenesis. These changes include transcriptional dysregulation, caspase activation, expanded polyQ repeat protein interactions with other CNS proteins, NMDAR activation, calcium dyshomeostatis, defective axonal trafficking, and abnormal mitochondrial dynamics (e.g. increased fission and decreased fusion) [19–23]. In studies focusing on abnormal mitochondrial dynamics in HD, defective transport of mitochondria in axons and selective synaptic degeneration were found to play a central role in HD progression and pathogenesis.
The purpose of this article was to summarize the roles of mitochondrial dynamics and axonal transport in HD neurons. We will also highlight the relationship between mutant Htt and mitochondrial dynamics, and between mutant Htt and dynamin-related protein 1 (Drp1) – a mitochondrial protein that has been implicated in causing mitochondrial fragmentation and abnormal distribution, defective axonal transport of mitochondria and selective synaptic degeneration in a variety of neurodegenerative diseases. We also will discuss the therapeutic strategies that decrease mitochondrial fragmentation and neuronal damage in HD.
2. Mutant Huntingtin Aggregates and Oligomers in HD Pathogenesis
HD was the first among nine neurodegenerative diseases that had an expanded polyglutamine (polyQ) repeats as a dominant mutation in the coding part of the gene [19]. HD is caused by expanded polyQ repeats within exon 1 of HD gene. In HD patients, the number of polyQ repeats ranges from 36–120, whereas in healthy individuals, it ranges from 6–35 [4,24]. In HD, polyQ repeats are highly polymorphic in general, and the onset of disease is inversely correlates with polyQ repeat length.
Htt is a 350 kDa protein, ubiquitously expressed in the peripheral and central nervous systems (CNS) [25]. WT Htt is a cytosolic protein. However, increasing evidence from postmortem HD brains and HD mouse models revealed that a small part of mutant Htt is present in subcellular organelles, including the nucleus, plasma membrane, mitochondria, lysosomes, and endoplasmic reticulum [26–33]. Mutant Htt is reported to interacts with a large number of CNS proteins, and this abnormal interaction ultimately leads to the gain of function of mutant Htt in the progression of HD.
2.1. Mutant Htt aggregates
Mutant polyQ aggregates have been extensively reported in HD and other polyQ repeat-associated diseases and responsible for disease progression [4,34]. More recently, formations of oligomers, fibrils, and protofibrils have been found in HD cell cultures and HD animal models, including, flies, worms and mice [35–39]. Mutant oligomeric proteins are toxic and accumulate in neurons in age-dependent manner. These aggregates have been found to enter subcellular organelles, such as mitochondria in neurons from patients with Alzheimer’s disease [40–45] and Parkinson’s disease [46–52].
2.2. Mutant Htt oligomers
Recently, using cell culture systems, several investigators have focused on developing molecules that inhibit oligomer formation and apoptotic cell death [35,38,53–61]. Tremendous efforts are underway in several laboratories across the world to reduce the oligomer formation and toxicity.
Using immunostaining analysis of mutant Htt oligomers (detected by the A11 antibody) [40], the Reddy lab recently found significantly increased numbers of mutant Htt oligomers in the cortical tissues from postmortem brains of patients at stages HD3 and HD4, relative to control subjects. These proteins ranged from 15–50 kDa [21]. Our immunostaining analysis of mutant Htt mitochondrial marker, cytochrome oxidase 1 (COX1) revealed that mutant Htt oligomers are colocalized with COX1, suggesting that mutant Htt oligomers may promote mitochondrial toxicity, oxidative stress and neuronal damage in HD patients [22]. Very recently, we (22) and others [23] found mutant Htt interaction with the mitochondrial protein, Drp1 selectively in affected regions of the brain from the postmortem brains of HD patients and BACHD transgenic mice, and in peripheral cells from the brain tissues of HD patients (see upcoming section for details).
3. Cellular Changes in Huntington’s Disease
The selective, premature death of striatal projection neurons has been reported in HD patients and HD transgenic mice [1,2,13–15,62]. As shown in Fig. 2, several cellular pathways have been proposed and extensively investigated to explain premature neuronal death in HD progression, but this neuronal death is not well understood. The following cellular changes have been reported to involve HD pathogenesis: 1) transcriptional dysregulation [63–66], 2) expanded polyglutamine repeat proteins interacting with other CNS proteins [67], 3) caspase activation [68–73], 4) NMDAR activation [74–78], 5) calcium dyshomeostasis [28, 79–85], and 6) abnormal mitochondrial bioenergetics and axonal trafficking [33,86–89]. The most compelling evidence from these studies implicates abnormal mitochondrial bioenergetics and defective axonal transport in the selective synaptic degeneration and neuronal damage in HD.
Figure 2.
Cellular changes that have been implicated in Huntington’s disease pathogenesis.
4. Defective mitochondrial bioenergetics
Multiple lines of evidence suggest that abnormal mitochondrial bioenergetics is involved in HD progression: 1) The loss of body weight is a major factor in HD progression in HD patients and HD mouse models [90–93]. 2) Studies using magnetic resonance imaging (MRIs) of postmortem brains from HD patients revealed a progressive atrophy of the striatum, compared to brain images of age-matched control subjects [94–95]. Several other studies of HD brains utilizing MRIs found atrophy in the caudate nucleus, putamen, globus pallidus, and thalamus [96–98]. 3) Using positron emission tomography in functional studies of the brains of HD patients and control subjects, researchers found a marked decrease in the amount of glucose utilized in the striatum [99–103]. These same studies found decreased glucose metabolism to correlate with reduced performance of HD patients on several cognitive tasks, including immediate recall memory, verbal associative learning, and executive functions, suggesting that cerebral glucose metabolism is defective in HD patients [99–103]. 4) Biochemical studies of mitochondria in striatal neurons from brain tissues of late-stage HD patients revealed reduced activity of several components of oxidative phosphorylation, including complexes II, III, and IV of the electron transport chain [86–87]. In studies of HD transgenic and knockin mice, and in experimental HD rodent models, decreases in enzyme activities of complexes I, II, III, and IV were found in brain tissues [104], suggesting that mitochondria are somehow involved in HD pathogenesis. And 5) recent studies of HD knock-in striatal cells and lymphoblasts from HD patients revealed expanded polyglutamine repeats associated with low levels of mitochondrial ATP and decreased mitochondrial ADP-uptake, suggesting that HD mutation is associated with mitochondrial functional defects [105].
Overall, findings from these studies suggest that defective mitochondrial bioenergetics plays a large role in the progression and pathogenesis of HD.
5. Mutant Htt and Abnormal Mitochondrial Dynamics
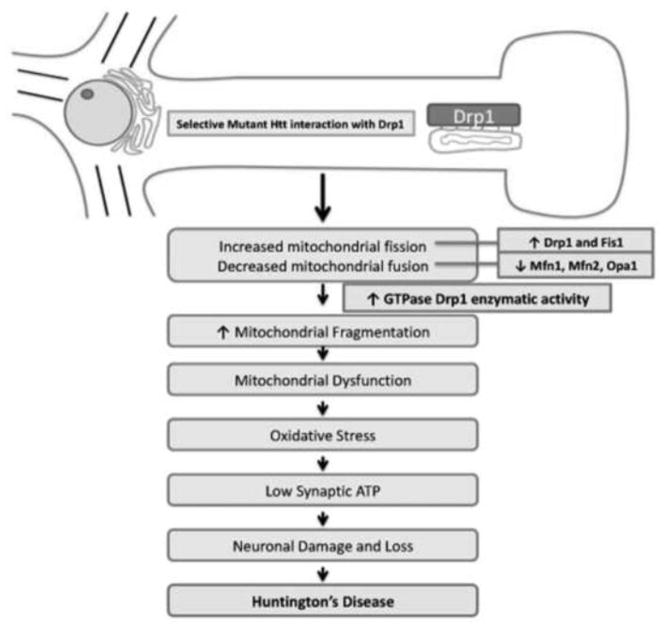
Mitochondrial shape and structure are maintained by two important but opposing forces: mitochondrial fission and mitochondrial fusion [106–110]. Mitochondrial fission is the division of single mitochondrion into two, and mitochondrial fusion is the integration of 2 mitochondria into single elongated mitochondrion. In a healthy neuron, fission and fusion balance equally. Mitochondria alter their shape and size to travel from the cell body to nerve terminals via anterograde movement, and back to the cell body via retrograde movement [19]. Mitochondrial fission and fusion are controlled by evolutionary conserved, large GTPases belonging to the family of dynamin. Mitochondrial fission is regulated by mitochondrial fission 1 (Fis1) and Drp1. Fis1 is localized to the outermembrane of mitochondria. Drp1 is localized in the cytoplasm, but a small part of Drp1 localized to the outermembrane, which promotes mitochondrial fragmentation [110]. Mitochondrial fusion is controlled by 3 GTPase proteins, 2 outermembrane proteins Mfn1 and Mfn2, and 1 innermembrane protein Opa1 [107]. The C-terminal part of Mfn1 mediates oligomerization among Mfn molecules of adjacent mitochondria and facilitates mitochondrial fusion.
