Abstract
The human immunodeficiency virus type 1 (HIV-1) Rev protein is essential for the virus because it promotes nuclear export of alternatively processed mRNAs, and Rev is also linked to translation of viral mRNAs and genome encapsidation. Previously, the human DEAD-box helicase DDX1 was suggested to be involved in Rev functions, but this relationship is not well understood. Biochemical studies of DDX1 and its interactions with Rev and model RNA oligonucleotides were carried out to investigate the molecular basis for association of these components. A combination of gel-filtration chromatography and circular dichroism spectroscopy demonstrated that recombinant DDX1 expressed in Escherichia coli is a well-behaved folded protein. Binding assays using fluorescently labeled Rev and cell-based immunoprecipitation analysis confirmed a specific RNA-independent DDX1–Rev interaction. Additionally, DDX1 was shown to be an RNA-activated ATPase, wherein Rev-bound RNA was equally effective at stimulating ATPase activity as protein-free RNA. Gel mobility shift assays further demonstrated that DDX1 forms complexes with Rev-bound RNA. RNA silencing of DDX1 provided strong evidence that DDX1 is required for both Rev activity and HIV production from infected cells. Collectively, these studies demonstrate a clear link between DDX1 and HIV-1 Rev in cell-based assays of HIV-1 production and provide the first demonstration that recombinant DDX1 binds Rev and RNA and has RNA-dependent catalytic activity.
Abbreviations: HIV-1, human immunodeficiency virus type 1; RRE, Rev response element; BSA, bovine serum albumin; siRNA, small interfering RNA; CTE, constitutive transport element; PBS, phosphate-buffered saline
Keywords: HIV, DEAD-box, Rev, DDX1, RNA trafficking
Introduction
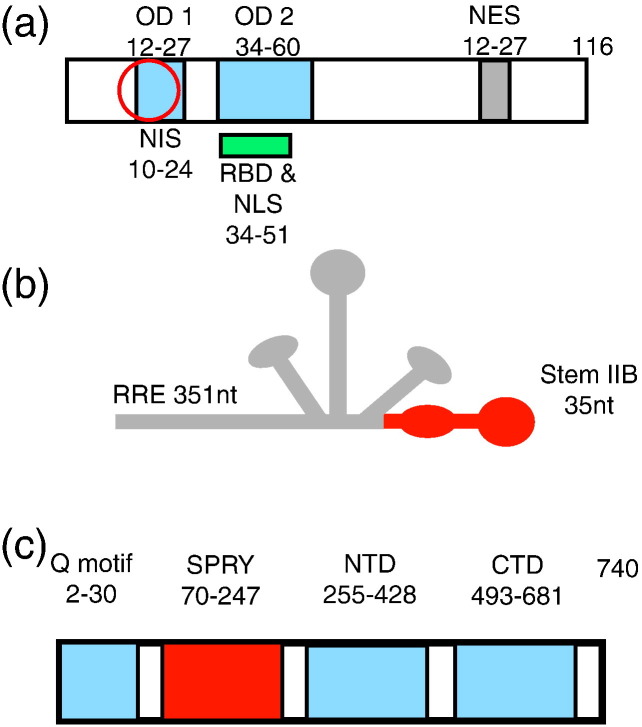
After cellular entry and genomic integration by the human immunodeficiency virus type 1 (HIV-1), the HIV-1 Rev protein regulates the timing of viral gene expression that ensures completion of the viral life cycle.1, 2, 3 Rev functions in part by promoting the nuclear export of unspliced and incompletely spliced viral mRNAs, resulting in the expression of the late-stage genes and providing genomic RNA for incorporation into virions. For this function, Rev overcomes cellular restrictions that prevent the nuclear export of incompletely processed RNA. Promotion of nuclear transport occurs through oligomeric binding of Rev on the Rev response element (RRE) (Fig. 1 ), a specific binding site within the viral RNA, which targets the RNA for nuclear export by the CRM-1 export receptor pathway. Specific oligomerization of Rev on the RRE involves well-defined oligomerization domains (OD 1 and OD 2 in Fig. 1a) involving a helix–turn–helix motif. However, it is unknown how viral RNA is released from nuclear retention sites, and it is not certain what other cellular factors are required for nuclear export of viral mRNA. Furthermore, Rev has also been linked to the translation of certain HIV-mRNAs4, 5, 6, 7, 8 and to encapsidation of the viral genome.6, 9, 10, 11, 12 The mechanisms of Rev activity in these functions and their required cellular factors are poorly understood. Furthermore, it remains to be determined how these Rev-accessory factors function in uninfected cells and how Rev utilizes their activities during infection.
Fig. 1.
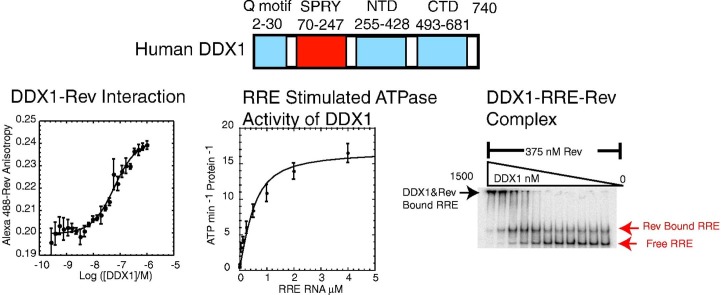
(a) HIV-1 Rev domain structure.3 The Rev oligomerization domains (OD 1 and OD 2) and the overlapping arginine-rich RNA binding domain (RBD) are highlighted in light blue and green, respectively. The overlapping NIS is indicated by a red circle. The Rev nuclear export sequence is highlighted in gray. (b) Secondary structure of the 351-nt RRE RNA from HIV-1. Stem IIB, the high-affinity site bound by HIV-1 Rev, is highlighted in red. (c) DDX1 domain structure. Domain boundaries were determined by sequence homology using Prosite from the ExPASy proteomics server (http://www.expasy.org/prosite/). DDX1 contains a unique SPRY domain inserted between the Q-motif and the N-terminal helicase domain, shown boxed here in red.
Members of the DEAD-box family of proteins are associated with all levels of RNA metabolism and function including transcription, pre-mRNA splicing, ribosome biogenesis, RNA transport, translation initiation, and RNA decay. Thus, it is not surprising that several DEAD-box proteins have been linked to Rev function in HIV-infected cells.13, 14, 15, 16, 17 However, the diverse cellular activities and biochemical properties of the DEAD-box family of proteins make it difficult to accurately predict the relationships between mechanism and function of its individual members in specific contexts. The human DEAD-box protein DDX1 has been proposed to contribute to viral replication by acting as a cofactor for Rev, and a region of Rev that might functionally interact with DDX1 has been identified.14 However, little is known about the intermolecular interactions and enzymatic activities of DDX1.
Because of their broad range of RNA-associated functions, there are many possible ways in which an individual DEAD-box protein could contribute to Rev and the HIV-1 life cycle. In particular, DDX1 is implicated in several host cell functions including RNA transcription,18, 19 pre-mRNA processing,20, 21 and mRNA translation.22, 23 DDX1 is expressed in essentially all cell types in mammals‡ , is primarily localized either within the nucleus20, 21 or in the cytoplasm13 depending on the cell type, and has been observed as a nucleocytoplasmic shuttling protein. Therefore, it is possible that DDX1 contributes to multiple aspects of Rev function.
In addition to their diverse cellular functions, DEAD-box proteins are associated with a broad range of biochemical activities. DEAD-box proteins commonly function as ATP-dependent RNA helicases, but the enzymatic activity of these proteins can vary widely.24, 25, 26 The typical DEAD-box protein contains a helicase core consisting of nine conserved motifs distributed between N-terminal (NTD) and C-terminal (CTD) RecA-like domains. Sequence diversity among DEAD-box proteins is found in N- and C-terminal extensions of the helicase core and in the flexible linker connecting the two RecA-like domains. DEAD-box proteins frequently bind nonspecifically to regions of double-stranded RNA and mediate conformational changes in the RNA in an ATP-dependent manner. RNA binding and conformational changes are coupled with protein conformational changes within the helicase core that are linked to a cycle of ATP binding and hydrolysis. While this is a general model for activity, there is significant variability in RNA specificity and catalytic parameters of ATP hydrolysis among DEAD-box family members. Values of k cat and K m (ATP) can range over 2 orders of magnitude,24, 26 and therefore, understanding a particular DEAD-box protein requires study of its own catalytic parameters, as well as investigation of other cellular factors that contribute to its function.
Studies of DDX1 enzymatic activity have thus far been disparate. Interestingly, while preparations enriched in recombinant DDX1 can hydrolyze ATP and unwind double-stranded RNA when extracted from cultured Sf21 cells,27 recombinant DDX1 has been reported to lack ATP-dependent RNA-unwinding activity when purified from Escherichia coli.27, 28 In addition, DDX1 is unique among DEAD-box proteins, as it includes an SPRY domain inserted between the Q-motif and the NTD, which may mediate protein–protein association29, 30 and enzymatic activity (Fig. 1). DDX1 interacts with an N-terminal sequence of the Rev oligomerization domain [termed NIS (Nuclear Diffusion Inhibitory Signal)] (Fig. 1) in yeast-two-hybrid experiments14 and has also been proposed to bind specifically to RRE RNA from interpretations of cell-based assays.14 However, quantitative binding data have not been obtained for direct interactions between DDX1 and Rev or DDX1 and RRE-containing RNA.
