Abstract
Multidomain proteins form in evolution by the concatenation of domains but structural domains may comprise multiple segments of the chain. In this work, we demonstrate that new multidomain architectures can evolve by an apparent 3D swap of segments between structurally similar domains within a single-chain monomer. By a comprehensive structural search of the current Protein Data Bank (PDB), we identified 32 well-defined segment-swapped proteins belonging to 18 structural families. Nearly 13% of all multidomain proteins in the PDB may have a segment-swapped evolutionary precursor as estimated by more permissive searching criteria. The formation of segment-swapped proteins can be explained by two principal evolutionary mechanisms: (i) domain swapping and fusion, (ii) circular permutation. By large-scale comparative analyses using structural alignment and Hidden Markov Model methods, it was found that the majority of segment-swapped proteins have evolved by the “domain swapping and fusion” mechanism, and a much smaller fraction by circular permutation. Functional analyses further revealed that segment swapping, which results in two linkers connecting the domains, may impart directed flexibility to multidomain proteins, and contributes to the development of new functions. Thus, inter-domain segment swapping represents a novel general mechanism by which new protein folds and multidomain architectures arise in evolution, and segment-swapped proteins have structural and functional properties that make them worth defining as a separate group.
Keywords: domain swapping, protein evolution, circular permutation, multidomain proteins, fold age
Introduction
How new protein structures arise during evolution is one of the most intriguing questions in molecular evolutionary biology1–4. Ancient single-domain protein folds may have arisen by the fusion of ancient short peptide ancestors5 or from dynamic, partially ordered polypeptides6. Existing folds may transform into different folds7 by various means including circular permutation8–10. Structural and functional diversity in proteins also arises by the appearance of multidomain proteins11–13. These arise in evolution by duplication, divergence, and recombination (“domain shuffling” or fusion) of individual domains14. At the level of quaternary structures, a major avenue for the generation of new architectures is through 3D domain swapping15,16. In domain-swapped homodimers, the interface simply arises by the two monomers exchanging equivalent parts between them, thus transforming intra-chain contacts into equivalent inter-chain ones17. Higher-order oligomers may also form by this mechanism18. 3D domain swapping is thought to have been involved in the evolutionary past of many present-day oligomers19.
Here, we focus on a group of multidomain proteins that has so far been paid little attention: proteins that have structurally similar domains with equivalent segments swapped between them. This is analogous to 3D domain swapping but the “swap” occurs between domains within a chain rather than between the subunits of an oligomer. We became aware of this phenomenon during the recent CASP blind protein structure prediction experiments where for some targets (T0504 in CASP8 and T0579 in CASP9), the highest-scoring templates turned out to have quite a different fold from the actual target structures20. While the target structures had two consecutive domains, the templates (e.g. PDB entry 2gf7A) had a domain formed from the middle of the chain and another one formed from the N- and C-termini. On closer examination, the target and template folds were found to be related to each other by a 3D domain swapping operation occurring between the two domains. We propose that the term “segment swapping” be used for this intra-chain, inter-domain swapping, keeping the widely accepted “(3D) domain swapping” term for inter-subunit swapping.
While 3D domain swapping has a massive literature15–19, segment swapping as a distinct phenomenon has not been widely recognized and studied. The phenomenon was briefly described in a 2006 review on protein fold evolution by Andreeva & Murzin who showed a few examples and discussed the implications of segment swapping and related phenomena in relation with protein function21. Here, for the first time to our knowledge, we compile a list of all known segment-swapped proteins (SSPs for short) and show that segment swapping occurs, as a general fold-generation mechanism, in a number of proteins previously not recognized as such. Because of the evolutionary mechanisms peculiar to them, these proteins are worth treating as a separate group. We show two principal mechanisms of how SSPs may be generated during evolution, and perform tests to find out which mechanism may have been at work for each particular protein. We argue that segment swapping has special functional implications, making the phenomenon worth studying for its own merits.
Results
Definitions
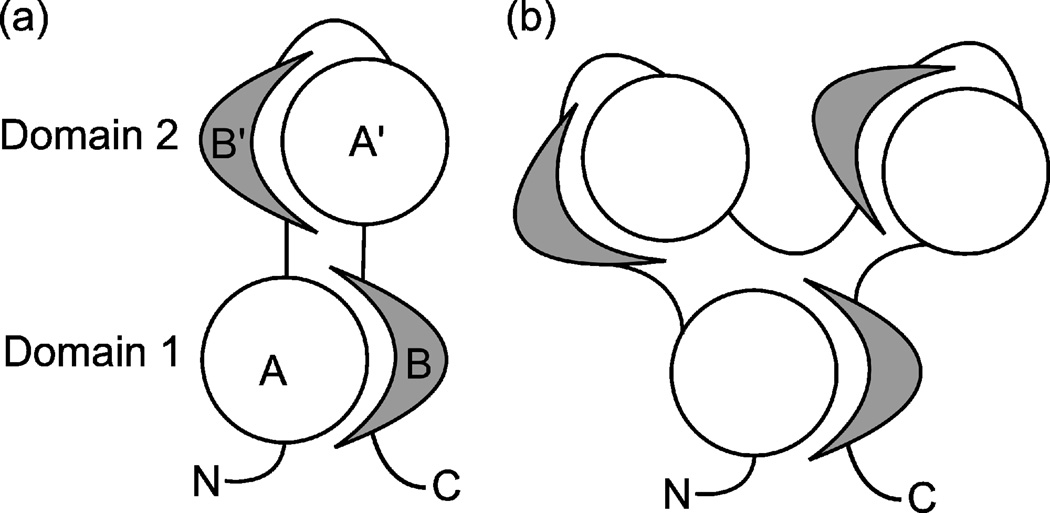
Fig. 1 shows the schematic representation of a two-domain segment-swapped protein (SSP). The protein consists of a discontinuous domain (Domain 1) and a continuous domain (Domain 2) that is inserted into Domain 1. Domain 1 is composed of an N-terminal and a C-terminal segment, denoted as segment A and segment B, respectively. Domain 2 contains structurally similar segments but in a reverse sequential order relative to Domain 1: segment B’ is similar to segment B, and segment A’ to segment A. SSPs may contain additional continuous domains inserted into Domain 1; in this case, each one must be structurally similar to Domain 2. The protein may also contain additional domains at the N- and C-termini, but those domains are ignored in this study; all structural analyses were done after cutting off any N- or C-terminal continuous domains. Following the notation (A, B) for Domain 1 and (B’, A’) for Domain 2, we will use the “AB-type domain” and “BA-type domain” designations for domains structurally similar to Domains 1 and 2, respectively.
Fig. 1.
Schematic representation of SSPs. Disks and crescents represent chain segments having a particular structure. (a) A 2-domain SSP. (b) A 3-domain SSP.
Segment-swapped structures in the PDB
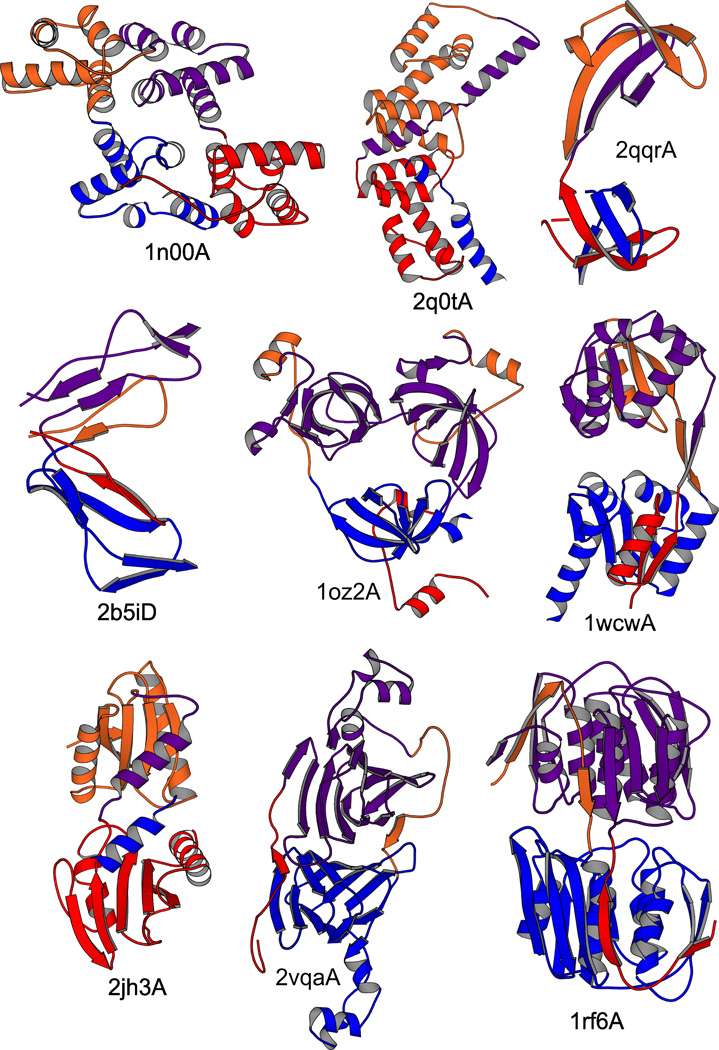
We scanned a structurally representative subset of the Protein Data Bank, herein named ReprPDB, for SSPs. By our definition, to qualify as an SSP, a protein must correspond to the structural scheme shown in Fig. 1, i.e. it must be multi-domain, containing exactly one discontinuous domain, and the two segments of the discontinuous domain, when swapped, must structurally match the corresponding regions of the other, continuous domain(s). Domains were identified with the Domainparser2 program, which provides ~82% accuracy as tested on the gold-standard manual domain decompositions22. Structural matching was assessed using structural alignment by TM-ALIGN23 with the criterion TM-score >0.5, which was shown to identify identical folds with high probability24. A segment-wise alignment coverage >2/3 is also required (see Methods for details). This procedure is general and identifies SSPs regardless of the particular structure of the swapped part and the extent of the swap. By this procedure, 32 SSPs were identified, and they were divided into 18 families (clusters) by structural similarity. The families, their central proteins, the names of the associated protein folds and the functions assigned to each family are summarized in Table 1; see Supplementary Table S1 for more detail. Cartoon representations of selected structures are shown in Fig. 2; a full gallery is shown in Supplementary Fig. S1.