Increasing evidence suggests that mitochondrial dynamics is imbalanced (increased fission and decreased fusion) in neurodegenerative diseases, including in AD [111–114], PD [110,115] and HD [20–23]. This imbalanced mitochondrial dynamics is caused by an altered expression of mitochondrial fission and fusion genes [110]. Further, in aged neurons, in neurons exposed to toxins, and in neurons that express mutant proteins, such as mutant Htt, an imbalance between fission and fusion leads to abnormalities in mitochondrial structure and function, and neuronal damage [110].
Several studies reported such abnormal mitochondrial dynamics in HD patients, HD mouse models, and cell lines that express mutant Htt [20–23]. Recently, the Reddy lab studied abnormal mitochondrial dynamics in tissues from postmortem brains of HD patients [21] and primary neurons and brain tissues from BACHD transgenic mice [22]. In a study of brain specimens from patients at HD3 and HD4 stages and from control subjects, we found increased expressions of Drp1 and Fis1 in HD4 than in HD3, and decreased levels of Mfn1, Mfn2, and Opa1 in HD4 than in HD3 [21] in the striatum and cortex (HD-affected brain regions), but not in the cerebellum (non-HD-affected brain region), indicating that abnormal mitochondrial dynamics may be related to HD [21]. Further, we also found significantly increased levels of cylophilin D (CypD) only in the striatum and cortex of the HD3 and HD4 patients relative to control subjects [21], again suggesting structurally damaged mitochondria in the HD-affected brain regions since increased CypD is known to damage mitochondrial structure.
Using recently developed BACHD mice that express the full-length (170 kb DNA) human Htt gene with 97 CAA and CAG (mixed repeats) [8], we studied mutant Htt and mitochondrial and synaptic genes. We found significantly increased mRNA levels of fission genes, Drp1 and Fis1 and matrix gene CypD, and decreased levels of fusion genes, Mfn1 and Mfn2 in 2-month-old BACHD mice relative to age-matched WT mice, suggesting that abnormal mitochondrial dynamics is an early event in HD progression [22].
Overall, findings from our lab together with earlier study by Kim et al [20] indicate a relationship between mutant Htt and impaired mitochondrial dynamics and dysfunction in HD.
5. Interaction between Mutant Htt and Drp1, and Elevated GTPase Drp1 Enzymatic Activity
To determine whether the interaction of Drp1 and mutant Htt increases as HD progresses, the Reddy lab performed co-immunoprecipitation analysis of Drp1 and mutant Htt, using a Drp1 antibody, and immunoblotting analysis, using the mutant Htt-specific antibody 1C2 and protein lysates of cortical tissues from control subjects and HD3 and HD4 patients and protein lysates from 2-month-old BACHD mice [22]. We found an 82 kDa and 40kDa mutant Htt protein in IP elutes from HD3 and HD4 patients. We also found Drp1 interaction with WT Htt, but lesser extent, in the control subjects, indicating that mutant Htt interaction with Drp1 is specific and related to disease progression.
Using cortical protein lysates from BACHD mice and WT mice, and Drp1 and 1C2 antibodies, we also performed co-IP analysis. Similar to HD brains, two bands of proteins, one with an 82 kDa and the other with 40 kDa mutant Htt proteins were found in the IP elutes of the BACHD mice, but not in those of the WT mice [22]. Overall, our lab findings together with results from Song et al [23], indicate that Drp1 interacts with mutant Htt and may participate in mitochondrial fragmentation and impaired mitochondrial biogenesis in HD neurons.
To determine whether Drp1 interaction with mutant Htt in affected and unaffected brain tissues from HD patients and BACHD mice enhance GTPase activity, we measured GTPase Drp1 enzymatic activity [22]. Interestingly, we found that significantly increased levels of Dp1 enzymatic activity in the cortex but not in the cerebellum of HD3 and HD4 patients relative to control subjects. We also found elevated levels of Drp1 enzymatic activity in the cerebral cortex and striatum in the BACHD mice relative to the WT mice [22]. Our findings are consistent with earlier findings from Song and colleagues who found that increased Dp1 enzymatic activity in HD neurons [23].
Overall, recent evidence indicates that Drp1 interacts with mutant Htt and enhances Drp1 enzymatic activity in HD-affected regions, leading to synaptic and neuronal damage (see Fig. 3 for summary). Further, we recently reported an increased interaction of Drp1 enzymatic activity, excessive mitochondrial fragmentation, and altered mitochondrial distribution in neurons affected by AD [113]. We also reported a similar finding in a study of HD, in which the interaction of mutant Htt with Drp1 had similar consequences and this abnormal interaction resulted in elevated Drp1 enzymatic activity, excessive mitochondrial fragmentation, and altered mitochondrial distribution in HD neurons [22]. These parallel findings for AD and HD suggest a common pathway that may be involved in triggering abnormal mitochondrial dynamics and selective neuronal damage.
Figure 3.
Schematic illustration showing mutant huntingtin interaction with mitochondrial protein, Drp1 and subsequent pathogenic changes in HD neuron.
6. Defective Axonal Transport of Mitochondria in HD neurons
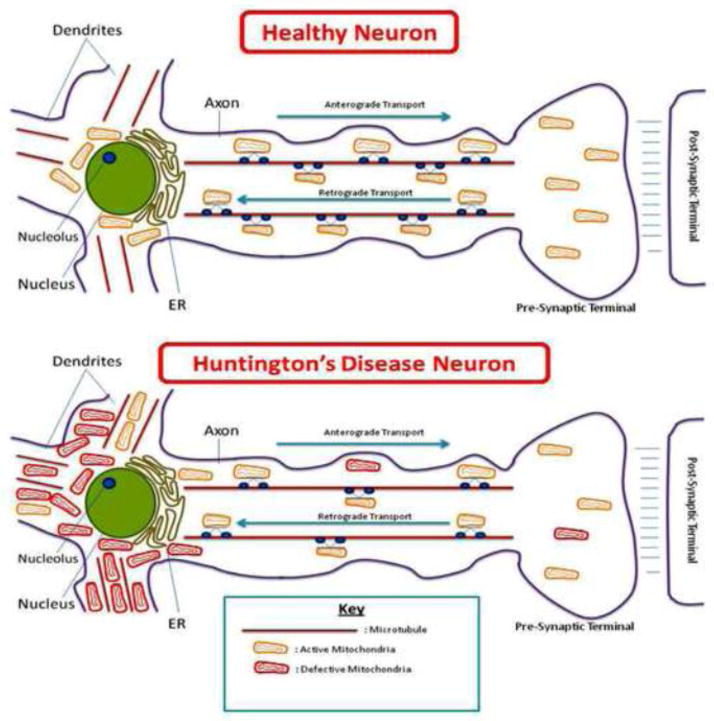
Normal axonal transport of organelles, including mitochondria, synaptic vesicles, and proteins, is essential for synaptic activities and neural communication. In diseased neurons, such as in HD neurons, axonal transport is impaired mostly due to mutant proteins that interact with proteins localized in axons. These abnormal interactions block the transport of organelles along the axons, ultimately leading to synaptic starvation. Studies of axonal transport reported impaired mitochondrial transport in cortical neurons overexpressed with mutant Htt [89], in HD striatal neurons [88], in rat cortical neurons, and in mouse striatal neurons transfected with N-terminal Htt [33].
Using live-cell imaging tools, primary neurons from BACHD mice and WT mice, and DsRed-mito transfections, we analyzed the mitochondrial motility in primary-neuron axonal projections from BACHD and WT mice [22]. We found significantly decreased motility in the primary neurons from BACHD mice (20.88 ± 4.86%) relative to WT neurons (with 36.73 ± 3.36%) (P = 0.015). We also found retrograde motility largely unaffected, and anterograde motility greatly affected (WT neurons 21.58±2.42 and BACHD neurons 10.04±1.78, P = 0.0009) [22].
Ours is the first study that investigated mitochondrial mass, trafficking, anterograde and retrograde movements and synaptic viability in primary neurons from BACHD mice. Our findings agree with a previous study [23], in which they studied mitochondrial transport in neurons transfected with exon 1 containing 17, 46, and/or 97 polyQ repeats. They found that neurons with exogenously expressed exon 1 Htt with 17 polyQ repeats exhibit filamentous, normal, and healthy mitochondria, whereas neurons expressing exon 1 Htt with 46 polyQ showed both elongated and round mitochondria. Neurons that exogenously expressed 97 polyQ repeats show mainly rounded, fragmented mitochondria. The findings from Song et al. [23] together with our observations of mitochondrial trafficking in BACHD mice, indicate that mutant Htt with expanded polyQ repeats are responsible for mitochondrial fragmentation in neurons affected by HD.