Because of its association with HIV-1, DDX1 could make an attractive target for the development of novel therapeutics. However, a more through understanding of the relationships between DDX1, Rev function, and the viral life cycle is necessary. Here, we describe how DDX1 interacts with the HIV-1 Rev protein by quantitative binding assays and quantify the enzymatic parameters of DDX1 ATPase activity in the presence of RRE-related RNAs. These RNAs included a 351-nt oligomer comprising the viral RRE sequence31 and a 35-nt oligomer representing stem IIB,32 the initial site of Rev binding33, 34 (Fig. 1). In addition, we describe quantitative measurements of both a Rev-dependent transactivation assay and a viral replication assay in cells where DDX1 is down-regulated by RNA interference. Collectively, the results of these experiments provide a clear link between DDX1, Rev function, and the HIV-1 lifecycle. Furthermore, they provide the most-detailed characterizations to date of molecular interactions with DDX1 and its enzymatic properties. These results will serve as an important platform for future studies of DDX1 mechanism and function.
Results
Expression, purification, and characterization of recombinant DDX1
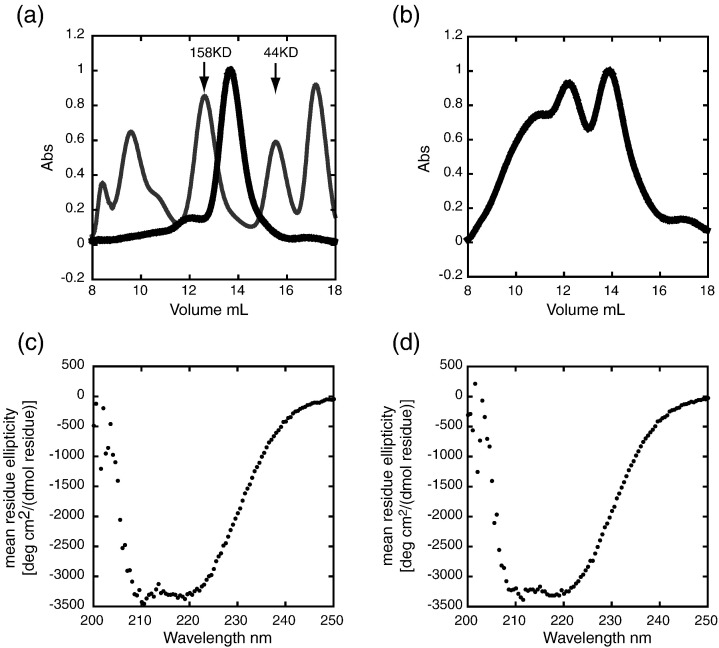
The human DDX1 was expressed in E. coli and purified as described in Materials and Methods. Because of the wide range in enzymatic activity observed for DEAD-box proteins, it was important to determine if the recombinant DDX1 was compactly folded prior to enzymatic characterization. Gel-filtration analysis revealed that DDX1 eluted from the column in a single peak in 500 mM KCl, with the retention expected for a monomeric ∼ 80-kDa protein (Fig. 2 ). A small shoulder consistent with dimeric DDX1 was also visible, but this represented only a small fraction of the total protein. In contrast, DDX1 eluted as several peaks in 150 mM KCl corresponding to monomer, dimer, and larger aggregate. This chromatogram demonstrated that 10 μM recombinant DDX1 self-associates in a salt-sensitive manner at 25 °C. However, in neither case did DDX1 primarily elute in the void volume as would be expected for a denatured protein, indicating that recombinant DDX1 is folded in a compact conformation.
Fig. 2.
Gel filtration and CD spectra of 10 μM DDX1. (a) DDX1 elution from a Superdex 200 column in 500 mM KCl at 25 °C. Absorption spectra were normalized to the highest point. The primary elution peak is consistent with the expected retention for an ∼ 80-kDa protein, corresponding to monomeric DDX1. Superimposed on this elution profile is the independent elution of the molecular mass standards used to calibrate the column (gray). The 158-kDa and 44-kDa standards are indicated by arrows. (b) DDX1 elution in 150 mM KCl at 25 °C. Peaks centered at 14-, 12-, and 11-ml retention times are consistent with monomeric DDX1, dimeric DDX1, and higher-order aggregates, respectively. (c) CD spectra of DDX1 in 500 mM KCl at 25 °C. The negative peaks near 210 nm and 219 nm are consistent with a protein containing mixed α-helix and β-sheet secondary structure. (d) CD spectra of DDX1 in 150 mM KCl at 25 °C. This spectrum is identical within error to that of DDX1 in 500 mM KCl.
Circular dichroism (CD) was used to further investigate the folded state of DDX1 at 25 °C using the same samples analyzed by gel-filtration chromatography (Fig. 2). The spectra of DDX1 in both 500 mM and 150 mM KCl are essentially identical, within experimental error. The negative signals at lower wavelengths are consistent with the presence of mixed α-helical and β-sheet content.35 These spectra are also similar to those reported for other DEAD-box proteins.36 The secondary structure of DDX1 is not KCl dependent, as judged by CD, indicating that the self-association observed at lower KCl concentration is not due to a gross unfolding of the protein. Thus, by both gel filtration and CD spectroscopy, recombinant DDX1 is folded at 25 °C.
Interaction between DDX1 and Rev proteins
Previous cell-based studies and yeast-two-hybrid experiments suggested that DDX1 and Rev form a complex in vivo.14 However, the interaction has not been characterized biochemically, and the binding affinity of the Rev–DDX1 complex has not been measured. As described in the previous section, DDX1 has a salt-sensitive tendency to self-associate above micromolar concentrations. Additionally, Rev has a well-known tendency for aggregation, precipitation, and fibril formation in vitro.37 These inconvenient properties of both proteins precluded assay of the protein–protein interaction by methods such as isothermal titration calorimetry or surface plasmon resonance. Instead, a direct titration assay was developed that monitored the change in fluorescence anisotropy of an Alexa-488-labeled Rev protein in response to DDX1 binding (Fig. 3 and Table 1 ). This assay had the distinct advantage of enabling direct detection of Rev protein at nanomolar concentrations that are below the threshold of self-association.
Fig. 3.
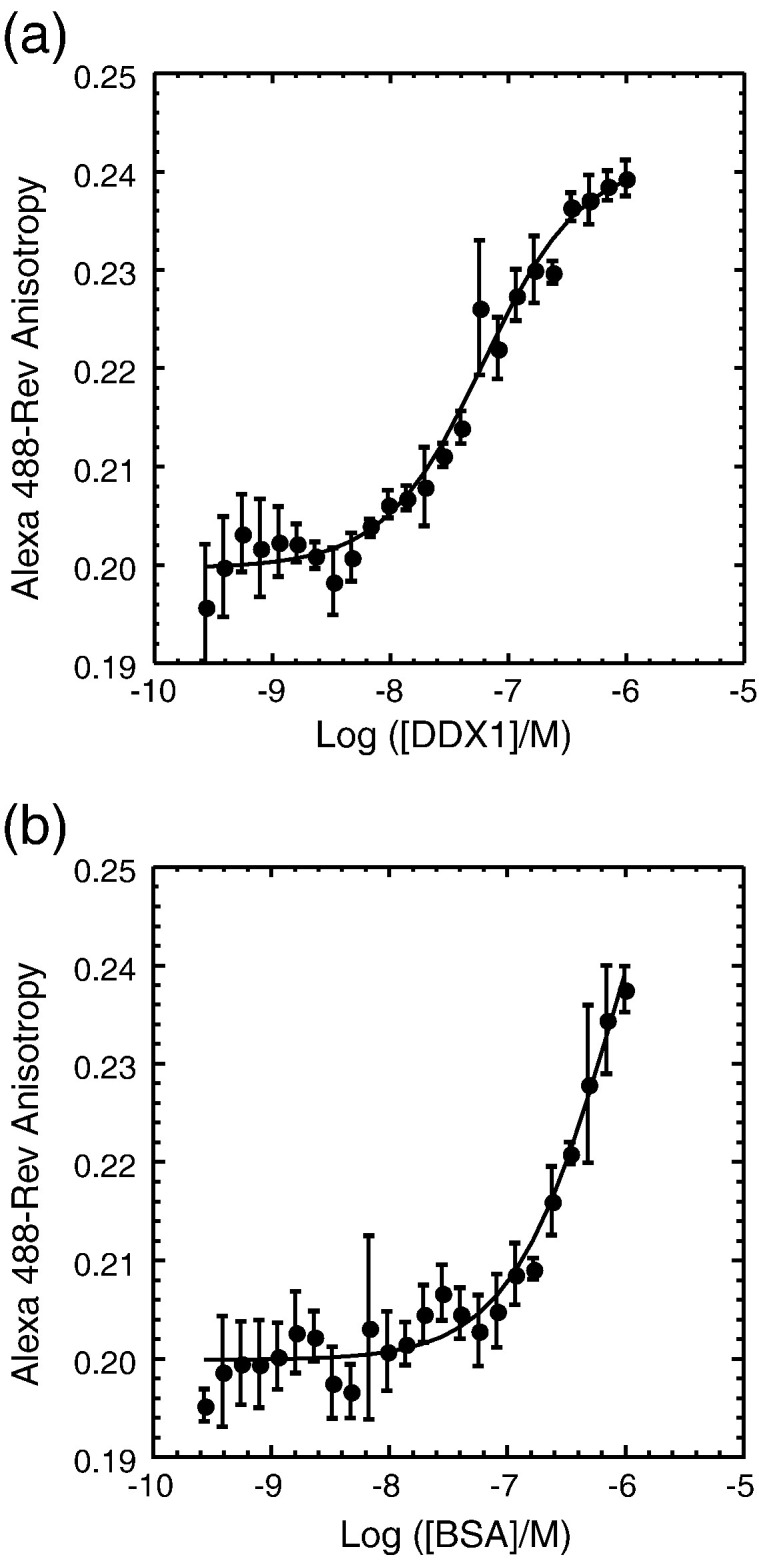
DDX1 and HIV-1 Rev form a specific complex in vitro. (a) Full-length DDX1 forms a specific complex with Alexa 488-Rev as monitored by fluorescence anisotropy. DDX1 binds Rev with a measured Kd = 36 nM. (b) BSA binds Rev relatively weakly, indicated by the isotherm that does not reach an upper baseline even at [BSA] > 500 nM.