Table 1.
Segment-swapped proteins and their properties
| Domain fold and family name | function | PDB chainsa | Avg. domain size / Extent of swapb |
|---|---|---|---|
| Mainly-α folds | |||
| Orthogonal bundle, annexin fold | phospholipid binding | 1n00A 1dk5A | 158 / 53% |
| Up-down bundle, AhpD-like fold | lyase, decarboxylase | 2q0tA | 127 / 70% |
| Mainly-β folds | |||
| SH3-like, two MBT repeats | transcriptional regulator | 2r58A 2bivA 1oz2Ac | 106 / 23% |
| SH3-like, Jumonji domains | oxidoreductase, demethylase |
2qqrA | 58 / 52% |
| 2-layer β-sandwich variantd | hydrolase, galactosidase | 3d3aA | 127 / 14% |
| Complement control module | receptor | 2b5iD | 61 / 30% |
| Double-stranded β-helix; RmlC-like Cupins |
oxidoreductase | 1y3tA | 165 / 12% |
| 3-layer αβα sandwiches | |||
| Periplasmic binding protein-like II | transcriptional regulator | 2ql3A 3hhfA 1ixcA | 100 / 75% |
| HemD-like | lyase | 1wcwA 1jr2A | 128 / 27% |
| PrpR receptor domain-like | transcriptional regulator | 2q5cA | 93 / 80% |
| NAD(P)-binding Rossmann fold | oxidoreductase, dehydrogenase |
2et6A | 291 / 80% |
| Rossmann fold variant 1d | transferase | 2hcrA | 152 / 87% |
| Rossmann fold variant 2d | unknown | 2jh3A | 132 / 82% |
| Other αβ proteins | |||
| αβ-prism (6 repeats of IF3 fold) | transferase | 1rf6A 2yvwA 1g6sA 1ejdA 2pqcA 2o0bA 2rl1A |
212 / 9% |
| CBS domain pair (αββα sandwich) | adenosyl binding | 1yavA 3hf7A 2emqA | 66 / 22% |
| Double-stranded β-helix + α-helicesd | metal binding | 2vqaA | 178 / 13% |
| αβ-roll, diaminopimelate epimerase- like |
unknown (isomerise?) | 2h9fA | 190 / 89% |
| WWE domain | signaling | 2a90A | 77 / 91% |
The first chain listed is the central protein in each family.
Measured as the average length of the N-terminal segment of Domain 1 divided by the average domain length.
The protein has 3 domains.
Fold names assigned by us.
Fig. 2.
Cartoon representations of selected SSP structures. The N- and C-terminal segments, forming Domain 1 are shown in red and blue, and the corresponding segments in Domain 2 in lighter red and blue, respectively. The cartoons were generated with MOLSCRIPT68; breaks in the chain are due to missing Cα atoms.
A more permissive definition was also used to define SSPs, resulting in 12 additional SSP families. However, these were found too ambiguous, and were not used for further analysis except when otherwise noted. See Methods for more details; the proteins are listed in Supplementary Table S2 and Supplementary Dataset S1.
SSPs were identified in all structural classes, i.e. mainly-α, mainly-β as well as αβ proteins. Three-layer αβα-sandwiches are especially prominent, with several varieties of the Rossmann fold. The functions of the SSPs are diverse; enzymes, transcriptional regulators, signaling and binding proteins are all represented. All except one SSP identified by our procedure consist of 2 domains; the exception is the PDB chain 1oz2A containing 3 domains: one discontinuous and 2 inserted continuous domains (see the cartoon in Fig. 2). Table 1 also shows the average lengths of the domains in each family (58 to 291 residues) and the extent of the “swap” relative to the domain size, which is nearly evenly distributed between 9% and 91%. In a few cases, the swapped motif consists of only a single α-helix or β-strand but typically, groups of several secondary structure elements are swapped. The sequence identities (not shown) between the 2 domains of each protein (based on a structural alignment between Domain 2 and Domain 1 after swapping its segments) are generally low, with an average of 19%, and exceeding 30% in only 4 cases. The highest sequence identity was found between the domains of 1oz2A (46%). Even the low sequence identities, however, suggest evolutionary relatedness between the domains (when calculated from structural alignment, a sequence identity >~11% already makes homology more probable than analogy25).
The prevalence of SSPs
We identified 32 SSPs in our representative PDB subset containing 21,938 protein chains (6,118 multi-domain proteins). Although this may seem a small number, this is partly due to the strict criteria we used to identify these proteins. In these proteins, the internal structural symmetries are pronounced enough to confidently recognize them as segment-swapped. For many other proteins, these symmetries may have broken down as the sequences and the structures of the domains diverged with evolution. In some proteins, a more extensive interface may have formed between the domains, thus obscuring the multidomain architecture and making the swap undetectable by our method. In fact, one of the SSPs presented by Andreeva & Murzin21, periplasmic nitrate reductase (PDB ID: 2nap26, another 3-layer αβα sandwich protein) could not be detected by our method because of the strong structural divergence of the domains and the presence of N- and C-terminal extensions that form closely associated subdomains. To estimate the number of proteins that cannot be clearly identified as segment-swapped but may have descended from a segment-swapped ancestor, we scanned ReprPDB to find structural analogs of the 32 identified SSPs, simply using a TM-score threshold of 0.5. This yielded 494 additional chains. Adding the analogs of the 12 more permissively defined SSPs (see Methods) increases the total number of such proteins to 788, which is 12.9% of all multi-domain structures in ReprPDB (full listings of the proteins are available in Supplementary Dataset S1).
Evolutionary mechanisms that may generate SSPs
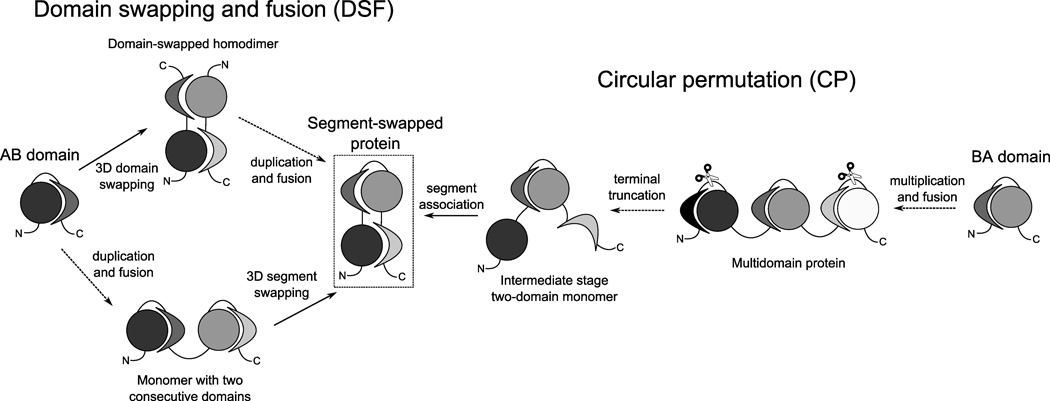
By definition, the N-terminal and C-terminal halves of a two-domain SSP structure are structurally similar, even though each half includes segments from both domains. This suggests that gene duplication may have been involved in generating these structures. Given the fact that SSPs look like a domain-swapped homodimer after fusing the monomers, domain swapping and fusion is obviously a plausible and likely mechanism for their generation. However, another, relatively simple mechanism, involving circular permutation, is also possible. Thus, we propose two evolutionary scenarios that are capable of producing SSPs (see Fig. 3):
Fig. 3.
Evolutionary scenarios generating segment-swapped structures. Dotted arrows indicate events occurring at the DNA level, while solid arrows represent protein structural rearrangements, possibly also favored by point mutations at the DNA level.