These findings also indicate that mutant Htt interaction with Drp1 may cause excessive mitochondrial fragmentation, leading to defective axonal transport and abnormal distribution of mitochondria in BACHD mice. Figure 4 summarizes data supporting excessive mitochondrial fragmentation and defective axonal transport of mitochondria.
Figure 4.
Huntington’s disease neuron showing excessive fragmentation of mitochondria, accumulation in cell soma, and abnormal distribution of mitochondria in neuronal processes and synapses.
7. Synaptic Damage in Huntington’s disease
Synaptic damage has been extensively reported in neurodegenerative diseases, including Alzheimer’s [111–114,116,117] Parkinson’s [118–122] and Huntington’s [22–23,123–127]. However, the precise factors that cause synaptic degeneration are not completely understood. Recent evidence from our lab [21,22] and others [20,23] revealed that abnormal interaction of mutant Htt with mitochondrial protein Drp1, cause excessive mitochondrial fragmentation that leads to defective axonal transport and abnormal mitochondrial distribution, particularly in neurites and synapse. Synapses are the sites of high ATP demand, and reduced number of mitochondria neurites and synapses produce low ATP and cause synaptic degeneration in HD neurons. These events are summarized above, and we now focus on synaptic damage and degeneration in HD neurons.
To determine the effect of mutant Htt on synapses in HD, using cortical tissues from BACHD mice and quantitative real-time RT-PCR, the Reddy lab measured mRNA levels of presynaptic protein, synaptophysin and postsynaptic protein, PSD95 [22]. They found a significant reduction in the mRNA fold changes for the synaptic genes, synaptophysin (-1.2 fold) and PSD 95 (-1.3 fold) relative to WT mice.
Further, to determine the effects of mutant Htt on synaptophysin and MAP2 levels, they also performed immunostaining analysis of BACHD neurons using 10 DIV neurons from BACHD mice and WT mice. They found significantly decreased immunoreactivity of synaptophysin in the BACHD neurons relative to WT neurons [22]. Similar to synaptophysin, immunoreactivity of MAP2 was significantly decreased in the BACHD neurons relative to WT neurons, indicating that mutant Htt may be involved in synaptic degeneration [22].
Overall, our recent studies of mitochondrial transport, mitochondrial dynamics, expression of synaptic genes in BACHD mice together with earlier studies, indicate that synaptic damage is an early event in disease process and may be linked to mutant Htt and abnormal mitochondrial distribution at synapses in HD.
8. Therapeutic Strategies for HD
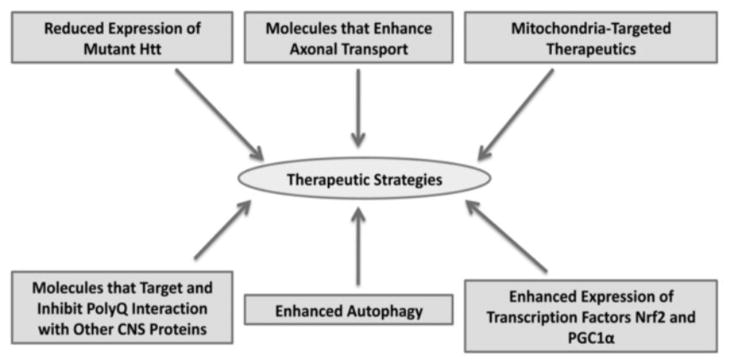
Based on cellular changes observed in HD pathogenesis and progression, multiple therapeutic strategies have been developed and tested in experimental cell, animal models, and even clinical trials, to stop or delay disease progression. As shown in Fig. 5, these strategies include: reducing mutant Htt allele expression (RNA silencing), inhibit oligomer formation, inhibiting the interaction of polyQ with other proteins in the central nervous system, enhancing the expression of endogenous transcription factors, enhancing autophagy, increasing molecules that enhance axonal transport, reducing abnormal mitochondrial dynamics, and increasing healthy mitochondrial biogenesis.
Figure 5.
Possible therapeutic approaches for Huntington’s disease patients.
8.1. Reducing mutant Htt allele expression
Extensive research reveled that soluble mutant Htt aggregates and oligomers are toxic, cause synaptic degeneration and neuronal damage in HD. Therefore, reducing the expression of mutant Htt allele is a direct and upstream approach to treat HD. This approach involves silencing the post-transcriptional mutant Htt allele using small, anti-sense RNA molecules. Currently, RNA silencing (shRNA) technology is being used to reduce mutant Htt allele [128–136]. RNA silencing is tested in cell and mouse models of HD [137–139].
Further, delivery of short hainpin RNA using adeno-associated viral (AAV) vectors is successful in HD truncated mice (N171-82Q). Mice that received shRNA showed decreased mutant Htt aggregates and improved rotorod performance and gait [140]. This approach has been tested in BACHD mice and other mouse models. Further research is still needed to target selectively mutant Htt allele in the presence of both WT and mutant Htt alleles in HD neurons.
8.2. Inhibition of mutant oligomer formation
Several researchers around the world are using cell and animal models, high throughput screening, and inhibitors of oligomers and apoptotic cell death to determine molecules that reduce and/or prevent mutant Htt oligomer formation in HD neurons [35,36,53–61].
8.3. Inhibiting the interaction between mutant Htt and mitochondrial protein, Drp1
As described above, polyQ protein is sticky and likely to interact with other CNS proteins [67] and participate in oligomers, fibrils, protofibrils and ultimately fibrillogenesis formation [35–39]. The soluble mutant Htt aggregates are toxic and interact with axonal proteins, including Drp1, impair axonal transport and cause synaptic degeneration. Regarding mutant soluble Htt aggregates and their interaction with Drp1, further research is still needed in order to determine which domain of Drp1 protein interacts with mutant Htt aggregates and cause mitochondrial fragmentation, and neuronal damage. Therefore, identifying the molecules that inhibit abnormal interactions between mutant Htt and Drp1 is an interesting therapeutic approach.
8.4. Enhancing endogenous transcription factors (PGC1α and Nrf2)
Increasing evidence suggests that transcription factors, including PGC1α and Nrf2, reduce mitochondrial oxidative damage and increase neuronal survival, in general, and HD neurons, in particular [141–143]. It is critical to increase endogenous levels of PGC1α and Nrf2 in neurons affected by HD. Efforts are underway to enhance endogenous transcription factors that protect neurons against mutant Htt aggregates and oligomers and other oxidative insults.
8.5. Inactivating mTOR and enhancing autophagy
Growing evidence suggests that autophagy plays a large role in clearing a cell’s degraded proteins and organelles in HD neurons. Further extensive literature on mammalian target of rapamycin (mTOR) suggests that cells pre-treated with rapamycin are protected against apoptotic cell death, particularly cells that express mutant Htt [144–151]. Rapamycin, a lipophilic, macrolide antibiotic, induces autophagy by inactivating the protein mTOR. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of HD [152]. Therefore, inactivation of mTOR is another approach to treat patients with HD. Further research is still needed to understand the role of autophagy, in HD.
8.6. Developing molecules capable of enhancing axonal transport
To improve axonal transport of mitochondria and other organelles, molecules could be developed to enhance microtubule binding capacity of ‘microtubule associated proteins with microtubules’ and increase both anterograde and retrograde movements of organelles. The tight microtubule binding of microtubule associated proteins may enhance axonal transport of organelles may provide and supply necessary components and organelles to synapses and increase synaptic outgrowth and neural communications.
8.7. Reduction of abnormal mitochondrial dynamics and increase of healthy mitochondrial biogenesis
As discussed above, abnormal mitochondrial dynamics is an emerging and key component in neuronal damage in HD [20–23,109–110]. Maintaining the balance of mitochondrial dynamics is an important therapeutic strategy to protect neurons against the toxicity of mutant Htt oligomers and oxidative insults [21]. It is equally important to maintain healthy mitochondrial biogenesis in HD neurons. Recent research using primary neurons from AD transgenic mice and mitochondria-targeted antioxidant, SS1 revealed that SS31 appears increase the numbers of healthy mitochondria and also anterograde movement of mitochondria in primary neurons treated with SS31 [114].
9. The Current Status of Experimental Therapeutics in HD
Recent studies suggest that mitochondrial dysfunction and oxidative stress are key players in HD progression and pathogenesis. To reduce mitochondrial toxicity and abnormal mitochondrial dynamics in HD neurons, we need to develop drugs that protect mitochondria.
In the last decade, several drugs have been tested in experimental animal models of HD and also clinical trials. Several mitochondrial drugs that act to protect neurons from mitochondrial damage, including Creatine, CoQ10 and resveratrol – have shown beneficial effects in cell and animal models. Creatine and CoQ10 have shown deceased HD pathology and rotorod performance in R6/2 and N171-82Q lines of HD mice. Findings from these studies suggest that Creatine and CoQ10 boost ATP levels and increase mitochondrial function [153–156]. Further, resveratrol also has shown beneficial effects in the worm model of HD, and increased survival of striatal neurons from knockin mice indicating that resveratrol protect against age-dependent mutant Htt toxicity in HD neurons [156].