Table 1.
Kd values for Rev protein interactions
| Rev–DDX1 (nM) | 36 ± 1 |
| Rev–DDX1 (residues 1–428) (nM) | 43 ± 6 |
| Rev–DDX1 (2 mM AMPPNP) (nM) | 53 ± 1 |
| Rev–DDX1 (2 mM ADP) (nM) | 14.7 ± 0.6 |
| Rev-BSA (nonspecific control) (nM) | > 500 |
Increasing concentrations of DDX1 were titrated into 10 nM Alexa-488-labeled Rev over a range of nanomolar to 1 μM at 25 °C. The resulting isotherm was analyzed using the quadratic form of a single-site binding model because the observed binding constant was close to the concentration of labeled Rev protein. Saturable binding was observed in this concentration range, and the dissociation constant (K d) for the Rev–DDX1 complex was measured as 36 nM using the fluorescence anisotropy method. In contrast, labeled Rev only weakly interacted with bovine serum albumin (BSA), and the binding isotherm did not saturate at concentrations up to 1 μM BSA. Titrations conducted in the presence of either the non-hydrolyzable ATP analog Adenylyl Imidodiphosphate or ADP revealed modest effects on DDX1 binding, with dissociation constants of 53 nM and 14 nM in the presence of Adenylyl Imidodiphosphate and ADP, respectively.
Direct titrations were also used to further map the interaction of labeled Rev with truncated DDX1 constructs by fluorescence spectroscopy. DDX1 (1–428), with the C-terminal RecA domain removed, binds to Rev with a K d of 43 nM, which is essentially the same K d observed for full-length DDX1, indicating that the Rev binding domain is contained entirely within the N-terminal region of DDX1.
Cell-based assays of DDX1–Rev interaction
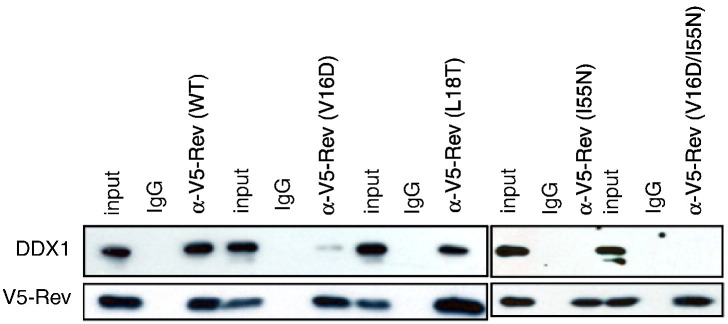
For us to demonstrate that DDX1 and Rev directly interact in a cellular environment without contact through a bridging RNA, HeLa cells were transfected with an expression plasmid for V5-tagged Rev protein, and cell lysates were treated with RNase prior to immunoprecipitation of Rev with anti-V5 antibody (Fig. 4 ). Western blotting with anti-Rev and anti-DDX1 clearly demonstrated that DDX1 and Rev were pulled down by anti-V5, whereas IgG did not adsorb either protein. Rev self-associates into higher-order oligomers through a helix–turn–helix oligomerization domain,38, 39 and oligomerization is proposed to contribute to both nuclear import of Rev from the cytoplasm1, 40 and nuclear export of Rev–RRE complexes via the Crm1 pathway.2 Additionally, the first helix of the Rev oligomerization domain (OD 1) overlaps with the NIS (Fig. 1). The effects of mutations of apolar residues to polar residues within OD 1 (V16D and L18T) on the interaction between DDX1 and Rev were determined using the immunoprecipitation assay. The mutation I55N in (OD 2) (Fig. 1) was chosen because it is known to affect Rev oligomerization41, 42 while being outside of the NIS. The double mutation V16D/I55N was chosen because of its well-characterized effects on Rev oligomerization.41, 42 The V16D and I55N mutations highly attenuate oligomerization, and the double mutant protein remains primarily monomeric at micromolar concentrations.43 In contrast, the L18T mutation results in a protein that is less prone to oligomerize than wild-type Rev but is still capable of forming higher-order oligomers.41 The V16D, I55N, and V16D/I55N variants of Rev did not coimmunoprecipitate a significant quantity of DDX1 in the assay. However, the L18T variant was indistinguishable from wild-type Rev.
Fig. 4.
Coimmunoprecipitation of endogenous DDX1 with transfected Rev-V5 (WT) versus oligomerization-incompetent Rev-V5 mutants from HeLa cells. Western blot analysis of whole-cell extract (input) and immunoprecipitates of RNase-treated cell extracts with anti-V5-antibody or a control IgG by anti-DDX1 antibody or anti-V5-antibody. DDX1 associates with Rev (WT), but not with the Rev oligomerization-incompetent mutants Rev (V16D), Rev (I55N), or Rev (V16D/I55N).
These results indicate that mutation of the Rev oligomerization domain can reduce the interaction between DDX1 and Rev. It is not possible to determine whether the effects of the Rev mutations on DDX1 binding result from a direct loss interaction with the mutated residues or from an indirect disruption of the Rev structure. However, the L18T mutation within the NIS had a modest effect on DDX1 binding, while the I55N mutation outside of the NIS had a strong effect on DDX1 binding. This observation suggests that the interaction may require oligomeric Rev as well as specific contacts to the NIS.
RNA-dependent hydrolysis of ATP by DDX1
The RNA-dependent ATPase activity of DDX1 was measured using a spectroscopic molybdate binding assay.44, 45 In this assay, free phosphate generated from ATP hydrolysis reacts with molybdate, resulting in a blue complex when reduced with ascorbic acid. The resulting color is used to directly quantify the amount of free phosphate by measurement of absorption at 850 nm. The observed hydrolysis curves were consistent with single-exponential kinetics, and the Michaelis–Menten kinetic constants were determined from fits to the Briggs–Haldane equation (Fig. 5 ).
Fig. 5.
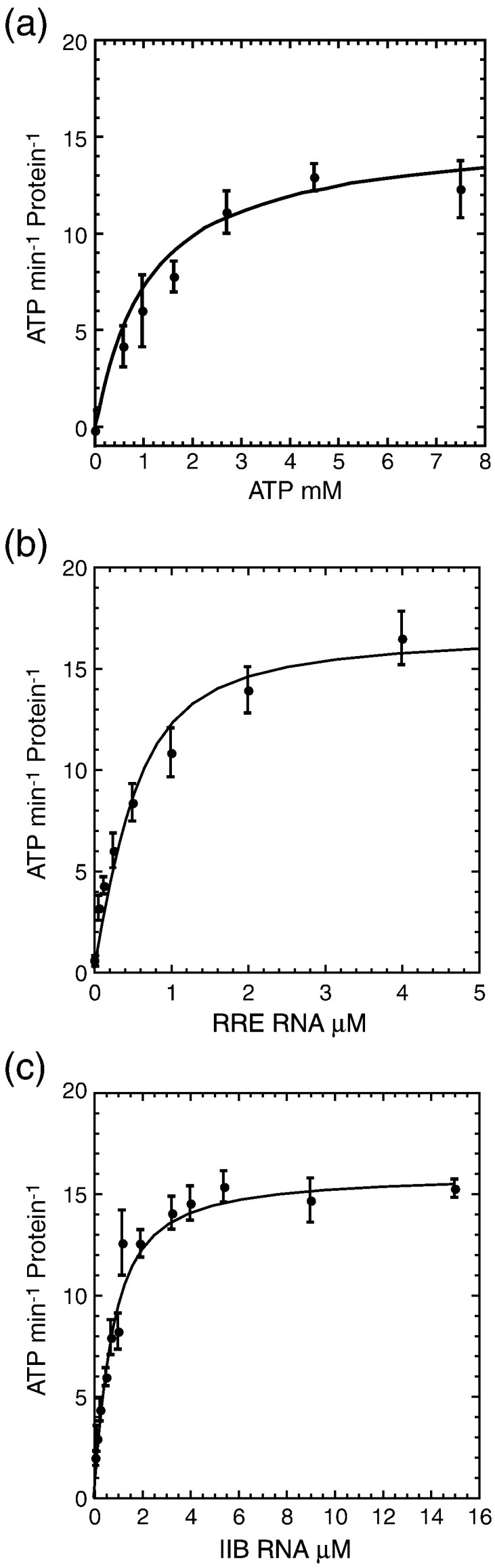
DDX1 ATPase activity is RNA dependent. (a) Spectroscopic ATPase activity of DDX1 in the presence of 15 μM stem IIB RNA measured as a function of ATP concentration. Michaelis kinetic hydrolysis constants (kcat = 15.9 ± 0.7 min− 1; Km = 1.0 ± 0.1 mM ATP) were calculated from a fit of the data to the Briggs–Haldane equation. Increasing concentrations of either (b) 351-nt RRE or (c) stem IIB RNA were titrated into DDX1 in the presence of 2.5 mM ATP. Hydrolysis curves plot the rate of ATP hydrolysis as a function of RNA concentration.
The rate of ATP hydrolysis for DDX1 was measured as a function of ATP concentration in the presence of 15 μM stem IIB RNA. DDX1 has a k cat value of 15.9 ± 0.7 min− 1 and an apparent K m (ATP) value of 1.0 ± 0.1 mM with this RNA. Similar values have been observed for several DEAD-box proteins.24 The determined K m (ATP) values are below cellular ATP concentrations, suggesting that ATP is saturating in vivo.