Domain swapping and fusion (DSF mechanism)
This scenario assumes that at an earlier point in the evolutionary history of a present-day SSP, an ancient gene corresponding to one half of the present-day protein existed. The protein encoded by this gene formed a single-domain monomer, with segment A at the N-terminus and segment B at the C-terminus. Then, two events happened: (i) the monomer dimerized by 3D domain swapping15,16, and (ii) tandem gene duplication and fusion occurred, resulting in 2 copies of the initial chain fused into a single chain. These events may also have occurred in a reverse order, i.e. gene duplication and fusion may have first generated a protein containing 2 consecutive domains, and an intra-chain 3D domain swapping event may have occurred between the domains. Regardless of the order of the events, subsequent point mutations caused the sequences of the duplicated copies to diverge, and stabilized the “swapped” conformation, preventing a “flipback” of the domains to a consecutive arrangement.
Circular permutation (CP mechanism)
In this scenario, an ancient single-domain monomer existed that corresponded to Domain 2 of the present-day protein, i.e. the middle part of the chain. In this domain, segment B is at the N-terminus and segment A is at the C-terminus. A circular permutation8 of this protein is generated as follows. Tandem gene multiplication and fusion generated a protein with at least 3 identical domains. Both termini of this protein were truncated (at the DNA level), removing a part of each terminal domain. Then, the remaining segments of the terminal domains associated, forming a new domain that is similar to the ancient domain but its segments are in a reverse order relative to the ancient monomer.
The principle of the CP mechanism is exemplified by β-propeller proteins. The blades of the propellers are motifs of 4 β-strands each. The N- and C-termini are connected within a “mixed” blade, with a varying number of the strands coming from the N- and C-terminus, which suggests that the variants are the result of circular permutation27.
In both scenarios, existing domains open up and their segments associate to form a new domain. The new domain will be a circular permutant of the “donor” domains. Because the two halves of the SSP are generated by gene duplication, their sequences are initially identical but they diverge due to the accumulation of mutations while the segment-swapped structure remains conserved.
We propose that DSF and CP are the two fundamental, most parsimonious mechanisms for producing SSPs. Although more complex mechanisms, consisting of multiple fusion or transposition events, may be imagined, we will assume in the following that each SSP was generated either by the DSF or the CP mechanism.
Distinguishing between the DSF and CP mechanisms
Even though the two evolutionary mechanisms, DSF and CP, generate a protein with the same present-day structure, there may be clues that allow one to infer the mechanism that generated each particular present-day protein. We propose three such clues and investigate them each.
The first clue is based on estimating the age of domain folds. As Fig. 3 shows, if the structure of the present-day segment-swapped protein is described as AB’A’B, then, in order to arrive at this final structure, the DSF mechanism must start with a single domain with segment A at the N-terminus and segment B at the C-terminus; i.e. an AB-type domain. The CP mechanism, however, must start with a domain with segment B at the N-terminus and segment A at the C-terminus, i.e. a BA-type domain. Essentially, the AB-type and the BA-type structures are different (although related) folds, which may occur independently in other proteins. There are ways to infer the relative age of a protein fold1,28, and if we determine which of the two folds is older, we can infer the origin of a particular SSP. If the AB-type fold is found more ancient than the BA-type fold then it may be concluded that DSF is more likely than CP to have generated the SSP, and vice versa. We estimated the relative ages of the AB- and BA-type folds by analyzing their occurrences in 22 complete proteomes (by threading) and in ReprPDB (by structural comparisons).
Another clue involves searching for homodimeric analogs of present-day (monomeric) SSPs. As an intermediate on the DSF pathway, a homodimeric analog is a strong indication of the DSF mechanism. We scanned the PDB for homodimeric analogs of segment-swapped monomers.
A third clue is based on detecting and comparing variants of SSPs that are based on the same fold but differ from each other in one of the domains. As shown later, the DSF and CP mechanisms generate different variants, and the type of variants can be used to infer the originating mechanism.
Scanning 22 complete genomes for analogs/homologs of domains of SSPs
A simple approach to estimate relative fold ages is by counting the genomes (proteomes) that a particular fold occurs in1,28; a higher occurrence implies a more ancient fold. For this purpose, we scanned 22 complete genomes for analogs/homologs of each domain of our SSPs by profile hidden Markov model (HMM) comparisons29. HMM-HMM comparison has been shown to be a very sensitive method capable of identifying related proteins even when sequence identity is low (distant homologs or analogs)29. We counted the hits containing a single AB-type or BA-type region; hits containing more than one recognizable region were excluded because it is impossible to know whether such hits contain segment-swapped or consecutive domains. For an additional analysis, we also took into account the phylogenetic distribution of the hits, based on a simple phylogenetic tree of the 22 genomes (see Methods for details). A fold is considered older when it appears earlier (i.e. at a lower position) in the phylogenetic tree28. The numbers of source organisms associated with the hits are shown in Table 2; more detailed data, including phylogenetic positions, are shown in Supplementary Table S3.
Table 2.
The number of source organisms of analogs of the two domains (Domain 1, AB-type; Domain 2, BA-type) of the SSPs listed in Table 1 in 22 genomes and in ReprPDB, respectively.
| Group | Central protein |
In 22 genomes AB-type / BA-type |
In ReprPDB AB-type / BA-type |
|---|---|---|---|
| Mainly-α | 1n00A | 0 / 1 | 0 / 0 |
| 2q0tA | 11 / 1 | 1 / 0 | |
| Mainly-β | 2r58A | 2 / 0 | 0 / 1 |
| 2qqrA | 6 / 0 | 43 / 0 | |
| 3d3aA | 0 / 0 | 37 / 11 | |
| 2b5iD | 4 / 3 | 4 / 0 | |
| 1y3tA | 5 / 0 | 2 / 0 | |
|
3-layer αβα sandwich |
2ql3A | 9 / 4 | 13 / 1 |
| 1wcwA | 21 / 10 | 137 / 98 | |
| 2q5cA | 20 / 8 | 43 / 24 | |
| 2et6A | 6 / 1 | 0 / 0 | |
| 2hcrA | 9 / 5 | 105 / 66 | |
| 2jh3A | 4 / 10 | 67 / 10 | |
| Other αβ | 1rf6A | 0 / 0 | 0 / 0 |
| 1yavA | 20 / 1 | 0 / 1 | |
| 2vqaA | 2 / 6 | 0 / 0 | |
| 2h9fA | 0 / 0 | 0 / 0 | |
| 2a90A | 2 / 1 | 0 / 0 | |
The analogs were pooled in each family.
In almost all cases, we get more hits to the AB-type than the BA-type domain, regardless of whether we count proteins or genomes. The phylogenetic positions of the folds can be compared for 7 families; in 4 cases, the BA-type fold turns out to be younger than the AB-type fold, and in 3 cases, they appear at the same height of the phylogenetic tree (Supplementary Table S3). These findings further support the proposal that most SSPs were generated by the DSF mechanism. There are 2 cases where the CP mechanism seems to be more supported. One of the 3-layer αβα sandwich proteins, 2jh3A, has more BA-type than AB-type homologs, supporting the CP mechanism. One of the other αβ type proteins, 2vqaA, has 35 AB- and 8 BA-type homologs but the AB-type homologs come from only 2 genomes while the BA-type ones from 6, thus tipping the balance towards the CP mechanism; phylogenetic positions, are, however, identical for the two domain folds.
Scanning the PDB for analogs of domains in SSPs
In addition to scanning 22 complete genomes for homologs of the two domains of each SSP, we also performed a structural similarity based scan on ReprPDB and counted the occurrences of the AB- and BA-type folds for each SSP. Naturally, the occurrence of a fold in the PDB (or ReprPDB) depends, besides fold age, on many factors such as crystallizability, the interest of biologists in the proteins with the fold, designability30,31, etc. Thus, the occurrence in the PDB is, in itself, is not a good indicator of fold age in general. However, when comparing the occurrences of folds that are related to each other by circular permutation (such as the two domains of SSPs), many of those factors are similar, and thus fold age becomes more significant. Thus, the occurrence of a fold in the PDB, and even better, the number of source organisms associated with those occurrences, appears to be useful as a rough estimate of relative fold age in our case.
Using the protein structure alignment algorithm TM-ALIGN23, we scanned ReprPDB for continuous domain structures similar to Domain 1 (i.e. AB-type folds) and Domain 2 (BA-type folds), respectively. (Recall that Domain 1 (2) refers to the discontinuous (continuous) domain of an SSP as shown in Fig. 1.) The numbers of the source organisms of the resulting analogs are shown in Table 2; more detailed data are shown in Supplementary Table S3. For 6 out of the 18 SSP families, no analogs are found either for Domain 1 or Domain 2. Out of the remaining 12 families, Domain 1 has significantly more analogs than Domain 2 in 10 cases, and the same inequality is found when the number of source organisms is considered. In fact, for 4 families, no analog is found for Domain 2, only for Domain 1. This suggests that the AB-type fold is older in most cases than the BA-type fold; thus, the DSF mechanism may have generated most present-day SSPs. One exception is the family with the central protein 2r58A, where (in contrast to the findings obtained by the complete genome searches) the CP mechanism is more supported. This family contains the chain 1oz2A, the only chain containing 3 domains, 2 of them BA-type. The other chain with more BA- than AB-type analogs is 1yavA, a hypothetical protein with unknown function; but the existence of only one analog does not allow one to make a firm conclusion about its origin.