Dimebon (or Dimebolin hydrochloride or atreperdine) is an antihistamine drug that has been used clinically in Russia to reduce cognitive deficits in AD patients [157]. Based on cell culture and murine models of AD, phase III clinical trial was conducted in US and initial findings were positive. However, a large-scale clinical trials did not show significant improved positive clinical symptoms in patients with AD. Scientists from Medication, Inc., in collaboration with clinicians and researchers in North America, conducted phase III clinical trials in HD patients, and the outcome was not positive [158–159]. These disappointing outcomes posing major challenge to HD patients and researchers.
10. Conclusions and Future Directions
Mitochondrial dysfunction and oxidative stress are critical factors in the development and progression of HD. Recent research revealed that mutant Htt aggregates and oligomers appear to interact with Drp1, to enhance GTPase Drp1 enzymatic activity, to increase mitochondrial fragmentation, and to cause abnormal mitochondrial distribution, ultimately leading to neuronal damage. These findings suggest that abnormal mitochondrial dynamics and defective axonal transport of mitochondria are mechanisms in HD pathogenesis.
In terms of therapeutic approaches, multiple approaches appear to be promising, including silencing of mutant Htt allele, reducing oligomer formation, enhancing endogenous levels of transcription factors (PGC1α and Nrf2) and balancing mitochondrial dynamics and axonal transport in neurons affected by HD. Recent experimental therapeutics in cell and animal models are promising but no definitive drugs/agents are available to treat HD patients. Further research is also needed to better understand the mechanisms how mutant Htt aggregates and oligomers cause mitochondrial fragmentation and impair axonal transport in HD neurons. Further, it is worth investigating and developing the molecules that promote healthy mitochondrial biogenesis, and increase axonal transport in neurons affected by HD.
Highlights.
Summarized the link between mutant Htt and mitochondrial dynamics in Huntington’s disease.
Discussed mutant Htt interaction with Drp1 in Huntington’s disease neurons.
Highlighted possible factors of defective axonal transport in Huntington’s disease neurons.
Discussed the therapeutic approaches for Huntington’s disease.
Acknowledgments
This research was supported by NIH grants AG028072, RR00163, and Alzheimer Association grant IIRG-09-92429.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP. Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Folstein SE. Huntington’s Disease. Johns Hopkins University Press; 1990. [Google Scholar]
- 3.Montoya A, Price BH, Menear M, Lepage M. Brain imaging and cognitive dysfunctions in Huntington’s disease. J Psychiatry Neurosci. 2006;31:21–29. [PMC free article] [PubMed] [Google Scholar]
- 4.Ross CA, Tabrizi SJ. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 5.Byers RK, Gilles FH, Fung C. Huntington’s disease in children: Neuropathologic study of four cases. Neurology. 1973;23:561–569. doi: 10.1212/wnl.23.6.561. [DOI] [PubMed] [Google Scholar]
- 6.Spargo E, Everall IP, Lantos PL. Neuronal loss in the hippocampus in Huntington’s disease: a comparison with HIV infection. J Neurol Neurosurg Psychiatry. 1993;56:487–491. doi: 10.1136/jnnp.56.5.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Politis M, Pavese N, Tai YF, Tabrizi SJ, Barker RA, Piccini P. Hypothalamic involvement in Huntington’s disease: an in vivo PET study. Brain. 2008;131:2860–2869. doi: 10.1093/brain/awn244. [DOI] [PubMed] [Google Scholar]
- 8.Soneson C, Fontes M, Zhou Y, Denisov V, Paulsen JS, Kirik D, Petersén A Huntington Study Group PREDICT-HD investigators. Early changes in the hypothalamic region in prodromal Huntington disease revealed by MRI analysis. Neurobiol Dis. 2010;40:531–543. doi: 10.1016/j.nbd.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray M, Shirasaki DI, Cepeda C, André VM, Wilburn B, Lu XH, Tao J, Yamazaki I, Li SH, Sun YE, Li XJ, Levine MS, Yang XW. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 11.DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 12.Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 13.Reddy PH, Williams M, Charles V, Garrett L, Pike-Buchanan L, Whetsell WO, Jr, Miller G, Tagle DA. Behavioural abnormalities and selective neuronal loss in HD transgenic mice expressing mutated full-length HD cDNA. Nat Genet. 1998;20:198–202. doi: 10.1038/2510. [DOI] [PubMed] [Google Scholar]
- 14.Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, Kotzuk JA, Slunt HH, Ratovitski T, Cooper JK, Jenkins NA, Copeland NG, Price DL, Ross CA, Borchelt DR. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;(8):397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 15.Hodgson JG, Agopyan N, Gutekunst CA, Leavitt BR, LePiane F, Singaraja R, Smith DJ, Bissada N, McCutcheon K, Nasir J, Jamot L, Li XJ, Stevens ME, Rosemond E, Roder JC, Phillips AG, Rubin EM, Hersch SM, Hayden MR. A YAC mouse model for Huntington’s disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron. 1999;23:181–192. doi: 10.1016/s0896-6273(00)80764-3. [DOI] [PubMed] [Google Scholar]
- 16.Levine MS, Klapstein GJ, Koppel A, Gruen E, Cepeda C, Vargas ME, Jokel ES, Carpenter EM, Zanjani H, Hurst RS, Efstratiadis A, Zeitlin S, Chesselet MF. Enhanced sensitivity to N-methyl-D-aspartate receptor activation in transgenic and knock-in mouse models of Huntington’s disease. J Neurosci Res. 1999;58:515–532. [PubMed] [Google Scholar]
- 17.Wheeler VC, Auerbach W, White JK, Srinidhi J, Auerbach A, Ryan A, Duyao MP, Vrbanac V, Weaver M, Gusella JF, Joyner AL, MacDonald ME. Length-dependent gametic CAG repeat instability in the Huntington’s disease knock-in mouse. Hum Mol Genet. 1999;8:115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 19.Reddy PH, Mao P, Manczak M. Mitochondrial structural and functional dynamics in Huntington’s disease. Brain Res Rev. 2009;61:33–48. doi: 10.1016/j.brainresrev.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, Moody JP, Edgerly CK, Bordiuk OL, Cormier K, Smith K, Beal MF, Ferrante RJ. Mitochondrial loss, dysfunction and altered dynamics in Huntington’s disease. Hum Mol Genet. 2010;19:3919–3935. doi: 10.1093/hmg/ddq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirendeb U, Reddy AP, Manczak M, Calkins MJ, Mao P, Tagle DA, Reddy PH. Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington’s disease: implications for selective neuronal damage. Hum Mol Genet. 2011;20:1438–1455. doi: 10.1093/hmg/ddr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirendeb UP, Calkins MJ, Manczak M, Anekonda V, Dufour B, McBride JL, Mao P, Reddy PH. Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum Mol Genet. 2011 Oct 13; doi: 10.1093/hmg/ddr475. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song W, Chen J, Petrilli A, Liot G, Klinglmayr E, Zhou Y, Poquiz P, Tjong J, Pouladi MA, Hayden MR, Masliah E, Ellisman M, Rouiller I, Schwarzenbacher R, Bossy B, Perkins G, Bossy-Wetzel E. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddy PH, Williams M, Tagle DA. Recent advances in understanding the pathogenesis of Huntington’s disease. Trends Neurosci. 1999;22:248–255. doi: 10.1016/s0166-2236(99)01415-0. [DOI] [PubMed] [Google Scholar]
- 25.The Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 26.Kegel KB, Meloni AR, Yi Y, Kim YJ, Doyle E, Cuiffo BG, Sapp E, Wang Y, Qin ZH, Chen JD, Nevins JR, Aronin N, DiFiglia M. Huntingtin is present in the nucleus, interacts with the transcriptional corepressor Cterminal binding protein, and represses transcription. J Biol Chem. 2002;277:7466–7476. doi: 10.1074/jbc.M103946200. [DOI] [PubMed] [Google Scholar]
- 27.Kegel KB, Sapp E, Yoder J, Cuiffo B, Sobin L, Kim YJ, Qin ZH, Hayden MR, Aronin N, Scott DL, Isenberg G, Goldmann WH, DiFiglia M. Huntingtin associates with acidic phospholipids at the plasma membrane. J Biol Chem. 2005;280:36464–36473. doi: 10.1074/jbc.M503672200. [DOI] [PubMed] [Google Scholar]