Three different RNA substrates were used to evaluate the RNA requirement for DDX1 ATPase activity. ATP hydrolysis was measured as a function of 351-nt RRE, stem IIB RNA, and yeast tRNAphe at 2.5 mM ATP. tRNA was used as a nonspecific control since it is highly structured and does not contain sequence homology to RRE RNA. Hydrolysis curves presented in Fig. 5 indicate that k max and K app (RNA) values for HIV-1-derived RNAs are similar within error, whereas tRNAphe is the less optimal RNA substrate for stimulating DDX1 ATPase activity than either RRE or stem IIB as k max (tRNA) is 77% of k max (RRE) and the K app (tRNA) was 10-fold larger than K app (RRE). This suggests that the virus-derived RNAs may represent specific RNA targets for DDX1. For all RNAs tested, the observed kinetic parameters are within the published range for other RNA-stimulated ATPase activity of DEAD-box proteins,5, 16, 46 although DDX1 is among the slowest reported. In terms of K app, the values for HIV-1-derived RNAs with DDX1 are near the middle values reported for DEAD-box protein–RNA values, while the K app values are near the high extreme values for DDX1-tRNAphe.5, 16, 46
The ATPase activity of DDX1 was measured in the context of the full 351-nt RRE from HIV-1; RNAs were incubated with DDX1 at three concentrations of Rev. The Rev–RRE interaction has been extensively characterized as Rev oligomers monomers that bind in a stepwise fashion to form oligomers with initial K d values in the sub-nanomolar range.33, 34, 41, 43, 47, 48 Under the assay conditions, multiple copies of Rev are bound to the RRE at any point where Rev is in excess of the RNA. The k max and K app (RRE) values at the various Rev concentrations are presented in Table 2 . Interestingly, Rev binding to RRE had no effect on DDX1 ATPase activity. This clearly indicates that Rev does not activate DDX1 activity, and this lack of inhibition demonstrates that Rev-bound RRE remains suitable for stimulating the RNA-dependent ATPase activity of DDX1. Furthermore, this implies that DDX1 forms a functional complex with both Rev and RRE.
Table 2.
Steady-state ATPase kinetics of DDX1
| ATP titration | kcat (min− 1) | Km (ATP) (mM) |
|---|---|---|
| 15.2 ± 0.6 | 1.0 ± 0.1 |
| RNA titration | kmax (min− 1) | Kapp (RNA) (μM) |
|---|---|---|
| IIB | 16.0 ± 0.4 | 0.50 ± 0.05 |
| RRE | 16.8 ± 0.6 | 0.24 ± 0.05 |
| RRE 0.5 μM Rev | 17 ± 1 | 0.2 ± 0.1 |
| RRE 1.0 μM Rev | 16 ± 1 | 0.3 ± 0.1 |
| RRE 2.0 μM Rev | 17 ± 1 | 0.3 ± 0.2 |
| tRNA | 13.5 ± 0.6 | 1.9 ± 0.3 |
Assembly of Rev–RRE–DDX1 complexes
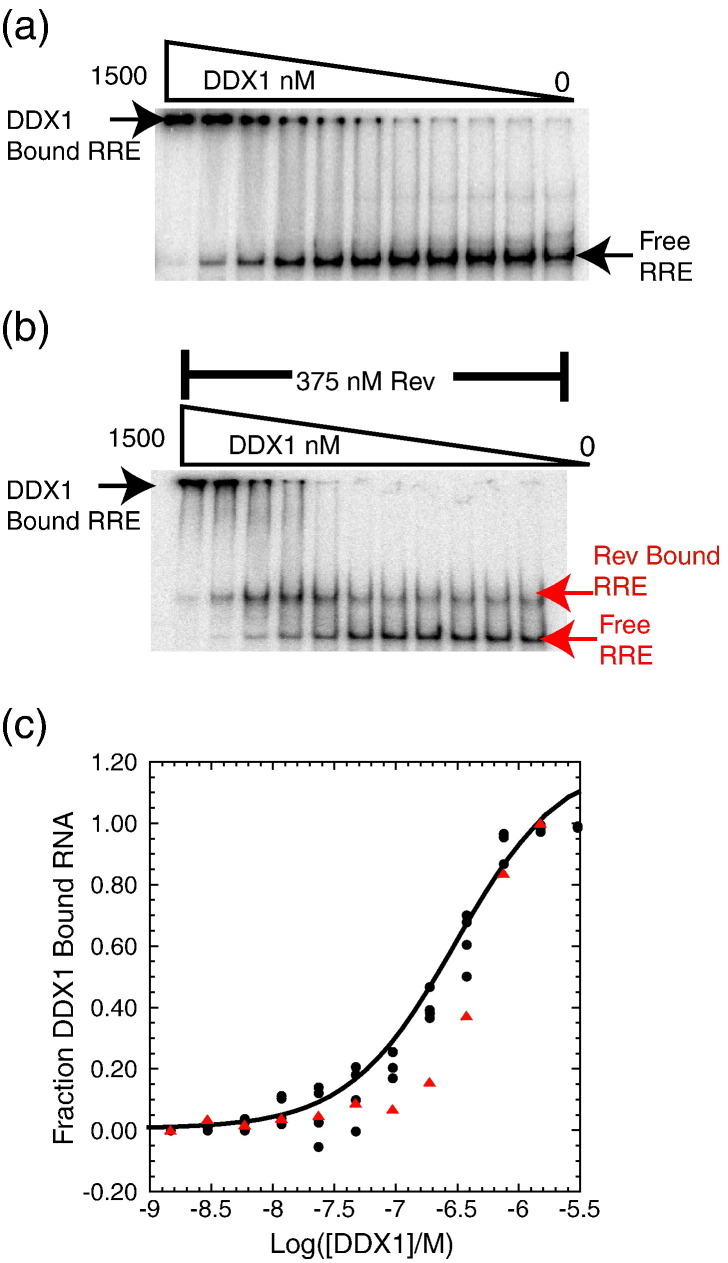
The formation of complexes between radiolabeled RRE RNA and DDX1 was monitored by an electrophoretic gel mobility shift assay. DDX1 was titrated over a concentration range from 0 to 1.5 μM into a trace amount of labeled RRE. Binding data were fit using a nonlinear least-squares regression to a single-transition binding model (Fig. 6 ). Because the DDX1–RRE complex did not migrate well in the gel, the reaction was quantified by following the disappearance of free RNA. The apparent K d for this interaction is 310 nM, which is similar to the K app (RNA) value of 240 nM observed for the RRE in the previous ATPase assay.
Fig. 6.
DDX1 and Rev bind independently to RRE RNA. (a) Electrophoretic gel mobility shift titration of DDX1 into radiolabeled RRE RNA, with the bands corresponding to free RRE, DDX1–RRE complex, and DDX1 concentration through the titration denoted. (b) DDX1 titration into radiolabeled RRE RNA in the presence of 375 nM Rev protein. Oligomeric Rev forms a tight complex with RRE RNA as indicated by the first shifted band. DDX1 binds independently to the Rev–RRE complex, seen as a supershifted band at increasing concentrations of DDX1. Bands corresponding to free RRE, Rev–RRE, and DDX1–Rev–RRE complexes are labeled. (c) Plot showing the fraction of DDX1 bound to labeled RRE RNA or Rev–RRE complex as a function of DDX1 concentration. The fraction of bound DDX1 was calculated as the disappearance of free RRE where black circles represent the direct titration of DDX1 in three independent trials described in (a) and red triangles denote the disappearance of free RRE and Rev–RRE bands described in (b).
To determine the effect of Rev on DDX1 binding to RRE RNA, we repeated gel mobility shift assays in the presence of 375 nM Rev (Fig. 6). Rev and RRE were incubated for 20 min prior to reaction with DDX1. In the absence of DDX1, two RRE populations are observable on the gel, including free RRE and a Rev–RRE complex consisting of multiple Rev copies. Reactions with DDX1 were quantified by summing the population of each RRE-containing band (Fig. 6). The observed DDX1–RRE interaction in the presence of Rev is within experimental variability for the interaction in the absence of Rev.
The results of the binding study support a model where Rev and DDX1 bind independently and simultaneously to RRE RNA through interactions between the RNA and the individual proteins. In the assay, DDX1 does not favor Rev-free RNA, it does not have an overall reduced affinity for RNA in the presence of Rev, and it does not reduce complex formation between Rev and the RRE. RNA binding that is independent of protein–protein interactions may arise because of the overlap between the NIS and OD 1 (Fig. 1). Under the conditions of our assay, Rev forms an oligomeric complex on the RRE that may sufficiently sequester the NIS and prevent the protein–protein interaction with DDX1.
Effect of DDX1 silencing on the cellular activity of Rev and viral replication
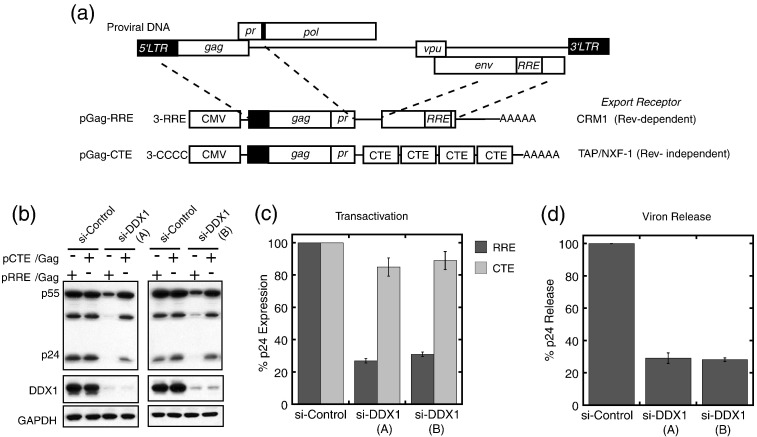
Previous studies have suggested a role for DDX1 in HIV-1 replication and Rev function in cultured cells using either antisense RNA or a single small interfering RNA (siRNA) targeting DDX1 mRNA.14 To exclude possible off-target effects from a single siRNA, we analyzed cells where DDX1 was silenced by either of two distinct siRNAs, which targeted sequences of DDX1 different from the one used in the previous studies.14 In addition, we simultaneously analyzed the effects of the siRNAs on expression of a control gene whose nuclear transport was not Rev dependent.