It should be noted that in a few cases, we found a few analogs that were structurally similar to both domains of a segment-swapped protein. In these cases, the analogous domain could be described as having three segments, i.e. (A,B,A’) or (B,A,B’); thus, it aligns well with both an (A,B) and a (B,A) domain. Such analogs were found for the segment-swapped proteins 1wcwA, 1jr2A, 2hcrA (3-layer αβα sandwiches) and 3d3aA (a 2-layer beta sandwich). Although such structures might be circular permutation intermediates32, there are many more AB-type analogs for these segment-swapped proteins, thus the data still favor the DSF mechanism.
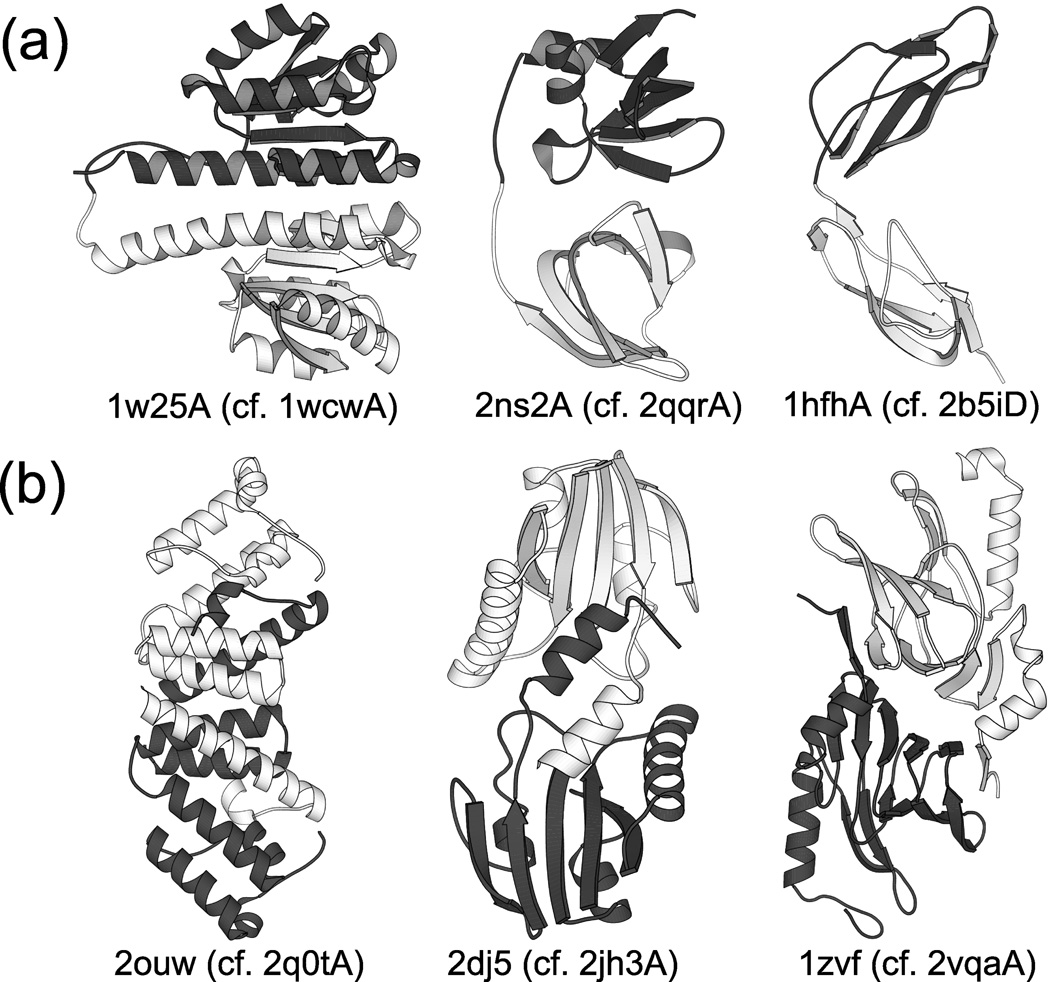
We also scanned ReprPDB for proteins containing more than one AB- or BA-type domain. A protein with 2 consecutive AB-type (BA-type) domains may be an intermediate in the DSF (CP) mechanism (see Fig. 3), and thus their existence supports the corresponding mechanism. Analogs containing 2 or more consecutive AB-type domains were found for the SSP families with these central proteins (the number of analogs given in parentheses): 1wcwA (10), 2hcrA (6), 2jh3A (1), 3d3aA (5), 2qqrA (2), 2b5iD (15). Fig. 4a presents 3 of these analogs compared with the corresponding segment-swapped structures. Analogs containing 2 or more consecutive BA-type domains were only found for the families 1wcwA (2) and 2hcrA (3).
Fig. 4.
Proteins having structures related to selected SSPs. (a) Protein structures containing 2 consecutive domains structurally similar to Domain 1 of selected SSPs from Fig. 2. The N- and C-terminal domains are shown in light and dark shades, respectively. The corresponding SSP is indicated for each structure. (b) Homodimeric analogs of selected SSPs from Fig. 2. The two subunits are shown in light and dark shades, respectively. The corresponding SSP is indicated for each structure. The existence of these structures supports the DSF mechanism for the formation of the corresponding SSPs because the DSF mechanism involves either a two-domain or a homodimeric intermediate as illustrated in Fig. 3.
The chain 2b5iD, a structure of the interleukin-2 receptor α chain33, is especially interesting as it consists of 2 segment-swapped complement control modules, also known as the sushi domain, which occurs in a number of complement and adhesion proteins as repeats (hence its other name: short consensus repeat or SCR)34. But Domain 2 of 2b5iD (a circularly permuted version of the sushi domain) represents a unique fold that has not been found in any other proteins, indicating that it is a novel form. Thus, the evidence for the DSF mechanism is strong in this case.
Similarly, the chain 2qqrA, a histone demethylase35, contains 2 segment-swapped Tudor domains, which occurs in several RNA-binding proteins, and the Drosophila Tudor protein contains 10 repeats of it36. Domain 2 of 2qqrA, corresponding to a circularly permuted Tudor domain, does not occur in any other known structures, which again strongly supports the DSF mechanism for this protein.
For SSPs containing two 3-layer αβα-sandwich domains, we find many analogs for both domains, but more for the first domain (AB-type). This indicates that the DSF mechanism probably generated many of these proteins, but the CP mechanism may also have occurred.
Searching for homodimeric analogs of SSPs
The DSF mechanism may involve a stage where two identical chains, each corresponding to a single domain, open up and form a domain-swapped homodimer. Thus, the existence of a homodimeric analog of a (monomeric) SSP supports the DSF mechanism for that particular protein because it shows that the AB-type fold is indeed capable of opening up and forming a 3D domain swapped complex, and this actually occurs. We scanned the PDB for homodimeric analogs of each central SSP listed in Table 1 (see Methods for details). We found homodimeric analogs for 5 SSPs (the best analog and its TM-score are shown in parentheses): 2q0tA (2ouw, 0.73), 2jh3A (2dj5, 0.83), 2et6A (1zbq, 0.92), 2vqaA (1zvf, 0.61), 1y3tA (1lrh, 0.65). For 3 of these 5 cases, the corresponding homodimers are shown in Fig. 4b. It should be noted that the biological unit for 2q0t is trimeric, and that of 2ouw is, correspondingly, hexameric. This pair is also one of the examples discussed by Andreeva & Murzin21.
In some cases, there is little or no functional similarity between the SSPs and their closest homodimeric analogs. The monomeric 2vqaA is a cyanobacterial metal binding protein while its analog 2zvf is a yeast enzyme; and 1y3tA is a bacterial enzyme while its analog 1lrh is a plant binding protein. In these cases, a direct evolutionary relationship between the segment-swapped proteins and their homodimeric analogs cannot be established. On the other hand, the segment-swapped monomer 2q0tA and the homodimeric 2ouw are both bacterial enzymes (although with different functions), and the monomeric 2et6A and its dimeric analog 1zbq are both dehydrogenases from eukaryotes. The second closest dimeric analog of the monomeric 2et6A is 1gz6, which has the same function (hydroxyacyl-CoA dehydrogenase), suggesting that 2et6A may have been generated by a recent fusion event while retaining the protein function. The monomeric 2jh3A is a bacterial protein of unknown function while its dimeric analog 2dj5 is an archaeal enzyme. However, they are structurally similar to 1qgoA, a known monomeric cobalt chelatase37 and 1tjn, a putative dimeric cobalt chelatase, respectively; this pair was also discussed by Andreeva & Murzin21.