- 28.Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, Greenamyre JT. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 29.Choo YS, Johnson GV, MacDonald M, Detloff PJ, Lesort M. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum Mol Genet. 2004;13:1407–1420. doi: 10.1093/hmg/ddh162. [DOI] [PubMed] [Google Scholar]
- 30.Truant R, Atwal R, Burtnik A. Hypothesis: Huntingtin may function in membrane association and vesicular trafficking. Biochem Cell Biol. 2006;84:912–917. doi: 10.1139/o06-181. [DOI] [PubMed] [Google Scholar]
- 31.Strehlow AN, Li JZ, Myers RM. Wild-type huntingtin participates in protein trafficking between the Golgi and the extracellular space. Hum Mol Genet. 2007;16:391–409. doi: 10.1093/hmg/ddl467. [DOI] [PubMed] [Google Scholar]
- 32.Atwal RS, Xia J, Pinchev D, Taylor J, Epand RM, Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 33.Orr AL, Li S, Wang CE, Li H, Wang J, Rong J, Xu X, Mastroberardino PG, Greenamyre JT, Li XJ. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci. 2008;28:2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 35.Wacker JL, Zareie MH, Fong H, Sarikaya M, Muchowski PJ. Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat Struct Mol Biol. 2004;11:1215–1222. doi: 10.1038/nsmb860. [DOI] [PubMed] [Google Scholar]
- 36.Poirier MA, Li H, Macosko J, Cai S, Amzel M, Ross CA. Huntingtin spheroids and protofibrils as precursors in polyglutamine fibrilization. J Biol Chem. 2002;277:41032–41037. doi: 10.1074/jbc.M205809200. [DOI] [PubMed] [Google Scholar]
- 37.Thakur AK, Jayaraman M, Mishra R, Thakur M, Chellgren VM, Byeon IJ, Anjum DH, Kodali R, Creamer TP, Conway JF, Gronenborn AM, Wetzel R. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat Struct Mol Biol. 2009;16:380–389. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Legleiter J, Lotz GP, Miller J, Ko J, Ng C, Williams GL, Finkbeiner S, Patterson PH, Muchowski PJ. Monoclonal antibodies recognize distinct conformational epitopes formed by polyglutamine in a mutant huntingtin fragment. J Biol Chem. 2009;284:21647–21658. doi: 10.1074/jbc.M109.016923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sathasivam K, Lane A, Legleiter J, Warley A, Woodman B, Finkbeiner S, Paganetti P, Muchowski PJ, Wilson S, Bates GP. Identical oligomeric and fibrillary structures captured from the brains of R6/2 and knock-in mouse models of Huntington’s disease. Hum Mol Genet. 2010;19:65–78. doi: 10.1093/hmg/ddp467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 41.Mezler M, Barghorn S, Schoemaker H, Gross G, Nimmrich V. Aβ oligomer directly modulates P/Q-type calcium currents in Xenopus oocytes. Br J Pharmacol. 2011 Aug 30; doi: 10.1111/j.1476–5381.2011.01646.x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veloso AJ, Yoshikawa H, Cheng XR, Tamiya E, Kerman K. Optical trapping for the characterization of amyloid-beta aggregation kinetics. Analyst. 2011 Aug 26; doi: 10.1039/c1an15480j. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 43.Hou L, Liu Y, Wang X, Ma H, He J, Zhang Y, Yu C, Guan W, Ma Y. The effects of amyloid-β(42) oligomer on the proliferation and activation of astrocytes in vitro. In Vitro Cell Dev Biol Anim. 2011 Aug 20; doi: 10.1007/s11626-011-9439-y. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 44.Klaver AC, Coffey MP, Smith LM, Bennett DA, Finke JM, Dang L, Loeffler DA. ELISA measurement of specific non-antigen-bound antibodies to Aβ1-42 monomer and soluble oligomers in sera from Alzheimer’s disease, mild cognitively impaired, and noncognitively impaired subjects. J Neuroinflammation. 2011;8:93. doi: 10.1186/1742-2094-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manczak M, Calkins MJ, Reddy PH. Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer’s disease: implications for neuronal damage. Hum Mol Genet. 2011;20:2495–2509. doi: 10.1093/hmg/ddr139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruggink KA, Kuiperij HB, Verbeek MM, El-Agnaf OM, Tokuda T. Detection of elevated levels of {alpha}-synuclein oligomers in CSF from patients with Parkinson disease. Neurology. 2011;77:510–511. doi: 10.1212/WNL.0b013e318219dd92. [DOI] [PubMed] [Google Scholar]
- 47.Kim YM, Jang WH, Quezado MM, Oh Y, Chung KC, Junn E, Mouradian MM. Proteasome inhibition induces α-synuclein SUMOylation and aggregate formation. J Neurol Sci. 2011;307:57–161. doi: 10.1016/j.jns.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gadad BS, Britton GB, Rao KS. Targeting oligomers in neurodegenerative disorders: lessons from α-synuclein, tau, and amyloid-β peptide. J Alzheimers Dis. 2011;24(Suppl 2):223–232. doi: 10.3233/JAD-2011-110182. [DOI] [PubMed] [Google Scholar]
- 49.Caruana M, Högen T, Levin J, Hillmer A, Giese A, Vassallo N. Inhibition and disaggregation of α-synuclein oligomers by natural polyphenolic compounds. FEBS Lett. 2011;585:1113–1120. doi: 10.1016/j.febslet.2011.03.046. [DOI] [PubMed] [Google Scholar]
- 50.Hüls S, Högen T, Vassallo N, Danzer KM, Hengerer B, Giese A, Herms J. AMPA-receptor-mediated excitatory synaptic transmission is enhanced by iron-induced α-synuclein oligomers. J Neurochem. 2011;117:868–878. doi: 10.1111/j.1471-4159.2011.07254.x. [DOI] [PubMed] [Google Scholar]
- 51.Lee HJ, Baek SM, Ho DH, Suk JE, Cho ED, Lee SJ. Dopamine promotes formation and secretion of non-fibrillar alpha-synuclein oligomers. Exp Mol Med. 2011;43:216–222. doi: 10.3858/emm.2011.43.4.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giehm L, Svergun DI, Otzen DE, Vestergaard B. Low-resolution structure of a vesicle disrupting α-synuclein oligomer that accumulates during fibrillation. Proc Natl Acad Sci U S A. 2011;108:3246–3251. doi: 10.1073/pnas.1013225108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Behrends C, Langer CA, Boteva R, Böttcher UM, Stemp MJ, Schaffar G, Rao BV, Giese A, Kretzschmar H, Siegers K, Hartl FU. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell. 2006;23:887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 54.Ellisdon AM, Thomas B, Bottomley SP. The two-stage pathway of ataxin-3 fibrillogenesis involves a polyglutamine-independent step. J Biol Chem. 2006;281:16888–16896. doi: 10.1074/jbc.M601470200. [DOI] [PubMed] [Google Scholar]
- 55.Kitamura A, Kubota H, Pack CG, Matsumoto G, Hirayama S, Takahashi Y, Kimura H, Kinjo M, Morimoto RI, Nagata K. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol. 2006;8:1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- 56.Nagai Y, Inui T, Popiel HA, Fujikake N, Hasegawa K, Urade Y, Goto Y, Naiki H, Toda T. A toxic monomeric conformer of the polyglutamine protein. Nat Struct Mol Biol. 2007;14:332–340. doi: 10.1038/nsmb1215. [DOI] [PubMed] [Google Scholar]
- 57.Sánchez I, Mahlke C, Yuan J. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature. 2003;421:373–379. doi: 10.1038/nature01301. [DOI] [PubMed] [Google Scholar]
- 58.Schaffar G, Breuer P, Boteva R, Behrends C, Tzvetkov N, Strippel N, Sakahira H, Siegers K, Hayer-Hartl M, Hartl FU. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol Cell. 2004;15:95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 59.Takahashi Y, Okamoto Y, Popiel HA, Fujikake N, Toda T, Kinjo M, Nagai Y. Detection of polyglutamine protein oligomers in cells by fluorescence correlation spectroscopy. J Biol Chem. 2007;282:24039–24048. doi: 10.1074/jbc.M704789200. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka M, Machida Y, Nishikawa Y, Akagi T, Hashikawa T, Fujisawa T, Nukina N. Expansion of polyglutamine induces the formation of quasi-aggregate in the early stage of protein fibrillization. J Biol Chem. 2003;278:34717–34724. doi: 10.1074/jbc.M209852200. [DOI] [PubMed] [Google Scholar]
- 61.Lajoie P, Snapp EL. Formation and toxicity of soluble polyglutamine oligomers in living cells. PLoS One. 2010;5:e15245. doi: 10.1371/journal.pone.0015245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Raamsdonk JM, Metzler M, Slow E, Pearson J, Schwab C, Carroll J, Graham RK, Leavitt BR, Hayden MR. Phenotypic abnormalities in the YAC128 mouse model of Huntington disease are penetrant on multiple genetic backgrounds and modulated by strain. Neurobiol Dis. 2007;26:189–200. doi: 10.1016/j.nbd.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 63.Huang CC, Faber PW, Persichetti F, Mittal V, Vonsattel JP, MacDonald ME, Gusella JF. Amyloid formation by mutant huntingtin: threshold, progressivity and recruitment of normal polyglutamine proteins. Somat Cell Mol Genet. 1998;24:217–233. doi: 10.1023/b:scam.0000007124.19463.e5. [DOI] [PubMed] [Google Scholar]