The Rev-dependent reporter plasmid (pGag-RRE; Fig. 7a) encodes the HIV-1 Gag/protease protein under the control of a cytomegalovirus promoter. Fused to this is a segment from the env gene that contains the RRE.31 Because the transcript contains a 5′ splice donor site but not a splice acceptor site, it accumulates in the nucleus in the absence of Rev.31 When cells are cotransfected with pGag-RRE and a plasmid encoding Rev protein, the binding of Rev to the RRE overcomes nuclear retention and specifies export by CRM1, a receptor that exports only a minor fraction of cellular mRNAs. Rev-dependent transactivation is monitored by the level of Gag protein expression. For comparison, a control plasmid was used containing four copies of the constitutive transport element (CTE) instead of the RRE (pGag-CTE). The CTE specifies nuclear export by the NXF-1 receptor that exports most cellular mRNAs and overcomes nuclear retention of the Gag transcript that normally occurs in the absence of Rev. Thus, in cells transfected with pGag-CTE, Gag production is independent of Rev.
Fig. 7.
DDX1 silencing in HeLa cells leads to selective loss of Rev–RRE-dependent Gag expression from an HIV-1 subgenomic reporter and inhibits virus production by infected cells. (a) Schematics of Gag constructs used for the screening of Rev–RRE and CTE transactivation. Both plasmids contain part of the 5′-long terminal repeat and the coding region for the Gag and PR domains of HIV-1. In addition, pGag-RRE contains part of the env gene containing the RRE, and pGag-CTE contains four copies of the CTE of Mason–Pfizer monkey virus. (b) Western blots of cells treated with control siRNA or two different siRNAs targeting DDX1 (a and b) and transfected with reporters. Silencing of DDX1 leads to strong reduction in Gag expression from pGag-RRE but not from the pGag-CTE construct, as compared to cells treated with control siRNA. (c) Quantification of Gag expression levels from pGag-RRE and pGag-CTE demonstrates a 5-fold reduction in Gag expression from the RRE construct in DDX1 silenced cells but only minor loss of Gag from the CTE construct. (d) Effects of DDX1 silencing on virus production. Down-regulation of DDX1 with each of two siRNAs strongly reduces p24 capsid release from cells, relative to control siRNA.
Silencing of DDX1 by each of the siRNAs was highly efficient, as seen by Western blot analysis (Fig. 7b). For both siRNAs, DDX1 silencing resulted in an > 85% loss of Gag expression from pGag-RRE, as compared to a scrambled control siRNA (Fig. 7b and c). In contrast, silencing DDX1 had only a minor effect on Gag expression from pGag-CTE. Taken together, these results indicate that DDX1 specifically promotes Rev-dependent expression of Gag in HeLa cells and that the effects of DDX1 silencing on gene expression are not the result of a general defect involving mRNA production or translation.
The effect of DDX1 silencing on Rev function was previously analyzed using a proviral replication assay.14 For direct analysis of a potential role of DDX1 in HIV-1 replication, DDX1 silenced HeLa cells as well as cells treated with a control siRNA were infected with virus, and levels of HIV-1 released into media were monitored by an anti-HIV-1 capsid, p24 ELISA. Consistent with the transactivation assay, DDX1 silencing by both siRNAs decreased de novo virus particle production by 75% relative to controls (Fig. 7d). Altogether, our results indicate that DDX1 is required for the production of HIV-1 virus particles and suggest that this involves, at least in part, Rev–RRE-mediated processes.
Discussion
System for studying function in DDX1 silenced cells
The prime motivation for investigating DDX1 is to develop a comprehensive understanding of this protein's contribution to the HIV-1 lifecycle. This requires a clear link between DDX1, HIV-1 Rev, and discrete steps of HIV-1 replication. It also requires a validated system for further cell-based studies of DDX1 function and molecular interactions. Both siRNAs used in our experiments inhibited production of HIV in the infection assay. Additionally, these siRNAs inhibited expression of a reporter gene whose production was driven by Rev–RRE but not expression of the same reporter driven by CTE. This effectively rules out off-target effects of the two siRNAs on general features of transcription, RNA metabolism, and translation. This approach, using two independent siRNAs that efficiently target DDX1 and that lack substantial nonspecific effects on gene expression, makes a decisive case for a role of DDX1 for HIV replication in the widely used model of HeLa cells.
Human cofactors are often considered desirable targets for pharmacological treatment of HIV-1 because they are not under evolutionary pressure to generate escape mutations. Additionally, variation in Rev activity has been suggested to contribute to both the establishment and escape of viral latency.41, 49, 50 Targeting DDX1 function could be useful for exploiting the regulation of HIV-1 by Rev. In addition to HIV-1, DDX1 plays a role in the progression of several human diseases. For example, DDX1 is overexpressed in retinoblastoma and has been found co-amplified with neuronal Myc in neuroblastoma cell lines and primary tumor specimens.30, 51 Overexpression of DDX1 has also been linked with stem cell-associated genes in human testicular germ cell tumors19 and as a prognostic marker for recurrence of breast cancer.52 Furthermore, DDX1 contributes to the life cycle of several viruses. It promotes the proliferation of human polyomavirus,53, 54 and it contributes to coronavirus replication in cell culture.55 DDX1 is also associated with the 3′ untranslated region of the hepatitis C virus genome in human liver cells.23 Understanding of DDX1 functions and molecular mechanisms may have implications for the treatment of multiple diseases. The development of a validated siRNA system for DDX1 silencing will promote future efforts to uncover these mechanisms.
Assembly of distinct DDX–Rev and DDX1–Rev–RRE complexes
When purified from E. coli, recombinant DDX1 is a soluble, folded protein that forms specific complexes with HIV-1 Rev. Under the conditions of the in vitro binding assay, the protein–protein interaction between DDX1 and Rev occurs with a disassociation constant on the order of tens of nanomolar. While this is relatively tight for a protein–protein interaction, it is considerably weaker than oligomeric binding of Rev on the RRE, which occurs in the sub-nanomolar range.41, 43, 48 The relative difference in these affinities is consistent with previous estimates from cell-based studies of the DDX1–Rev interaction.14 These observations demonstrate that the purified RRE complexes containing both Rev and DDX1 are mediated through protein–RNA contacts and not protein–protein interactions. The lack of cooperativity between a protein–protein interaction and RNA binding by Rev and DDX1 may result from sequestering the NIS within Rev oligomers on the RRE. Sequestering of protein–protein interaction domains within the oligomeric and RNA binding interfaces could be a general method of regulating protein cofactor binding to Rev. For instance, the interaction between nuclear import factors and Rev has been observed to occur through interactions within the nuclear localization signal inside arginine-rich domain of RBD/OD 2.3, 56
There are several possible roles for a DDX1–Rev protein complex that forms independently of RNA. One possibility may be to prevent diffusion of Rev from the nucleus prior to binding on the RRE. At steady state, Rev displays a nuclear and nucleolar distribution in cells.34, 57 The diffuse cytoplasmic and nuclear localization of Rev observed in DDX1 silenced cells is very similar to the distribution that has been observed for double mutants containing the V16D variant of Rev58 and for Rev variants that have been constructed to lack the NIS sequence.59 It is possible that the disruption of the DDX1–Rev interaction caused by the V16D mutation results in the same change in Rev localization that is observed in DDX1 silenced cells. Another possible function may be to assist in the formation of complexes containing Rev–RRE RNA and DDX1. In the crowded and compartmentalized environment of the nucleus, a preloaded Rev–RRE complex may have a kinetic advantage in forming the ternary complex. It is conceivable that Rev interaction with DDX1 could prevent other nonproductive interactions with competing cellular components or nonproductive Rev oligomers. The relatively tighter Rev–RRE interaction could assist in disrupting the Rev–DDX1 complex while still allowing for a productive Rev–RRE–DDX1 complex assembled through protein–RNA interactions. In this manner, the Rev–DDX1 complex could serve to specifically target DDX1 to unspliced and incompletely spliced HIV transcripts in the nucleus. This could promote CRM1-mediated export of the RRE-containing transcripts by several mechanisms, which could include hypothetical DDX1-facilitated release of the transcripts from nuclear retention sites. Notwithstanding our findings, it is possible that the in vitro system is missing levels of posttranslational regulation that could influence the relativity dissociation constants of the complex. All of these possibilities warrant further investigation, and the in vitro experiments using recombinant proteins and synthetic RNAs should be considered the first step in this process.
In the pull-down assays, both the V16D and I55N mutations to the Rev oligomerization domain reduced the interaction with DDX1. It is tempting to infer from these results that Rev oligomerization is required for functional association with DDX1. However, the oligomerization state of Rev is dependent on several factors including solution conditions,37 Rev secondary structure,41 and, as recently demonstrated by alternative crystal structures of dimers of the Rev oligomerization domain, both differences in protein sequence60 and interaction with other proteins.61 Additionally, the interaction between the NIS sequence of Rev and DDX1 was identified in a yeast-two-hybrid assay with a NIS-based peptide that would not preserve the oligomerization state of the Rev protein but could still retain helical propensity. Both V16D and I55N are highly disruptive of Rev secondary and tertiary structures.41 It is possible that disruptions of the helical structure of the oligomerization domain in these variants contributes more to the loss of interaction with DDX1 than is due to either the loss of specific residue contacts between Rev and DDX1 or the loss of Rev oligomerization. Structural and DDX1 binding studies with synthetic peptides based on the NIS sequence could be very valuable for investigating the mechanism of the DDX1–Rev interaction.