Detecting and comparing variants of SSPs
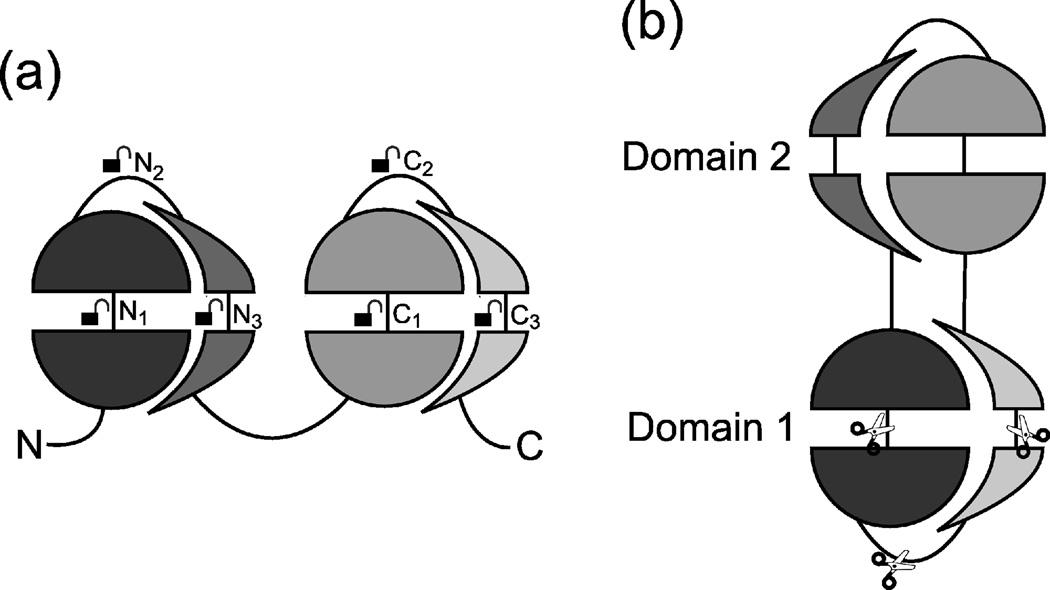
As Fig. 5 illustrates, both the CP and the DSF mechanisms may generate several variants of SSPs based on the same fold; see also Supplementary Fig. S3. The DSF mechanism may generate different variants depending on where the initial N-terminal and C-terminal domains open up to form a new, middle domain. Thus, variants generated by the DSF mechanism will have identical “Domain 1”s (apart from the location of the discontinuity), and their “Domain 2”s will be circular permutants of each other (Fig. 5a). The CP mechanism involves terminal truncation of a chain containing at least 3 domains. Depending on the site of truncation, different variants may arise, which, however, will all have the same middle domain (Domain 2); their discontinuous domains (“Domain 1”s) will be circular permutants of each other (Fig. 5b).
Fig. 5.
Generation of SSP variants by different mechanisms. Here, each domain of an SSP is divided into 4 segments, indicated by the half-disks and half-crescents. (a) DSF variants are generated from a protein with 2 consecutive domains by opening up the N-terminal domain at various sites Ni (i=1, 2, 3) and opening up the C-terminal domain at corresponding sites Ci; the sites are indicated by the open padlock symbols. Domain 2 of the SSP is then formed from the segment between Ni and Ci, while Domain 1 is formed from the N to Ni and the Ci to C stretches of the chain. The resulting variants have different “Domain 2”s but (structurally) identical “Domain 1”s (ignoring the discontinuity). (b) CP variants can be construed as generated by cutting the circularized chain at different sites, as indicated by the scissors symbols. The resulting variants have identical “Domain 2”s but different “Domain 1”s. Supplementary Fig. S3 depicts all variants separately for greater clarity.
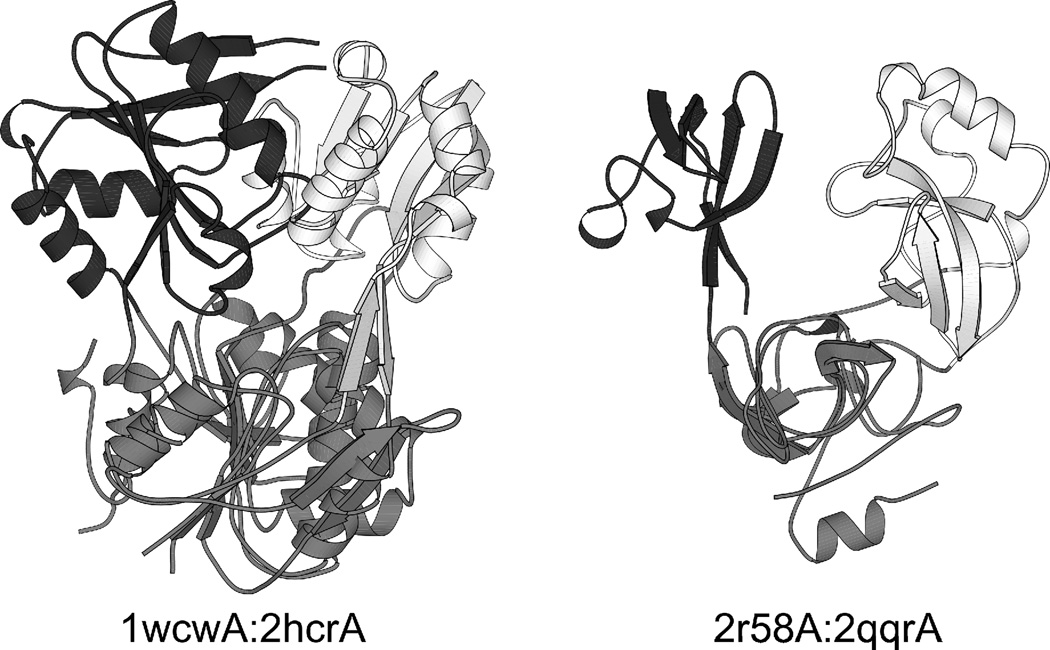
In order to identify pairs of SSPs with similar discontinuous or continuous domains, we performed a pairwise structural comparison of all domains constituting the 18 central proteins as described in Methods. We found 2 pairs meeting the criteria, 1wcwA:2hcrA and the similar 1wcwA:2jh3A with a stricter criterion, and another pair, 2r58A:2qqrA, with a looser criterion for structural similarity. Two of these 3 pairs are presented in Fig. 6. In both cases, the discontinuous domains (superimposed and shown in gray) are structurally similar but the continuous domains are inserted in them at quite different sites (shown in white and black, respectively). In 1wcwA, the continuous domain is inserted into the discontinuous one near the N-terminus (at position 31) while in 2hcrA, the same occurs near the C-terminus (at position 144). In the other pair, 2r58A:2qqrA, the size of the domains is quite different because of additional inserted helices in 2r58A. In 2r58A, the continuous domain is inserted into the discontinuous one near the N-terminus (after the N-terminal helix) while in 2qqrA, the same occurs near the middle of the chain. Sequence identities within the pairs are negligible, and there is little functional similarity, although 1wcwA and 2hcrA are both synthetases.
Fig. 6. Pairs of related segment-swapped proteins, representing variants generated by the DSF mechanism.
In these pairs, the discontinuous domains (gray) are structurally similar and are shown superimposed, but the continuous domains (shown in white for the first protein and in black for the second) are inserted into the discontinuous domain at different sites, and are therefore circular permutants of each other.
To find additional pairs, the search was extended to the 12 more permissively defined SSPs. Although two cases meeting the formal criteria were found (2q5cA:3i04A with similar continuous domains, and 2q5cA:2rkbA with similar continuous domains), these are unconvincing because of the high divergence of the structures (long insertions, slightly different β-strand order). In summary, we found 3 convincing examples of SSPs with similar discontinuous domains, and no convincing example for ones with similar continuous domains. Thus, in the light of the mechanism illustrated in Fig. 5a, these results support the DSF rather than the CP mechanism of SSP generation.
The functional implications of segment swapping
What functional advantages may be associated with segment swapping? We examined the available literature data on the function of the SSPs we identified, and found that two main types can be discerned:
The substrate or binding partner binds in a cleft between the two domains. In most such cases, a hinge-type relative motion of the two domains is known or assumed to be significant for the function of the protein. In these proteins, an advantage of having segment-swapped rather than sequential domains may be that the resulting two domains are connected by 2 linker regions rather than only one. Thus, a well-directed hinge motion becomes possible by constraining the relative domain motions to around a single axis (provided that the linkers are sufficiently short). This would be more difficult if there was only a single linker which still allows 3-axis motion of the domain moieties. Examples for such SSPs include the enzyme 1rf6A38 (and its orthologs 1g6sA, 2pqcA, 2o0bA), the enzyme 1ejdA39 (and its orthologs 2yvwA, 2rl1A), the transcriptional regulator 1ixcA40 (and the similar 2ql3A, 3hhfA), the enzyme 1jr2A41 (and its ortholog 1wcwA), the membrane-associated binding protein 1n00A42 (and the similar 1dk5A), the enzyme 2hcrA43, and the signaling protein 2a90A44. The facilitation of a well-directed hinge motion by the presence of two linkers seems especially plausible in 1n00A where the linkers are 25 Å from each other, and in 1wcwA where they form a two-stranded β-sheet.
The two domains each have their own binding sites for the ligand or binding partner and perform their (similar) functions independently, but the ligand or substrate specificities of the two domains are different. Clearly, gene duplication allows the substrate specificities to diverge. Here, the advantage of the segment-swapped topology may simply be a further rigidification of the overall structure in addition to the non-covalent contacts at the domain-domain interface. Examples of such SSPs include the histone binding protein 2r58A45 (along with the similar 2bivA and 1oz2A) and 2qqrA35 as well as the enzymes 2et6A46 and 1y3tA47.