- 64.Perez MK, Paulson HL, Pendse SJ, Saionz, Bonini NM, Pittman RN. Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation. J Cell Biol. 1998;143:1457–1470. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shimohata T, Nakajima T, Yamada M, Uchida C, Onodera O, Naruse S, Kimura T, Koide R, Nozaki K, Sano Y, Ishiguro H, Sakoe K, Ooshima T, Sato A, Ikeuchi T, Oyake M, Sato T, Aoyagi Y, Hozumi I, Nagatsu T, Takiyama Y, Nishizawa M, Goto J, Kanazawa I, Davidson I, Tanese N, Takahashi H, Tsuji S. Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat Genet. 2000;26:29–36. doi: 10.1038/79139. [DOI] [PubMed] [Google Scholar]
- 66.Sayer JA, Manczak M, Akileswaran L, Reddy PH, Coghlan VM. Interaction of the nuclear matrix protein NAKAP with HypA and huntingtin: implications for nuclear toxicity in Huntington’s disease pathogenesis. Neuromolecular Med. 2005;7:297–310. doi: 10.1385/NMM:7:4:297. [DOI] [PubMed] [Google Scholar]
- 67.Borrell-Pagès M, Zala D, Humbert S, Saudou F. Huntington’s disease: from huntingtin function and dysfunction to therapeutic strategies. Cell Mol Life Sci. 2006;63:2642–2660. doi: 10.1007/s00018-006-6242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rigamonti D, Bauer JH, De-Fraja C, Conti L, Sipione S, Sciorati C, Clementi E, Hackam A, Hayden MR, Li Y, Cooper, Ross CA, Govoni S, Vincenz C, Cattaneo CE. J Neurosci. 2000;20:3705–3713. doi: 10.1523/JNEUROSCI.20-10-03705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jana NR, Zemskov EA, Wang GH, Nukina N. Altered proteasomal function due to the expression of polyglutamine-expanded truncated N-terminal huntingtin induces apoptosis by caspase activation through mitochondrial cytochrome c release. Hum Mol Genet. 2001;10:1049–1059. doi: 10.1093/hmg/10.10.1049. [DOI] [PubMed] [Google Scholar]
- 70.Hermel E, Gafni J, Propp SS, Leavitt BR, Wellington CL, Young JE, Hackam AS, Logvinova AV, Peel AL, Chen SF, Hook V, Singaraja R, Krajewski S, Goldsmith PC, Ellerby HM, Hayden MR, Bredesen DE, Ellerby LM. Specific caspase interactions and amplification are involved in selective neuronal vulnerability in Huntington’s disease. Cell Death Differ. 2004;11:424–438. doi: 10.1038/sj.cdd.4401358. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y, Engelman J, Friedlander RM. Allele-specific silencing of mutant Huntington’s disease gene. J Neurochem. 2009;108:82–90. doi: 10.1111/j.1471-4159.2008.05734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Majumder P, Raychaudhuri S, Chattopadhyay B, Bhattacharyya NP. Increased caspase-2, calpain activations and decreased mitochondrial complex II activity in cells expressing exogenous huntingtin exon 1 containing CAG repeat in the pathogenic range. Cell Mol Neurobiol. 2007;27:1127–1145. doi: 10.1007/s10571-007-9220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Warby SC, Doty CN, Graham RK, Carroll JB, Yang YZ, Singaraja RR, Overall CM, Hayden MR. Hum Mol Genet. 2008;17:2390–2404. doi: 10.1093/hmg/ddn139. [DOI] [PubMed] [Google Scholar]
- 74.Sun Y, Savanenin A, Reddy PH, Liu YF. Polyglutamine-expanded huntingtin promotes sensitization of N-methyl-D-aspartate receptors via post-synaptic density 95. J Biol Chem. 2001;276:24713–24718. doi: 10.1074/jbc.M103501200. [DOI] [PubMed] [Google Scholar]
- 75.Laforet GA, Sapp E, Chase K, McIntyre C, Boyce FM, Campbell M, Cadigan BA, Warzecki L, Tagle DA, Reddy PH, Cepeda C, Calvert CR, Jokel ES, Klapstein GJ, Ariano MA, Levine MS, DiFiglia M, Aronin N. Changes in cortical and striatal neurons predict behavioral and electrophysiological abnormalities in a transgenic murine model of Huntington’s disease. J Neurosci. 2001;21:9112–9123. doi: 10.1523/JNEUROSCI.21-23-09112.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Centonze D, Gubellini P, Picconi B, Saulle E, Tolu M, Bonsi P, Giacomini P, Calabresi P. An abnormal striatal synaptic plasticity may account for the selective neuronal vulnerability in Huntington’s disease. Neurol Sci. 2001;22:61–62. doi: 10.1007/s100720170047. [DOI] [PubMed] [Google Scholar]
- 77.Lee WT, Chang C. Magnetic resonance imaging and spectroscopy in assessing 3-nitropropionic acid-induced brain lesions: an animal model of Huntington’s disease. Prog Neurobiol. 2004;(72):87–110. doi: 10.1016/j.pneurobio.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 78.Fernandes HB, Baimbridge KG, Church J, Hayden MR, Raymond LA. Mitochondrial sensitivity and altered calcium handling underlie enhanced NMDA induced apoptosis in YAC128 model of Huntington’s disease. J Neurosci. 2007;27:13614–13623. doi: 10.1523/JNEUROSCI.3455-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Panov AV, Lund S, Greenamyre JT. Ca2+-induced permeability transition in human lymphoblastoid cell mitochondria from normal and Huntington’s disease individuals. Mol Cell Biochem. 2005;269:143–152. doi: 10.1007/s11010-005-3454-9. [DOI] [PubMed] [Google Scholar]
- 80.Oliveira JM, Chen S, Almeida S, Riley R, Gonçalves J, Oliveira CR, Hayden MR, Nicholls DG, Ellerby LM, Rego AC. Mitochondrial-dependent Ca2+ handling in Huntington’s disease striatal cells: effect of histone deacetylase inhibitors. J Neurosci. 2006;26:11174–11186. doi: 10.1523/JNEUROSCI.3004-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Milakovic T, Quintanilla RA, Johnson GV. Mutant huntingtin expression induces mitochondrial calcium handling defects in clonal striatal cells: functional consequences. J Biol Chem. 2006;281:34785–34795. doi: 10.1074/jbc.M603845200. [DOI] [PubMed] [Google Scholar]
- 82.Rockabrand E, Slepko N, Pantalone A, Nukala VN, Kazantsev A, Marsh JL, Sullivan PG, Steffan JS, Sensi SL, Thompson LM. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum Mol Genet. 2007;16:61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- 83.Gellerich FN, Gizatullina Z, Nguyen HP, Trumbeckaite S, Vielhaber S, Seppet E, Zierz S, Landwehrmeyer B, Riess O, von Hörsten S, Striggow F. Impaired regulation of brain mitochondria by extramitochondrial Ca2+ in transgenic Huntington disease rats. J Biol Chem. 2008;283:30715–30724. doi: 10.1074/jbc.M709555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lim D, Fedrizzi L, Tartari M, Zuccato C, Cattaneo E, Brini M, Carafoli E. Calcium homeostasis and mitochondrial dysfunction in striatal neurons of Huntington disease. J Biol Chem. 2008;283:5780–5789. doi: 10.1074/jbc.M704704200. [DOI] [PubMed] [Google Scholar]
- 85.Oliveira JM, Gonçalves J. In situ mitochondrial Ca2+ buffering differences of intact neurons and astrocytes from cortex and striatum. J Biol Chem. 2009;284:5010–5020. doi: 10.1074/jbc.M807459200. [DOI] [PubMed] [Google Scholar]
- 86.Browne SE, Bowling AC, MacGarvey U, Baik MJ, Berger SC, Muqit MM, Bird ED, Beal MF. Oxidative damage and metabolic dysfunction in Huntington’s disease: selective vulnerability of the basal ganglia. Ann Neurol. 1997;41:646–653. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- 87.Tabrizin SJ, Cleeter MW, Xuereb J, Taanman JW, Cooper JM, Schapira AH. Biochemical abnormalities and excitotoxicity in Huntington’s disease brain. Ann Neurol. 1999;45:25–32. doi: 10.1002/1531-8249(199901)45:1<25::aid-art6>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 88.Trushina E, Dyer RB, Badger JD, Ure D, Eide L, Tran DD, Vrieze BT, Legendre-Guillemin V, McPherson PS, Mandavilli BS, Van Houten B, Zeitlin S, McNiven M, Aebersold R, Hayden M, Parisi JE, Seeberg E, Dragatsis I, DoylE K, Bender A, Chacko C, McMurray CT. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24:8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chang DT, Rintoul GL, Pandipati S, Reynolds IJ. Mutant huntingtin aggregates impair mitochondrial movement and trafficking in cortical neurons. Neurobiol Dis. 2006;22:388–400. doi: 10.1016/j.nbd.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 90.Kirkwood SC, Su JL, Conneally P, Foroud T. Progression of symptoms in the early and middle stages of Huntington disease. Arch Neurol. 2001;58:273–278. doi: 10.1001/archneur.58.2.273. [DOI] [PubMed] [Google Scholar]