ATPase activity of DDX1
DDX1 is an RNA-stimulated ATPase that binds viral RNA substrates. DDX1 self-associates to form multiple oligomeric states at 25 °C and concentrations near 10 μM; however, both the Rev binding thermodynamics and ATPase activities of DDX1 are consistent with monomeric behavior at concentrations below 1 μM. Previous reports for the ATPase activities of DEAD-box proteins range from k cat values of 1–600 min− 1 and K m (ATP) values from 0.06 to 2.5 mM.26 When compared to other recombinant DEAD-box proteins, DDX1 hydrolyzes ATP relatively slowly with a moderately high apparent K m. Although well within the range of DEAD-box protein behavior, the particular enzymology of recombinant DDX1 likely contributed to previous reports where the ATPase activity was lacking.27, 28 Previous enzyme assays were also conducted at 50 μM ATP concentrations, which would have been too low to provide measurable activity.27
As recently outlined,24 several molecular mechanisms may account for the particular enzymatic properties of an individual DEAD-box protein. The simplest source of variability among DEAD-box proteins is differences in the structure and dynamics of the enzyme active sites that affect ATP binding primarily through conserved residues in the NTD. Additional residues in the N-terminal Q-motif also contribute to nucleotide recognition. DDX1 is unique among DEAD-box proteins due to the inclusion of an SPRY domain between the Q-motif and the NTD. It seems reasonable that the SPRY domain could dramatically alter the relationship between the NTD and Q-motif and therefore affect ATPase activity. However, because the rates and K m values for DDX1 are within the range of other DEAD-box proteins, structural differences in the active site are likely to be similar to those of other slow DEAD-box proteins and not due to a gross alteration of the protein conformation. It remains possible that specific protein cofactors could interact with the SPRY domain and induce conformational changes in DDX1 that result in a more optimal enzyme. As the list of possible cellular partners for DDX1 grows, it will be interesting to determine if this mechanism provides a means of regulating activity.
The enzymatic properties of DEAD-box proteins can also be highly dependent on the specificity for a particular RNA target. The most thoroughly studied examples are members of the DbpA subfamily whose optimal ATP hydrolysis occurs only with the 23S rRNA as a substrate.62 Because it had not been previously determined if RRE RNA was a specific substrate for DDX1, the ATPase activity was compared using two synthetic RNA constructs based on the RRE and tRNAphe as a nonspecific control at a single ATP concentration. There is a modest enhancement of k max and a more significant decrease in K app (RNA) when HIV-1-derived RNAs are compared to the less optimal tRNAphe. tRNA is a highly structured RNA with a compact tertiary structure. In contrast, stem IIB consists only of short stretches of duplex RNA, bulged base pairs, and an unpaired loop. Given the modest k max values for both RRE and stem IIB, it is likely that the enhancements seen for the HIV-1-derived RNAs reflect the difficulty in binding a highly folded tRNA as a substrate. The observed kinetics may also reflect a preference for either double-stranded or partially unpaired double-stranded substrate. Such a preference would be consistent with the recent observation of DDX1 cellular localization at the site of double-stranded DNA breaks that result from ionizing radiation.28
A sophisticated mechanism for the regulation of DEAD-box proteins also comes from the interaction with protein cofactors. For example, the ATPase activity of the E. coli DEAD-box protein RhlB is highly stimulated by interactions with RNase E.63 Down-regulation of DEAD-box protein activity by protein cofactors has also been observed, as in the case of the competitive inhibition of DBP5 by the nucleoporin NUP214.64 The effects of Rev on the ATPase properties of DDX1 were an open question. Rev neither activates nor down-regulates DDX1 in the range of the tested conditions.
When compared to highly active DEAD-box proteins, recombinant DDX1 has a relatively low k max and moderate K app (RNA) RRE-stimulated ATPase activity. This may reflect a cellular function that uses catalytic activity for a cycle of binding and release rather than for processive disruption of complex RNA structure. Such cellular function has been suggested for Rok1p and Rrp3p in yeast ribosome biogenesis.24 Furthermore, eIF4AIII is a well-documented example that does not require ATP hydrolysis for its role as an accessory protein in the Exon Junction Complex.65, 66 Alternatively, the modest k max value of DDX1 may reflect that of an enzyme requiring posttranslational modification for full activity. This would be consistent with the observation that epitope-tagged DDX1 purified from human cell culture was more active than E. coli purified DDX1.27 In either case, the enzymatic activity of recombinant DDX1 stimulated by Rev-bound RRE indicates that the recombinant enzyme retains some functionality of native DDX1. It is therefore reasonable to use recombinant DDX1 as a starting point for future investigations of the specific molecular mechanisms that regulate DDX1 enzymatic activity and to discover how these mechanisms contribute to the HIV-1 lifecycle.
Materials and Methods
Reporter assays of Rev activity in DDX1 silenced cells
siRNA oligonucleotides 5′-GGCAATTTCCCTGGTGGCAACAGAA-3′ (si-DDX1-A) and 5′-GGAGTTAGCTGAACAAACTTTGAA-3′ (si-DDX1-B) and GC-content-matched control siRNA oligo were purchased from Invitrogen. Silencing of DDX1 was accomplished by two sequential transfections with siRNAs. Initially, HeLa cells were transfected with siRNA (at 20 nM) with RNAiMAX (Invitrogen). After 48 h, the cells were cotransfected with siRNAs (at 20 nM) and either pGag-RRE with the RRE-containing fragment of the env gene together with an expression plasmid encoding HIV-1-1 Rev (pRev) or the control plasmid pGag-CTE, in which the RRE was replaced with four copies of the CTE.31 The second transfection was performed with Lipofectamine 2000 (Invitrogen). After 24 h of incubation, the cells were collected for analysis by Western blotting. Knockdown efficiency and Gag expression were evaluated using rabbit anti-DDX1 (Proteinteck Group) and mouse anti-p24 (Chemicon), respectively.
HIV-1 infection assay
HeLa TZM-bl cells were obtained through the National Institutes of Health AIDS Research and Reference Reagent Program. Similar to the procedure used for the reporter transactivation assay, silencing of DDX1 was accomplished by two sequential transfections with siRNAs. TZM-bl cells in 6-well plates (1.7 × 105) were transfected with si-DDX1-A, si-DDX1-B, or si-Control (at 20 nM) with RNAiMAX (Invitrogen). After 48 h, the cells were transfected with siRNAs (at 20 nM) with Lipofectamine 2000 (Invitrogen). After another 24 h, the cells were infected with 50 ng R9 (NL4.3) virus (Gallay et al. 1997) for 18 h. Cells were subsequently washed 2× with phosphate-buffered saline (PBS), and fresh media were added. De novo production of virus released into the media was monitored 48 h postinfection via p24 ELISA (Alliance HIV-1 p24 Antigen ELISA kit; PerkinElmer). Total p24 production shown is normalized to cell numbers and is scored as a percentage relative to siRNA control cells. DDX1 knockdown efficiency was evaluated by Western blotting after collecting the cells 48 h postinfection.
Protein expression and purification
A plasmid containing DDX1 was initially provided by the Godbout Lab (University of Alberta). The DDX1 gene was amplified using PCR and inserted into a modified pET22b expression vector (Novagen) for expression of full-length DDX1 protein with an N-terminal 6-histidine tag. BL21(DE3) gold E. coli cells transformed with pET22-HT-DDX1 plasmid were grown in LB broth to an OD600 = 0.8 at 37 °C. Cell cultures were transferred to 30 °C, and DDX1 expression was induced by addition of 1 mM IPTG for 3 h. Cells were harvested by centrifugation and stored at − 80 °C until ready for use. Harvested E. coli cells were resuspended in nickel column buffer [20 mM Tris (pH 7.9), 1 M NaCl, 10% glycerol, 0.1% Triton X-100, 12 mM imidazole, and 10 mM β-mercaptoethanol], and the cell membranes were disrupted by sonication. Cell debris was removed by centrifugation, and the resulting lysate was treated with 5% polyethyleneimine (Sigma) to remove contaminating nucleic acids prior to affinity column purification using Ni-NTA resin (QIAGEN). Pooled fractions containing DDX1 protein were dialyzed against Q column buffer [25 mM Tris (pH 8.8), 25 mM NaCl, 2 mM DTT, and 1 mM ethylenediaminetetraacetic acid] and run over a 5-ml Hi-Trap Q HP Sepharose column (GE Healthcare). Purified DDX1 was dialyzed against 4 l of storage buffer [25 mM Hepes (pH 7.5), 175 mM KCl, and 2 mM tris(2-carboxyethyl)phosphine] and stored at 4 °C. A plasmid coding for the DDX1 C-terminal truncation (residues 1–428) was created using PCR from the full-length pET22-HT-DDX1 plasmid as a template. The amplified DDX1 (1–428) gene fragment was inserted into pET22-HT and expressed in BL21(DE3) E. coli cells. DDX1 (1–428) protein was purified on a Ni-NTA affinity column followed by a Q HP Sepharose column as described above. Pure DDX1 (1–428) was dialyzed against storage buffer [25 mM Tris (pH 7.5), 200 mM NaCl, 2 mM DTT, and 1 mM ethylenediaminetetraacetic acid] and stored at 4 °C for use. HIV-1 Rev expression, purification, and labeling were conducted as described previously.41, 43
Analytical gel-filtration chromatography and CD
The oligomeric state of DDX1 was assessed by analytical size-exclusion chromatography on a Superdex 2000 column (Amersham). DDX1 (10 μM) was loaded and eluted from the column in K2HPO4 (pH 7.5), 500 mM KCl or 150 mM KCl, and 1 mM DTT at 25 °C. The molecular weight of DDX1 was estimated by comparing elution volumes to size-exclusion standards (Biorad).