Discussion
We identified 32 well-defined segment-swapped proteins in 18 families, and estimated that ~12.9% of all multi-domain proteins may have a segment-swapped evolutionary ancestor. Thus, we propose that segment swapping is one of the common mechanisms by which new protein folds or multi-domain architectures arise in evolution. This higher-level mechanism is based on lower-level mechanisms (DSF and CP) which in turn are based on known phenomena such as gene duplication, fusion, 3D domain swapping, circular permutation, and sequence divergence.
Looking at the source publications for the PDB entries of SSPs, we observe that generally little attention has been devoted to the segment-swapped nature of the structure, especially in the more ambiguous cases like the otherwise well-studied enzymes isocitrate dehydrogenase and isopropylmalate dehydrogenase48 where large C-terminal additions and long loop insertions obscure the swapped architecture49. Segment-swapped structures have been described using various terms such as “interdigitated”50,51 or “hybrid”50 domains, domain-swapped modules33 or helices52, or “crossing back”44.
Like 3D domain swapping is a simple way to form a subunit-subunit interface, segment swapping is a simple way to form a domain-domain interface within a monomer: instead of evolving a new interface, two domains can be efficiently assembled by exchanging equivalent parts between them. Thus, segment swapping can quickly generate a new multidomain architecture. But an SSP can also be an intermediate step in an evolutionary process that results in a single-domain protein with a new fold. As Fig. 5a shows, the DSF mechanism can generate a variety of proteins with continuous domains that are circular permutants of each other. If the continuous domain gets cut out (i.e. the N- and C-terminal segments forming Domain 1 are cut off), a circular permutant of the ancient domain (corresponding to the structure of Domain 1) is obtained. Some of the SSP domains indeed have a large number of circular permutants; e.g. we identified 9 major circular permutants of the 1wcwA domain fold (see Supplementary Fig. S2).
We used the known protein structures as a starting point for our study. There are, however, proteins whose structures are not known but can be suspected to be segment-swapped. One example is dUTPase, which is usually a homotrimer53, but the dUTPase gene also occurs in tandemly triplicated form in some organisms (e.g. Caenorhabditis elegans), whose product is thought to form a segment-swapped, 3-domain protein54, although a 3D structure is not yet available. The structure of a related monomeric dUTPase from the Epstein-Barr virus is known but it is very distorted in comparison with trimeric dUTPases55 and cannot be recognized as segment-swapped.
There is an important difference between 3D domain swapping and segment swapping. In many cases, 3D domain swapping is a dynamic phenomenon, i.e. the chains forming a domain-swapped homodimer can, depending on the external conditions, exist in an “unswapped”, monomeric conformation as well56. In contrast, the two domains of an SSP can no longer “flip back” to the “unswapped” conformation because the sequences of the two halves of the chain, which were originally identical, have largely diverged during evolution. As a result, residue-residue contacts can only stabilize the domains (A,B) and (A’,B’) and not the mixed domains (A,B’) and (A’,B). This is attested by the fact that we found no “unswapped” conformation for any SSP; the “unswapped” structures in Fig. 4a have very different sequences from the corresponding SSPs. Thus, duplication and fusion allows the domain-swapped conformation to become genetically fixed. (Another, unrelated, way to genetically fix a domain-swapped structure is shortening the loop where one subunit opens up, as suggested for the histone fold57.) Because of this fixing, the domains of an SSP can lose the ability to “open up”, and thus may be less prone to forming domain-swapped aggregates than a corresponding domain-swapped homodimer.
We posited two basic evolutionary mechanisms, domain swapping and fusion (DSF) and circular permutation (CP), that may generate SSP architectures, and we have shown 3 ways to test which mechanism generated each particular SSP. Our tests indicate that DSF is the more common mechanism generating the segment-swapped topology. In those cases where a homodimeric analog is present or where the Domain 2 fold does not occur in any other protein, the evidence for the DSF mechanism is particularly strong. In the few cases where the CP mechanism seems to be more supported, data are scarce to make a firm conclusion; the only family where CP seems somewhat likely is that of MBT repeat proteins where the presence of 3 domains in 1oz2A makes CP plausible.
The fact that we found little support for the CP mechanism is in accord with the observation that circular permutation of proteins is relatively rare58, although this claim is somewhat controversial59. A plausible explanation why DSF rather than CP seems to be the dominant mechanism is that DSF requires only a single gene duplication while CP requires at least 2 gene duplications and 2 truncations, i.e. it involves more operations at the DNA level.
The DSF mechanism is discussed by Andreeva & Murzin21 as an evolutionary process leading to multi-domain proteins through “transient oligomerization”; they do not raise the possibility that besides domain swapping, CP may also generate a segment-swapped structure. Abraham et al.60 present the evolution of proteins with structurally similar domains as an alternative to homooligomerization, but they do not investigate segment swapping as a common scenario associated with it.
Our investigations regarding the evolutionary mechanisms of SSPs are primarily based on structure comparisons rather than conventional sequence-based phylogenetic analysis. The main reason for this is that sequence information alone is not sufficient to decide whether a two-domain protein has segment-swapped or consecutive domains, and thus sequence-based alignment does not guarantee that only proteins with similar structures are aligned. Although similar structures can arise by convergent evolution61, it has been argued that most folds are monophyletic62, and it has been shown that protein evolution can be studied using structural similarities, constructing “structure-based phylogenies”63. For some of the larger SSP families, like the Rossmann fold variants, where a large number of known structures are available, a sequence-based analysis may also be possible.
Examining the possible functional implications of segment swapping, we found that the presence of two linkers connecting the domains in SSPs may facilitate hinge-type relative domain motion around a single axis, which is often important for function. This may be an advantage of the segment-swapped topology over a simple concatenation of domains.
Segment swapping may also be an interesting subject for experimental protein design studies, e.g. to address the question whether and how an existing protein with consecutive domains could be switched over to a segment-swapped topology or vice versa.
In this paper, we have shown that segment-swapped proteins form a rich and diverse group of proteins that is worth defining as a separate group. The evolutionary mechanisms generating these proteins are peculiar to the group, and the segment-swapped architecture is associated with special functional advantages.
Methods
Creating ReprPDB, a structurally representative PDB Subset
The Protein Data Bank included more than 150,000 polypeptide chains at the time of our study, which is too large for practical use, and has redundant entries. We selected a subset of the PDB that is structurally representative. We started with a precompiled list of 21,650 PDB chains with a pairwise identity <90% as provided by the PISCES server64 (release 090905), and extended it by adding structures that were significantly different (TM-score <0.6 from TM-ALIGN23) from all structures already in the set despite being similar in sequence (>90% sequence identity) to one or more entries. (Here, the threshold 0.6 was used instead of the standard 0.5 in order to minimize missed folds at the price of allowing some redundancy.) The final size of the structure set was 21,938. Any pair of structures in the set either has a sequence identity <90% (with no TM-score limit) or a TM-score<0.6 (with no sequence identity limit).We will refer to this data set as ReprPDB. A listing of ReprPDB is available in Supplementary Dataset S1.
Searching for segment-swapped proteins
The chains in the set were divided into domains by the Domainparser2 program22, setting the minimum domain size to 20 and minimum segment length to 10, and allowing two β-strands to connect domains (option “-mbpass 2”). The proteins containing exactly one discontinuous domain (a domain having 2 separate segments) and at least one inserted, continuous domain were selected (1,757 such proteins were found). Any continuous domains at the N- and C-termini were cut off (394 cases). For two-domain proteins whose sequence corresponds to the scheme (A,M,B), with the segments A and B forming the discontinuous and M forming the inserted, continuous domain, the structures AB and BA were each aligned to M by TM-ALIGN23, resulting in 2 TM-scores: TM(AB,M) and TM(BA,M). In an SSP, BA must be structurally similar to M, so the proteins with TM(BA,M) > 0.5 and TM(BA,M) > TM(AB,M) were selected for further consideration. Based on the BA to M structural alignment, the M domain was split into 2 segments B’ and A’, and the alignment coverages were calculated for both segment pairs. The protein was accepted as segment-swapped if both alignment coverages were >2/3 and at least 4 pairs of residues were aligned in each segment. A protein with 2 or more inserted (continuous) domains was defined as segment-swapped if each inserted domain was found to be in a segment-swapping relationship with the discontinuous domain.
The proteins were divided into clusters by structural similarity using a variation of the QT (quality threshold) clustering algorithm65, selecting a central protein in each cluster so that the lower of the two per-domain TM-scores between the central protein and any other protein in the cluster is >0.5. We refer to these clusters as “families” throughout this paper.
This procedure resulted in 32 SSPs in 18 structural families (Table 1 and Supplementary Table S1). To identify more potential SSPs, a more permissive definition was also applied where segment-wise alignment coverages were not required to be >2/3. This resulted in 12 additional SSP families; see Supplementary Table S2.