- 91.Mahant N, McCusker EA, Byth K, Graham S Huntington Study Group, Huntington’s disease: clinical correlates of disability and progression. Neurology. 2003;61:1085–1092. doi: 10.1212/01.wnl.0000086373.32347.16. [DOI] [PubMed] [Google Scholar]
- 92.Phan J, Hickey MA, Zhang P, Chesselet MF, Reue K. Adipose tissue dysfunction tracks disease progression in two Huntington’s disease mouse models. Hum Mol Genet. 2009;18:1006–1016. doi: 10.1093/hmg/ddn428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Aziz NA, van der Burg JM, Landwehrmeyer GB, Brundin P, Stijnen T, Roos RA EHDI Study Group. Weight loss in Huntington disease increases with higher CAG repeat number. Neurology. 2008;71:1506–1513. doi: 10.1212/01.wnl.0000334276.09729.0e. [DOI] [PubMed] [Google Scholar]
- 94.Bamford KA, Caine ED, Kido DK, Plassche WM, Shoulson I. Clinical-pathologic correlation in Huntington’s disease: a neuropsychological and computed tomography study. Neurology. 1989;39:796–801. doi: 10.1212/wnl.39.6.796. [DOI] [PubMed] [Google Scholar]
- 95.Aylward EH, Brandt J, Codori AM, Mangus RS, Barta PE, Harris GJ. Reduced basal ganglia volume associated with the gene for Huntington’s disease in asymptomatic at-risk persons. Neurology. 1994;44:823–828. doi: 10.1212/wnl.44.5.823. [DOI] [PubMed] [Google Scholar]
- 96.Jernigan TL, Salmon DP, Butters N, Hesselink JR. Cerebral structure on MRI, part II: specific changes in Alzheimer’s and Huntington’s diseases. Biol Psychiatry. 1991;29:68–81. doi: 10.1016/0006-3223(91)90211-4. [DOI] [PubMed] [Google Scholar]
- 97.Harris GJ, Aylward EH, Peyser CE, Pearlson GD, Brandt J, Roberts-Twillie JV, Barta PE, Folstein SE. Single photon emission computed tomographic blood flow and magnetic resonance volume imaging of basal ganglia in Huntington’s disease. Arch Neurol. 1996;53:316–324. doi: 10.1001/archneur.1996.00550040044013. [DOI] [PubMed] [Google Scholar]
- 98.Aylward EH, Li Q, Stine OC, Ranen N, Sherr M, Barta PE, Bylsma FM, Pearlson GD, Ross CA. Longitudinal change in basal ganglia volume in patients with Huntington’s disease. Neurology. 1997;48:394–349. doi: 10.1212/wnl.48.2.394. [DOI] [PubMed] [Google Scholar]
- 99.Kuhl DE, Phelps ME, Markham CH, Metter EJ, Riege WH, Winter JC. erebral metabolism and atrophy in Huntington’s disease determined by 18FDG and computed tomographic scan. Ann Neurol. 1982;12:425–434. doi: 10.1002/ana.410120504. [DOI] [PubMed] [Google Scholar]
- 100.Young AB, Penney JB, Starosta-Rubinstein S, Markel DS, Berent S, Giordani B, Ehrenkaufer R, Jewett D, Hichwa R. PET scan investigations of Huntington’s disease: cerebral metabolic correlates of neurological features and functional decline. Ann Neurol. 1986;20:296–303. doi: 10.1002/ana.410200305. [DOI] [PubMed] [Google Scholar]
- 101.Hayden MR, Martin WR, Stoessl AJ, Clark C, Hollenberg S, Adam MJ, Ammann W, Harrop R, Rogers J, Ruth T. Positron emission tomography in the early diagnosis of Huntington’s disease. Neurology. 1986;36:888–894. doi: 10.1212/wnl.36.7.888. [DOI] [PubMed] [Google Scholar]
- 102.Kuwert T, Lange HW, Langen KJ, Herzog H, Aulich A, Feinendegen LE. Cortical and subcortical glucose consumption measured by PET in patients with Huntington’s disease. Brain. 1990;113:1405–1423. doi: 10.1093/brain/113.5.1405. [DOI] [PubMed] [Google Scholar]
- 103.Powers WJ, Videen TO, Markham J, McGee-Minnich L, Antenor-Dorsey JV, Hershey T, Perlmutter JS. Selective defect of in vivo glycolysis in early Huntington’s disease striatum. Proc Natl Acad Sci U S A. 2007;104:2945–2949. doi: 10.1073/pnas.0609833104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pandey M, Varghese M, Sindhu KM, Sreetama S, Navneet AK, Mohanakumar KP, Usha R. Mitochondrial NAD+-linked State 3 respiration and complex-I activity are compromised in the cerebral cortex of 3-nitropropionic acid-induced rat model of Huntington’s disease. J Neurochem. 2008;104:420–434. doi: 10.1111/j.1471-4159.2007.04996.x. [DOI] [PubMed] [Google Scholar]
- 105.Seong IS, Ivanova E, Lee JM, Choo YS, Fossale E, Anderson M, Gusella JF, Laramie JM, Myers RH, Lesort M, MacDonald ME. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum Mol Genet. 2005;14:2871–2880. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- 106.Chan DC. Mitochondrial fusion and fission in mammals. Annu Rev Cell Dev Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- 107.Reddy PH. Mitochondrial dysfunction in aging and Alzheimer’s disease: strategies to protect Neurons. Antioxidants & Redox Signaling. 2007;9:1647–1658. doi: 10.1089/ars.2007.1754. [DOI] [PubMed] [Google Scholar]
- 108.Knott AB, Bossy-Wetzel E. Impairing the mitochondrial fission and fusion balance: a new mechanism of neurodegeneration. Ann N Y Acad Sci. 2008;1147:283–292. doi: 10.1196/annals.1427.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reddy PH, Mao P, Manczak M. Mitochondrial structural and functional dynamics in Huntington’s disease. Brain Res Rev. 2009;61:33–48. doi: 10.1016/j.brainresrev.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Reddy PH, Reddy TP, Manczak M, Calkins MJ, Shirendeb U, Mao P. Dynamin-related protein 1 and mitochondrial fragmentation in neurodegenerative diseases. Brain Res Rev. 2011;67:103–118. doi: 10.1016/j.brainresrev.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang X, Su B, Siedlak SL, Moreira PI, Fujioka H, Wang Y, Casadesus G, Zhu X. Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc Natl Acad Sci U S A. 2008;105:19318–19323. doi: 10.1073/pnas.0804871105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang X, Su B, Lee HG, Li X, Perry G, Smith MA, Zhu X. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J Neurosci. 2009;29:9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Manczak M, Calkins MJ, Reddy PH. Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer’s disease: implications for neuronal damage. Hum Mol Genet. 2011;20:2495–2509. doi: 10.1093/hmg/ddr139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Calkins MJ, Manczak M, Mao P, Shirendeb U, Reddy PH. Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of Alzheimer’s disease. Hum Mol Genet. 2011 Sep 13; doi: 10.1093/hmg/ddr381. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang X, Su B, Liu W, He X, Gao Y, Castellani RJ, Perry G, Smith MA, Zhu X. DLP1-dependent mitochondrial fragmentation mediates 1-methyl-4-phenylpyridinium toxicity in neurons: implications for Parkinson’s disease. Aging Cell. 2011;10:807–823. doi: 10.1111/j.1474-9726.2011.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reddy PH, Mani G, Park BS, Jacques J, Murdoch G, Whetsell W, Jr, Kaye J, Manczak M. Differential loss of synaptic proteins in Alzheimer’s disease: implications for synaptic dysfunction. J Alzheimers Dis. 2005;7:103–117. doi: 10.3233/jad-2005-7203. [DOI] [PubMed] [Google Scholar]
- 117.Du H, Guo L, Yan S, Sosunov AA, McKhann GM, Yan SS. Early deficits in synaptic mitochondria in an Alzheimer’s disease mouse model. Proc Natl Acad Sci U S A. 2010;107:18670–18675. doi: 10.1073/pnas.1006586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bate C, Gentleman S, Williams A. α-synuclein induced synapse damage is enhanced by amyloid-β 1-42. Mol Neurodegener. 2010;5:55. doi: 10.1186/1750-1326-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nakamura T, Lipton SA. Redox regulation of mitochondrial fission, protein misfolding, synaptic damage, and neuronal cell death: potential implications for Alzheimer’s and Parkinson’s diseases. Apoptosis. 2010:1354–1363. doi: 10.1007/s10495-010-0476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cookson MR, van der Brug M. Cell systems and the toxic mechanism(s) of alpha-synuclein. Exp Neurol. 2008;209:5–11. doi: 10.1016/j.expneurol.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Testa CM, Sherer TB, Greenamyre JT. Rotenone induces oxidative stress and dopaminergic neuron damage in organotypic substantia nigra cultures. Brain Res Mol Brain Res. 2005;134:109–118. doi: 10.1016/j.molbrainres.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 122.Stephens B, Mueller AJ, Shering AF, Hood SH, Taggart P, Arbuthnott GW, Bell JE, Kilford L, Kingsbury AE, Daniel SE, Ingham CA. Evidence of a breakdown of corticostriatal connections in Parkinson’s disease. Neuroscience. 2005;132:741–754. doi: 10.1016/j.neuroscience.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 123.Rozas JL, Gómez-Sánchez L, Tomás-Zapico C, Lucas JJ, Fernández-Chacón R. Presynaptic dysfunction in Huntington’s disease. Biochem Soc Trans. 2010;38:488–492. doi: 10.1042/BST0380488. [DOI] [PubMed] [Google Scholar]