CD spectra were recorded on an Aviv 202SP spectropolarimeter with 10 μM DDX1 at 25 °C in K2HPO4 (pH 7.5), 500 mM KCl or 150 mM KCl, and 500 μM tris(2-carboxyl-ethyl)phosphine hydrochloride. Ellipticity was averaged for 3 s at 1-nm intervals.
Fluorescence anisotropy
The fluorescence anisotropy titration assay was conducted with 15 nM Alexa-488-labeled Rev protein in pH 7.5, 10 mM Hepes, 150 mM KCl, 2 mM MgCl2, 5 mM β-mercaptoethanol, and 10% glycerol at 25 °C. The final sample volume was 500 ml per well. Samples were equilibrated in 96-well opaque microtiter plates (Grenier), and the anisotropy was determined using a Fusion α-FP plate reader (Packard). The average anisotropy value for each DDX1 concentration was calculated from 10 consecutive readings of each plate. These values were averaged over at least three separate titrations, and the reported error is 1 SD calculated from the averaged titrations. Equilibrium K d values were determined by nonlinear least-squares fitting of the binding data to a model for a single-transition isotherm using Igor (Wavemetrics). The titration data were fit to the following equations:
where D t is the total concentration of DDX1 and R t is the total concentration of labeled Rev at each step. r d is the anisotropy of free Rev, and r pd is the anisotropy of DDX1-bound Rev.
Analysis of DDX1–Rev interaction by immunoprecipitation
The V5-tagged Rev or Rev mutants were coexpressed with the pGag-RRE in HeLa cells for 24 h in 10-cm dishes. After washing with 1× PBS, we lysed transfected cells in buffer containing 50 mM Tris (pH 7.4), 75 mM NaCl, 1 mM MgCl2, 1% NP40, 1 mM DTT, 1 mM PMSF, and protease inhibitor cocktails (Roche Diagnostics). Cell lysates were treated with 50 units of RNase I for 15 min at 37 °C. Cell debris was removed by centrifugation at 10,000g at 4 °C, and an aliquot of clarified supernatant was kept as “input.” The clarified cell lysate was divided and incubated for 30 min at 4 °C with either 4 μg anti-V5 antibodies (Invitrogen) or 4 μg control rabbit IgG. Immunoglobulin complexes were collected by incubating with protein G-agarose Dynabeads (Invitrogen) for 30 min at room temperature. The beads were washed five times with 1 ml of lysis buffer and three times with 1 ml PBS buffer. Protein complexes were recovered by boiling in Laemmli sample buffer and were analyzed by SDS-PAGE and Western blotting.
Steady-state ATPase assay
The rate of ATP hydrolysis was measured by quantifying the concentration of free Pi using a molybdate binding assay. Reactions (25 μl) containing 500 nM DDX1 were incubated with varying amounts of either RNA or ATP. The final concentrations of oligonucleotide and ATP at each step are reported in Table 2. Reactions were conducted in 10 mM Tris (pH 7.5), 100 mM NaCl, 4 mM MgCl2, 0.1% Triton x-100, 1% glycerol, and 0.1 mg/ml BSA at 37 °C and incubated for 90 min. The reaction was quenched by addition of 25 μl of 12% SDS. Following the quench, 50 μl of 6% ascorbic acid and 1% molybdate was added to the reaction and then incubated for 10 min at 37 °C. Subsequently, 75 μl of 2% sodium citrate, 2% sodium meta-arsenate, and 2% sodium acetate was added to the reaction and incubated for an additional 10 min at 37 °C. The quantity of reduced phosphomolybdate complex was determined by absorbance (λ = 850 nm) using a Molecular Devices Spectra Max M2. The steady-state ATPase constants were determined to fits of the quadratic form of the Briggs–Haldane equation.67
RNA transcription and purification and electrophoretic gel mobility analysis
RRE transcription and purification was conducted as previously described.40 The stem IIB oligo has been previously described32 and was purchased from Thermo Scientific. High-purity yeast tRNAphe was purchased from Sigma. The interaction of DDX1 with RRE and Rev–RRE was measured as previously described.41
Acknowledgements
The authors thank Dr. Roseline Godbout for the original DDX1 clone.
This work was funded by a grant from the National Institutes of Health, P50 GM082545 (to J.R.W. and L.G.). S.N. was supported by a California HIV/AIDS Research Program postdoctoral fellowship.
Edited by M. F. Summers
Footnotes
References
- 1.Hope T.J. The ins and outs of HIV. Rev. Arch. Biochem. Biophys. 1999;365:186–191. doi: 10.1006/abbi.1999.1207. [DOI] [PubMed] [Google Scholar]
- 2.Malim M.H., Hauber J., Le S.Y., Maizel J.V., Cullen B.R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 3.Pollard V.W., Malim M.H. The HIV-1 Rev protein. Annu. Rev. Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 4.Dagostino D.M., Felber B.K., Harrison J.E., Pavlakis G.N. The Rev protein of human-immunodeficiency-virus type-1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol. Cell. Biol. 1992;12:1375–1386. doi: 10.1128/mcb.12.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greatorex J., Gallego J., Varani G., Lever A. Structure and stability of wild-type and mutant RNA internal loops from the SL-1 domain of the HIV-1 packaging signal. J. Mol. Biol. 2002;322:543–557. doi: 10.1016/s0022-2836(02)00776-3. [DOI] [PubMed] [Google Scholar]
- 6.Groom H.C., Anderson E.C., Lever A.M. Rev: beyond nuclear export. J. Gen. Virol. 2009;90:1303–1318. doi: 10.1099/vir.0.011460-0. [DOI] [PubMed] [Google Scholar]
- 7.Groom H.C., Anderson E.C., Dangerfield J.A., Lever A.M. Rev regulates translation of human immunodeficiency virus type 1 RNAs. J. Gen. Virol. 2009;90:1141–1147. doi: 10.1099/vir.0.007963-0. [DOI] [PubMed] [Google Scholar]
- 8.Perales C., Carrasco L., Gonzalez M.E. Regulation of HIV-1 env mRNA translation by Rev protein. Biochim. Biophys. Acta. 2005;1743:169–175. doi: 10.1016/j.bbamcr.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 9.Anson D.S., Fuller M. Rational development of a HIV-1 gene therapy vector. J. Gene. Med. 2003;5:829–838. doi: 10.1002/jgm.415. [DOI] [PubMed] [Google Scholar]
- 10.Brandt S., Blissenbach M., Grewe B., Konietzny R., Grunwald T., Uberla K. Rev proteins of human and simian immunodeficiency virus enhance RNA encapsidation. PLoS Pathog. 2007;3:e54. doi: 10.1371/journal.ppat.0030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lucke S., Grunwald T., Uberla K. Reduced mobilization of Rev-responsive element-deficient lentiviral vectors. J. Virol. 2005;79:9359–9362. doi: 10.1128/JVI.79.14.9359-9362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson J.H., Child L.A., Lever A.M. Packaging of human immunodeficiency virus type 1 RNA requires cis-acting sequences outside the 5′ leader region. J. Virol. 1993;67:3997–4005. doi: 10.1128/jvi.67.7.3997-4005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang J., Acheampong E., Dave R., Wang F., Mukhtar M., Pomerantz R.J. The RNA helicase DDX1 is involved in restricted HIV-1 Rev function in human astrocytes. Virology. 2005;336:299–307. doi: 10.1016/j.virol.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Fang J., Kubota S., Yang B., Zhou N., Zhang H., Godbout R., Pomerantz R.J. A DEAD box protein facilitates HIV-1 replication as a cellular co-factor of Rev. Virology. 2004;330:471–480. doi: 10.1016/j.virol.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 15.Krishnan V., Zeichner S.L. Alterations in the expression of DEAD-box and other RNA binding proteins during HIV-1 replication. Retrovirology. 2004;1:42. doi: 10.1186/1742-4690-1-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma J., Rong L., Zhou Y., Roy B.B., Lu J., Abrahamyan L. The requirement of the DEAD-box protein DDX24 for the packaging of human immunodeficiency virus type 1 RNA. Virology. 2008;375:253–264. doi: 10.1016/j.virol.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 17.Yedavalli V.S., Neuveut C., Chi Y.H., Kleiman L., Jeang K.T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev–RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 18.Ishaq M., Ma L., Wu X., Mu Y., Pan J., Hu J. The DEAD-box RNA helicase DDX1 interacts with RelA and enhances nuclear factor kappaB-mediated transcription. J. Cell. Biochem. 2009;106:296–305. doi: 10.1002/jcb.22004. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka K., Okamoto S., Ishikawa Y., Tamura H., Hara T. DDX1 is required for testicular tumorigenesis, partially through the transcriptional activation of 12p stem cell genes. Oncogene. 2009;28:2142–2151. doi: 10.1038/onc.2009.89. [DOI] [PubMed] [Google Scholar]