Searching for analogs/homologs in 22 genomes
We used hidden Markov models (HMMs) to detect remote homologs of the domains of our SSPs in 22 genomes. The method is based on HMM-HMM comparison as implemented in the HHsearch package (version 1.5.1)29. Pre-built HMMs for all proteins in 22 genomes (Agrobacterium tumefaciens, Arabidopsis thaliana, Bacillus subtilis, Bartonella henselae, Corynebacterium diphtheriae, Desulfitobacterium hafniense, Drosophila melanogaster, Escherichia coli, Homo sapiens, Lactobacillus casei, Mus musculus, Neisseria meningitidis, Plasmodium falciparum, Pseudomonas aeruginosa, Saccharomyces cerevisiae, Staphylococcus aureus, Streoptococcus pneumoniae, Streptomyces coelicolor, Sulfolobus solfataricus, Synechococcus C9311, Thermoplasma acidophilum, Yersinia pestis) were downloaded from the HHsearch FTP site (toolkit.lmb.uni-muenchen.de/HHsearch/). For each SSP family, 3 HMMs were built based on the NCBI “nr” (non-redundant) database (downloaded 10.27.2009): one for the full chain, one for the AB-type domain, and one for the BA-type domain, initializing by a multiple structural alignment by MUSTANG66. The 22 genomes were scanned by the 3 HMMs as queries. Hits to the AB- and BA-type domain HMMs with a probability >80% as reported by HHsearch were accepted as true hits unless the full chain HMM also hit the same protein with >50% probability. The accepted hits were subjected to further filtering based on the alignment provided by HMMsearch: in order to pass, at least 4 residues of both segments (A and B) of the domain had to be aligned and the alignment coverage had to be >2/3 for each segment. Hits that were found to be homologous to the entire segment-swapped protein (i.e. an ABAB pattern) were excluded. Also, hits containing patterns other than just AB or BA (e.g. BAB, ABBA, etc.) were excluded. One reason for doing so is that we work with sequence information here, and if a complex pattern such as BAB is seen, there is no way to tell what type of structure (i.e. AB- or BA-type fold) the pattern is associated with.
The phylogenetic distribution of the hits was analyzed using a phylogenetic tree built using the NCBI Taxonomy database. The earliest appearance of a fold is assigned to the highest level of the tree corresponding to taxonomic terms common to all hits associated with the fold. A fold at higher level of the tree is considered younger.
Searching for structural analogs in the PDB
To estimate relative fold ages of AB- and BA-type domains of SSPs, we scanned ReprPDB for continuous domains that are structurally similar to the domains under study. Each two-domain SSP was divided into 4 segments (A, B, A’, B’) as described earlier. The structures formed by segments (A, B) and (A’, B’) were considered as AB-type domains and (B, A) and (B’, A’) as BA-type domains. These 4 structures were matched against the continuous domains in ReprPDB using TM-ALIGN23.
A domain from ReprPDB was accepted as similar to a particular AB- or BA-type domain if the TM-score was >0.5 and the coverage of the alignment was >2/3 in each segment, with at least 4 residues aligned in each segment. Proteins that were found to be structurally similar (TM-score >0.5, coverage >2/3) to the entire query SSP were removed from the hits because they do not carry new information.
To detect circularly permuted versions of the domains, a similar procedure was applied but one of the structures was duplicated before structural alignment. A sliding window of length equal to that of the original structure was used to select the best alignment; a circular permutation was identified when the TM-score was >0.5 and the alignment coverage was >2/3 in both the N- and C-terminal segments.
Searching for homodimeric analogs in the PDB
To find homodimeric analogs of segment-swapped monomers in the PDB, we first constructed a data set containing homodimeric proteins in the PDB. This data set is a union of 2 sets: (1) PDB entries containing exactly 2 polypeptide chains with >95% sequence identity (13,774 entries), and (2) PDB entries identified as homodimers by the PISA online service67 (12,141 entries). The unified data set contained 19,906 PDB entries. To find homodimeric analogs of SSPs, the central proteins of the 18 families were matched against the homodimers using TM-ALIGN, normalizing by the average length of the 2 structures. Hits with TM-score >0.5 were ranked and the top hits were visually inspected.
Detecting and comparing variants of SSPs
To detect variants of SSPs based on the same fold, a pairwise comparison of all domains of the central proteins of the 18 families was performed. The goal was to find protein pairs where the continuous domains are similar (TM-score > 0.5) and the discontinuous domains are circular permutants of each other (i.e. the TM-score is below 0.5 for a direct comparison but above 0.5 after circular permutation) or vice versa. The TM-score was normalized by the mean (shorter) length of the 2 compared domains for a stricter (looser) criterion for structural similarity.
Supplementary Material
Acknowledgments
We thank László Barna and Dániel Györffy for helpful discussions. This work was supported by the Hungarian Scientific Research Fund (PD73096, NK77978); the Hungarian National Office for Research and Technology (NKFP_07_01_-MASPOK07); the National Science Foundation (DBI 1027394); and the National Institute of General Medical Sciences (GM083107, GM084222).
Abbreviations used
- SSP
segment-swapped protein
- DSF
domain swapping and fusion
- CP
circular permutation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abeln S, Deane CM. Fold usage on genomes and protein fold evolution. Proteins. 2005;60:690–700. doi: 10.1002/prot.20506. [DOI] [PubMed] [Google Scholar]
- 2.Deeds EJ, Shakhnovich EI. A structure-centric view of protein evolution, design, and adaptation. Adv Enzymol Relat Areas Mol Biol. 2007;75:133–191. doi: 10.1002/9780471224464.ch2. xi. [DOI] [PubMed] [Google Scholar]
- 3.Koonin EV. The Biological Big Bang model for the major transitions in evolution. Biol Direct. 2007;2:21. doi: 10.1186/1745-6150-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rost B. Did evolution leap to create the protein universe? Curr Opin Struct Biol. 2002;12:409–416. doi: 10.1016/s0959-440x(02)00337-8. [DOI] [PubMed] [Google Scholar]
- 5.Lupas AN, Ponting CP, Russell RB. On the evolution of protein folds: are similar motifs in different protein folds the result of convergence, insertion, or relics of an ancient peptide world? J Struct Biol. 2001;134:191–203. doi: 10.1006/jsbi.2001.4393. [DOI] [PubMed] [Google Scholar]
- 6.Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 7.Taylor WR. Evolutionary transitions in protein fold space. Curr Opin Struct Biol. 2007;17:354–361. doi: 10.1016/j.sbi.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Lindqvist Y, Schneider G. Circular permutations of natural protein sequences: structural evidence. Curr Opin Struct Biol. 1997;7:422–427. doi: 10.1016/s0959-440x(97)80061-9. [DOI] [PubMed] [Google Scholar]
- 9.Lo W-C, Lee C-C, Lee C-Y, Lyu P-C. CPDB: a database of circular permutation in proteins. Nucleic Acids Res. 2009;37:D328–D332. doi: 10.1093/nar/gkn679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogel C, Morea V. Duplication, divergence and formation of novel protein topologies. Bioessays. 2006;28:973–978. doi: 10.1002/bies.20474. [DOI] [PubMed] [Google Scholar]
- 11.Bashton M, Chothia C. The geometry of domain combination in proteins. J Mol Biol. 2002;315:927–939. doi: 10.1006/jmbi.2001.5288. [DOI] [PubMed] [Google Scholar]
- 12.Basu MK, Carmel L, Rogozin IB, Koonin EV. Evolution of protein domain promiscuity in eukaryotes. Genome Res. 2008;18:449–461. doi: 10.1101/gr.6943508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiner J, 3rd, Moore AD, Bornberg-Bauer E. Just how versatile are domains? BMC Evol Biol. 2008;8:285. doi: 10.1186/1471-2148-8-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. Structure, function and evolution of multidomain proteins. Curr Opin Struct Biol. 2004;14:208–216. doi: 10.1016/j.sbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Bennett MJ, Choe S, Eisenberg D. Domain swapping: entangling alliances between proteins. Proc Natl Acad Sci U S A. 1994;91:3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gronenborn AM. Protein acrobatics in pairs--dimerization via domain swapping. Curr Opin Struct Biol. 2009;19:39–49. doi: 10.1016/j.sbi.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaskólski M. 3D domain swapping, protein oligomerization, and amyloid formation. Acta Biochim Pol. 2001;48:807–827. [PubMed] [Google Scholar]
- 19.Bennett MJ, Eisenberg D. The evolving role of 3D domain swapping in proteins. Structure. 2004;12:1339–1341. doi: 10.1016/j.str.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y. I-TASSER: fully automated protein structure prediction in CASP8. Proteins. 2009;77 Suppl 9:100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreeva A, Murzin AG. Evolution of protein fold in the presence of functional constraints. Curr Opin Struct Biol. 2006;16:399–408. doi: 10.1016/j.sbi.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Guo J-t, Xu D, Kim D, Xu Y. Improving the performance of DomainParser for structural domain partition using neural network. Nucleic Acids Res. 2003;31:944–952. doi: 10.1093/nar/gkg189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, Zhang Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics. 2010;26:889–895. doi: 10.1093/bioinformatics/btq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell RB, Saqi MA, Sayle RA, Bates PA, Sternberg MJ. Recognition of analogous and homologous protein folds: analysis of sequence and structure conservation. J Mol Biol. 1997;269:423–439. doi: 10.1006/jmbi.1997.1019. [DOI] [PubMed] [Google Scholar]