- 124.Pérez-De La Cruz V, Elinos-Calderón D, Robledo-Arratia Y, Medina-Campos ON, Pedraza-Chaverrí J, Ali SF, Santamaría A. Targeting oxidative/nitrergic stress ameliorates motor impairment, and attenuates synaptic mitochondrial dysfunction and lipid peroxidation in two models of Huntington’s disease. Behav Brain Res. 2009;199:210–217. doi: 10.1016/j.bbr.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 125.Richards P, Didszun C, Campesan S, Simpson A, Horley B, Young KW, Glynn P, Cain K, Kyriacou CP, Giorgini F, Nicotera P. Dendritic spine loss and neurodegeneration is rescued by Rab11 in models of Huntington’s disease. Cell Death Differ. 2011 Feb;18(2):191–200. doi: 10.1038/cdd.2010.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tebano MT, Martire A, Chiodi V, Ferrante A, Popoli P. Role of adenosine A(2A) receptors in modulating synaptic functions and brain levels of BDNF: a possible key mechanism in the pathophysiology of Huntington’s disease. Scientific World Journal. 2010;10:1768–1782. doi: 10.1100/tsw.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.André VM, Cepeda C, Fisher YE, Huynh M, Bardakjian N, Singh S, Yang XW, Levine MS. Differential electrophysiological changes in striatal output neurons in Huntington’s disease. J Neurosci. 2011;31:1170–11782. doi: 10.1523/JNEUROSCI.3539-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Boudreau RL, Monteys AM, Davidson BL. Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs. RNA. 2008;14:1834–1844. doi: 10.1261/rna.1062908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Boudreau RL, McBride JL, Martins I, Shen S, Xing Y, Carter BJ, Davidson BL. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, Carter BJ, Davidson BL. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U SA. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pfister EL, Kennington L, Straubhaar J, Wagh S, Liu W, DiFiglia M, Landwehrmeyer B, Vonsattel JP, Zamore PD, Aronin N. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington’s disease patients. Curr Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu W, Kennington LA, Rosas HD, Hersch S, Cha JH, Zamore PD, Aronin N. Linking SNPs to CAG repeat length in Huntington’s disease patients. Nat Methods. 2008;5:951–953. doi: 10.1038/nmeth.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M, Yoder J, Reeves P, Pandey RK, Rajeev KG, Manoharan M, Sah DW, Zamore PD, Aronin N. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci U S A. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J, Burchard J, Linsley PS, Aronin N, Xu Z, Zamore PD. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Aronin N. Target selectivity in mRNA silencing. Gene Ther. 2006;13:509–516. doi: 10.1038/sj.gt.3302726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sah DW, Aronin N. Oligonucleotide therapeutic approaches for Huntington disease. J Clin Invest. 2011;121:500–507. doi: 10.1172/JCI45130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Southwell AL, Patterson PH. Gene therapy in mouse models of huntington disease. Neuroscientist. 2011;17:153–162. doi: 10.1177/1073858410386236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Boudreau RL, Rodríguez-Lebrón E, Davidson BL. RNAi medicine for the brain: progresses and challenges. Hum Mol Genet. 2011;(20):R21–R27. doi: 10.1093/hmg/ddr137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc Natl Acad Sci U S A. 2005;(102):5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 142.Weydt P, Pineda VV, Torrence AE, Libby RT, Satterfield TF, Lazarowski ER, Gilbert ML, Morton GJ, Bammler TK, Strand AD, Cui L, Beyer RP, Easley CN, Smith AC, Krainc D, Luquet S, Sweet IR, Schwartz MW, La Spada AR. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1alpha in Huntington’s disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 143.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jäger S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 144.Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 2009;16:46–56. doi: 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
- 145.Ravikumar B, Rubinsztein DC. Role of autophagy in the clearance of mutant huntingtin: a step towards therapy? Mol Aspects Med. 2006;27:520–527. doi: 10.1016/j.mam.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 146.Williams A, Jahreiss L, Sarkar S, Saiki S, Menzies FM, Ravikumar B, Rubinsztein DC. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- 147.Menzies FM, Ravikumar B, Rubinsztein DC. Protective roles for induction of autophagy in multiple proteinopathies. Autophagy. 2006;2:224–225. doi: 10.4161/auto.2696. [DOI] [PubMed] [Google Scholar]
- 148.Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, Schmitt I, Wullner U, Evert BO, O’Kane CJ, Rubinsztein DC. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 149.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 150.Ravikumar B, Stewart A, Kita H, Kato K, Duden R, Rubinsztein DC. Raised intracellular glucose concentrations reduce aggregation and cell death caused by mutant huntingtin exon 1 by decreasing mTOR phosphorylation and inducing autophagy. Hum Mol Genet. 2003;12:985–994. doi: 10.1093/hmg/ddg109. [DOI] [PubMed] [Google Scholar]
- 151.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 152.Ravikumar B, Berger Z, Vacher C, O’Kane CJ, Rubinsztein DC. Rapamycin pre-treatment protects against apoptosis. Hum Mol Genet. 2006;15:1209–1216. doi: 10.1093/hmg/ddl036. [DOI] [PubMed] [Google Scholar]
- 153.Andreassen QA, Dedeoglu A, Ferrante RJ, Jenkins BG, Ferrante KL, Thomas M, Friedlich A, Browne SE, Schilling G, Borchelt DR, Hersch SM, Ross CA, Beal MF. Creatine increase survival and delays motor symptoms in a transgenic animal model of Huntington’s disease. Neurobiol Dis. 2001;8:479–491. doi: 10.1006/nbdi.2001.0406. [DOI] [PubMed] [Google Scholar]
- 154.Ferrante RJ, Andreassen OA, Jenkins BG, Dedeoglu A, Kuemmerle S, Kubilus JK, Kaddurah-Daouk R, Hersch SM, Beal MF. Neuroprotective effects of creatine in a transgenic mouse model of Huntington’s disease. J Neurosci. 2000;20:4389–4397. doi: 10.1523/JNEUROSCI.20-12-04389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Schilling G, Coonfield ML, Ross CA, Borchelt DR. Coenzyme Q10 and remacemide hydrochloride ameliorate motor deficits in a Huntington’s disease transgenic mouse model. Neurosci Lett. 2001;315:149–153. doi: 10.1016/s0304-3940(01)02326-6. [DOI] [PubMed] [Google Scholar]
- 156.Parker JA, Arango M, Abderrahmane S, Lambert E, Tourette C, Catoire H, Néri C. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. 2005;37:349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 157.Doody RS, Gavrilova SI, Sano M, Thomas RG, Aisen PS, Bachurin SO, Seely L, Hung D Dimebon investigators. Effect of dimebon on cognition, activities of daily living, behaviour, and global function in patients with mild-to-moderate Alzheimer’s disease: a randomised, double-blind, placebo-controlled study. Lancet. 2008;372:207–215. doi: 10.1016/S0140-6736(08)61074-0. [DOI] [PubMed] [Google Scholar]
- 158.Bezprozvanny I. The rise and fall of Dimebon. Drug News Perspect. 2010;23:518–523. doi: 10.1358/dnp.2010.23.8.1500435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kieburtz K, McDermott MP, Voss TS, Corey-Bloom J, Deuel LM, Dorsey ER, Factor S, Geschwind MD, Hodgeman K, Kayson E, Noonberg S, Pourfar M, Rabinowitz K, Ravina B, Sanchez-Ramos J, Seely L, Walker F, Feigin A. Huntington Disease Study Group DIMOND Investigators, A randomized, placebo-controlled trial of latrepirdine in Huntington disease. Arch Neurol. 2010;67:154–160. doi: 10.1001/archneurol.2009.334. [DOI] [PMC free article] [PubMed] [Google Scholar]