- 26.Dias JM, Than ME, Humm A, Huber R, Bourenkov GP, Bartunik HD, Bursakov S, Calvete J, Caldeira J, Carneiro C, Moura JJ, Moura I, Romao MJ. Crystal structure of the first dissimilatory nitrate reductase at 1.9 A solved by MAD methods. Structure. 1999;7:65–79. doi: 10.1016/s0969-2126(99)80010-0. [DOI] [PubMed] [Google Scholar]
- 27.Stevens TJ, Paoli M. RCC1-like repeat proteins: a pangenomic, structurally diverse new superfamily of beta-propeller domains. Proteins. 2008;70:378–387. doi: 10.1002/prot.21521. [DOI] [PubMed] [Google Scholar]
- 28.Winstanley HF, Abeln S, Deane CM. How old is your fold? Bioinformatics. 2005;21 Suppl 1:449–458. doi: 10.1093/bioinformatics/bti1008. [DOI] [PubMed] [Google Scholar]
- 29.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 30.Govindarajan S, Goldstein RA. Why are some proteins structures so common? Proc Natl Acad Sci U S A. 1996;93:3341–3345. doi: 10.1073/pnas.93.8.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Helling R, Tang C, Wingreen N. Emergence of preferred structures in a simple model of protein folding. Science (80-) 1996;273:666–669. doi: 10.1126/science.273.5275.666. [DOI] [PubMed] [Google Scholar]
- 32.Weiner J, 3rd, Bornberg-Bauer E. Evolution of circular permutations in multidomain proteins. Mol Biol Evol. 2006;23:734–743. doi: 10.1093/molbev/msj091. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Rickert M, Garcia KC. Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science. 2005;310:1159–1163. doi: 10.1126/science.1117893. [DOI] [PubMed] [Google Scholar]
- 34.Norman DG, Barlow PN, Baron M, Day AJ, Sim RB, Campbell ID. Three-dimensional structure of a complement control protein module in solution. J Mol Biol. 1991;219:717–725. doi: 10.1016/0022-2836(91)90666-t. [DOI] [PubMed] [Google Scholar]
- 35.Lee J, Thompson JR, Botuyan MV, Mer G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat Struct Mol Biol. 2008;15:109–111. doi: 10.1038/nsmb1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ponting CP. Tudor domains in proteins that interact with RNA. Trends Biochem Sci. 1997;22:51–52. doi: 10.1016/s0968-0004(96)30049-2. [DOI] [PubMed] [Google Scholar]
- 37.Schubert HL, Raux E, Wilson KS, Warren MJ. Common chelatase design in the branched tetrapyrrole pathways of heme and anaerobic cobalamin synthesis. Biochemistry. 1999;38:10660–10669. doi: 10.1021/bi9906773. [DOI] [PubMed] [Google Scholar]
- 38.Park H, Hilsenbeck JL, Kim HJ, Shuttleworth WA, Park YH, Evans JN, Kang C. Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state. Mol Microbiol. 2004;51:963–971. doi: 10.1046/j.1365-2958.2003.03885.x. [DOI] [PubMed] [Google Scholar]
- 39.Eschenburg S, Schönbrunn E. Comparative X-ray analysis of the un-liganded fosfomycin-target murA. Proteins. 2000;40:290–298. doi: 10.1002/(sici)1097-0134(20000801)40:2<290::aid-prot90>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 40.Muraoka S, Okumura R, Ogawa N, Nonaka T, Miyashita K, Senda T. Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J Mol Biol. 2003;328:555–566. doi: 10.1016/s0022-2836(03)00312-7. [DOI] [PubMed] [Google Scholar]
- 41.Mathews MA, Schubert HL, Whitby FG, Alexander KJ, Schadick K, Bergonia HA, Phillips JD, Hill CP. Crystal structure of human uroporphyrinogen III synthase. EMBO J. 2001;20:5832–5839. doi: 10.1093/emboj/20.21.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hofmann A, Delmer DP, Wlodawer A. The crystal structure of annexin Gh1 from Gossypium hirsutum reveals an unusual S3 cluster. Eur J Biochem. 2003;270:2557–2564. doi: 10.1046/j.1432-1033.2003.03612.x. [DOI] [PubMed] [Google Scholar]
- 43.Li S, Lu Y, Peng B, Ding J. Crystal structure of human phosphoribosylpyrophosphate synthetase 1 reveals a novel allosteric site. Biochem J. 2007;401:39–47. doi: 10.1042/BJ20061066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zweifel ME, Leahy DJ, Barrick D. Structure and Notch receptor binding of the tandem WWE domain of Deltex. Structure. 2005;13:1599–1611. doi: 10.1016/j.str.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 45.Grimm C, de Ayala Alonso AG, Rybin V, Steuerwald U, Ly-Hartig N, Fischle W, Müller J, Müller CW. Structural and functional analyses of methyl-lysine binding by the malignant brain tumour repeat protein Sex comb on midleg. EMBO Rep. 2007;8:1031–1037. doi: 10.1038/sj.embor.7401085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ylianttila MS, Pursiainen NV, Haapalainen AM, Juffer AH, Poirier Y, Hiltunen JK, Glumoff T. Crystal structure of yeast peroxisomal multifunctional enzyme: structural basis for substrate specificity of (3R)-hydroxyacyl-CoA dehydrogenase units. J Mol Biol. 2006;358:1286–1295. doi: 10.1016/j.jmb.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 47.Gopal B, Madan LL, Betz SF, Kossiakoff AA. The crystal structure of a quercetin 2,3-dioxygenase from Bacillus subtilis suggests modulation of enzyme activity by a change in the metal ion at the active site(s) Biochemistry. 2005;44:193–201. doi: 10.1021/bi0484421. [DOI] [PubMed] [Google Scholar]
- 48.Lunzer M, Miller SP, Felsheim R, Dean AM. The biochemical architecture of an ancient adaptive landscape. Science. 2005;310:499–501. doi: 10.1126/science.1115649. [DOI] [PubMed] [Google Scholar]
- 49.Imada K, Sato M, Tanaka N, Katsube Y, Matsuura Y, Oshima T. Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 A resolution. J Mol Biol. 1991;222:725–738. doi: 10.1016/0022-2836(91)90508-4. [DOI] [PubMed] [Google Scholar]
- 50.Huang Y, Fang J, Bedford MT, Zhang Y, Xu R-M. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 51.Wang WK, Tereshko V, Boccuni P, MacGrogan D, Nimer SD, Patel DJ. Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets. Structure. 2003;11:775–789. doi: 10.1016/s0969-2126(03)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leiros H-KS, McSweeney SnM. The crystal structure of DR2241 from Deinococcus radiodurans at 1.9 A resolution reveals a multi-domain protein with structural similarity to chelatases but also with two additional novel domains. J Struct Biol. 2007;159:92–102. doi: 10.1016/j.jsb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 53.Cedergren-Zeppezauer ES, Larsson G, Nyman PO, Dauter Z, Wilson KS. Crystal structure of a dUTPase. Nature. 1992;355:740–743. doi: 10.1038/355740a0. [DOI] [PubMed] [Google Scholar]
- 54.Baldo AM, McClure MA. Evolution and horizontal transfer of dUTPase-encoding genes in viruses and their hosts. J Virol. 1999;73:7710–7721. doi: 10.1128/jvi.73.9.7710-7721.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tarbouriech N, Buisson M, Seigneurin JM, Cusack S, Burmeister WP. The monomeric dUTPase from Epstein-Barr virus mimics trimeric dUTPases. Structure. 2005;13:1299–1310. doi: 10.1016/j.str.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 56.Ding F, Prutzman KC, Campbell SL, Dokholyan NV. Topological determinants of protein domain swapping. Structure. 2006;14:5–14. doi: 10.1016/j.str.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 57.Alva V, Ammelburg M, Söding J, Lupas AN. On the origin of the histone fold. BMC Struct Biol. 2007;7:17. doi: 10.1186/1472-6807-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uliel S, Fliess A, Unger R. Naturally occurring circular permutations in proteins. Protein Eng. 2001;14:533–542. doi: 10.1093/protein/14.8.533. [DOI] [PubMed] [Google Scholar]
- 59.Lo W-C, Lyu P-C. CPSARST: an efficient circular permutation search tool applied to the detection of novel protein structural relationships. Genome Biol. 2008;9:R11. doi: 10.1186/gb-2008-9-1-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abraham A-L, Pothier J, Rocha EPC. Alternative to homo-oligomerisation: the creation of local symmetry in proteins by internal amplification. J Mol Biol. 2009;394:522–534. doi: 10.1016/j.jmb.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 61.Dokholyan NV, Shakhnovich EI. Understanding hierarchical protein evolution from first principles. J Mol Biol. 2001;312:289–307. doi: 10.1006/jmbi.2001.4949. [DOI] [PubMed] [Google Scholar]
- 62.Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. doi: 10.1038/nature01256. [DOI] [PubMed] [Google Scholar]
- 63.Balaji S, Srinivasan N. Comparison of sequence-based and structure-based phylogenetic trees of homologous proteins: Inferences on protein evolution. J Biosci. 2007;32:83–96. doi: 10.1007/s12038-007-0008-1. [DOI] [PubMed] [Google Scholar]
- 64.Wang G, Dunbrack RL., Jr PISCES: a protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 65.Heyer LJ, Kruglyak S, Yooseph S. Exploring expression data: identification and analysis of coexpressed genes. Genome Res. 1999;9:1106–1115. doi: 10.1101/gr.9.11.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 67.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 68.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.