- 20.Bleoo S., Sun X., Hendzel M.J., Rowe J.M., Packer M., Godbout R. Association of human DEAD box protein DDX1 with a cleavage stimulation factor involved in 3′-end processing of pre-MRNA. Mol. Biol. Cell. 2001;12:3046–3059. doi: 10.1091/mbc.12.10.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L., Roy K., Katyal S., Sun X., Bleoo S., Godbout R. Dynamic nature of cleavage bodies and their spatial relationship to DDX1 bodies, Cajal bodies, and gems. Mol. Biol. Cell. 2006;17:1126–1140. doi: 10.1091/mbc.E05-08-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanai Y., Dohmae N., Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 23.Tingting P., Caiyun F., Zhigang Y., Pengyuan Y., Zhenghong Y. Subproteomic analysis of the cellular proteins associated with the 3′ untranslated region of the hepatitis C virus genome in human liver cells. Biochem. Biophys. Res. Commun. 2006;347:683–691. doi: 10.1016/j.bbrc.2006.06.144. [DOI] [PubMed] [Google Scholar]
- 24.Garcia I., Uhlenbeck O.C. Differential RNA-dependent ATPase activities of four rRNA processing yeast DEAD-box proteins. Biochemistry. 2008;47:12562–12573. doi: 10.1021/bi8016119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pyle A.M. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 26.Rocak S., Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev., Mol. Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 27.Chen H.C., Lin W.C., Tsay Y.G., Lee S.C., Chang C.J. An RNA helicase, DDX1, interacting with poly(A) RNA and heterogeneous nuclear ribonucleoprotein K. J. Biol. Chem. 2002;277:40403–40409. doi: 10.1074/jbc.M206981200. [DOI] [PubMed] [Google Scholar]
- 28.Li L., Monckton E.A., Godbout R. A role for DEAD box 1 at DNA double-strand breaks. Mol. Cell. Biol. 2008;28:6413–6425. doi: 10.1128/MCB.01053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godbout R., Hale M., Bisgrove D. A human DEAD box protein with partial homology to heterogeneous nuclear ribonucleoprotein U. Gene. 1994;138:243–245. doi: 10.1016/0378-1119(94)90816-8. [DOI] [PubMed] [Google Scholar]
- 30.Godbout R., Li L., Liu R.Z., Roy K. Role of DEAD box 1 in retinoblastoma and neuroblastoma. Future Oncol. 2007;3:575–587. doi: 10.2217/14796694.3.5.575. [DOI] [PubMed] [Google Scholar]
- 31.Wodrich H., Schambach A., Krausslich H.G. Multiple copies of the Mason–Pfizer monkey virus constitutive RNA transport element lead to enhanced HIV-1 Gag expression in a context-dependent manner. Nucleic Acids Res. 2000;28:901–910. doi: 10.1093/nar/28.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Battiste J.L., Mao H., Rao N.S., Tan R., Muhandiram D.R., Kay L.E. Alpha helix-RNA major groove recognition in an HIV-1 rev peptide–RRE RNA complex. Science. 1996;273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 33.Bogerd H., Greene W.C. Dominant negative mutants of human T-cell leukemia virus type I Rex and human immunodeficiency virus type 1 Rev fail to multimerize in vivo. J. Virol. 1993;67:2496–2502. doi: 10.1128/jvi.67.5.2496-2502.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malim M.H., Bohnlein S., Hauber J., Cullen B.R. Functional dissection of the HIV-1 Rev trans-activator–derivation of a trans-dominant repressor of Rev function. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 35.Sreerama N., Venyaminov S.Y., Woody R.W. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999;8:370–380. doi: 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shankar J., Pradhan A., Tuteja R. Isolation and characterization of Plasmodium falciparum UAP56 homolog: evidence for the coupling of RNA binding and splicing activity by site-directed mutations. Arch. Biochem. Biophys. 2008;478:143–153. doi: 10.1016/j.abb.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 37.Cole J.L., Gehman J.D., Shafer J.A., Kuo L.C. Solution oligomerization of the rev protein of HIV-1: implications for function. Biochemistry. 1993;32:11769–11775. doi: 10.1021/bi00095a004. [DOI] [PubMed] [Google Scholar]
- 38.Auer M., Gremlich H.U., Seifert J.M., Daly T.J., Parslow T.G., Casari G. Helix–loop–helix motif in HIV-1 Rev. Biochemistry. 1994;33:2988–2996. doi: 10.1021/bi00176a031. [DOI] [PubMed] [Google Scholar]
- 39.Blanco F.J., Hess S., Pannell L.K., Rizzo N.W., Tycko R. Solid-state NMR data support a helix–loop–helix structural model for the N-terminal half of HIV-1 Rev in fibrillar form. J. Mol. Biol. 2001;313:845–859. doi: 10.1006/jmbi.2001.5067. [DOI] [PubMed] [Google Scholar]
- 40.Szilvay A.M., Brokstad K.A., Boe S.O., Haukenes G., Kalland K.H. Oligomerization of HIV-1 Rev mutants in the cytoplasm and during nuclear import. Virology. 1997;235:73–81. doi: 10.1006/viro.1997.8671. [DOI] [PubMed] [Google Scholar]
- 41.Edgcomb S.P., Aschrafi A., Kompfner E., Williamson J.R., Gerace L., Hennig M. Protein structure and oligomerization are important for the formation of export-competent HIV-1 Rev–RRE complexes. Protein Sci. 2008;17:420–430. doi: 10.1110/ps.073246608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jain C., Belasco J.G. Structural model for the cooperative assembly of HIV-1 Rev multimers on the RRE as deduced from analysis of assembly-defective mutants. Mol. Cell. 2001;7:603–614. doi: 10.1016/s1097-2765(01)00207-6. [DOI] [PubMed] [Google Scholar]
- 43.Pond S.J., Ridgeway W.K., Robertson R., Wang J., Millar D.P. HIV-1 Rev protein assembles on viral RNA one molecule at a time. Proc. Natl Acad. Sci. USA. 2009;106:1404–1408. doi: 10.1073/pnas.0807388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carpousis A.J., Khemici V., Poljak L. Assaying DEAD-box RNA helicases and their role in mRNA degradation in Escherichia coli. Methods Enzymol. 2008;447:183–197. doi: 10.1016/S0076-6879(08)02210-6. [DOI] [PubMed] [Google Scholar]
- 45.Chifflet S., Torriglia A., Chiesa R., Tolosa S. A method for the determination of inorganic phosphate in the presence of labile organic phosphate and high concentrations of protein: application to lens ATPases. Anal. Biochem. 1988;168:1–4. doi: 10.1016/0003-2697(88)90002-4. [DOI] [PubMed] [Google Scholar]
- 46.Groom H.C.T., Anderson E.C., Dangerfield J.A., Lever A.M.L. Rev regulates translation of human immunodeficiency virus type 1 RNAs. J. Gen. Virol. 2009;90:1141–1147. doi: 10.1099/vir.0.007963-0. [DOI] [PubMed] [Google Scholar]
- 47.Daugherty M.D., Booth D.S., Jayaraman B., Cheng Y., Frankel A.D. HIV Rev response element (RRE) directs assembly of the Rev homooligomer into discrete asymmetric complexes. Proc. Natl Acad. Sci. USA. 2010;107:12481–12486. doi: 10.1073/pnas.1007022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daugherty M.D., D'Orso I., Frankel A.D. A solution to limited genomic capacity: using adaptable binding surfaces to assemble the functional HIV Rev oligomer on RNA. Mol. Cell. 2008;31:824–834. doi: 10.1016/j.molcel.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bobbitt K.R., Addo M.M., Altfeld M., Filzen T., Onafuwa A.A., Walker B.D. Rev activity determines sensitivity of HIV-1-infected primary T cells to CTL killing. Immunity. 2003;18:289–299. doi: 10.1016/s1074-7613(03)00031-1. [DOI] [PubMed] [Google Scholar]
- 50.Poropatich K., Sullivan D.J. Human immunodeficiency virus type 1 long-term non-progressors: the viral, genetic and immunological basis for disease non-progression. J. Gen. Virol. 2011;92:247–268. doi: 10.1099/vir.0.027102-0. [DOI] [PubMed] [Google Scholar]
- 51.Godbout R., Squire J. Amplification of a DEAD box protein gene in retinoblastoma cell lines. Proc. Natl Acad. Sci. USA. 1993;90:7578–7582. doi: 10.1073/pnas.90.16.7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Germain D.R., Graham K., Glubrecht D.D., Hugh J.C., Mackey J.R., Godbout R. DEAD box 1: a novel and independent prognostic marker for early recurrence in breast cancer. Breast Cancer Res. Treat. 2010;127:53–63. doi: 10.1007/s10549-010-0943-7. [DOI] [PubMed] [Google Scholar]
- 53.Sunden Y., Semba S., Suzuki T., Okada Y., Orba Y., Nagashima K. DDX1 promotes proliferation of the JC virus through transactivation of its promoter. Microbiol. Immunol. 2007;51:339–347. doi: 10.1111/j.1348-0421.2007.tb03907.x. [DOI] [PubMed] [Google Scholar]
- 54.Sunden Y., Semba S., Suzuki T., Okada Y., Orba Y., Nagashima K., Umemura T., Sawa H. Identification of DDX1 as a JC virus transcriptional control region-binding protein. Microbiol. Immunol. 2007;51:327–337. doi: 10.1111/j.1348-0421.2007.tb03915.x. [DOI] [PubMed] [Google Scholar]
- 55.Xu L., Khadijah S., Fang S., Wang L., Tay F.P., Liu D.X. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J. Virol. 2010;84:8571–8583. doi: 10.1128/JVI.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henderson B.R., Percipalle P. Interactions between HIV Rev and nuclear import and export factors: the Rev nuclear localisation signal mediates specific binding to human importin-beta. J. Mol. Biol. 1997;274:693–707. doi: 10.1006/jmbi.1997.1420. [DOI] [PubMed] [Google Scholar]
- 57.Cochrane A.W., Perkins A., Rosen C.A. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J. Virol. 1990;64:881–885. doi: 10.1128/jvi.64.2.881-885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trikha R., Brighty D.W. Phenotypic analysis of human immunodeficiency virus type 1 Rev trimerization-interface mutants in human cells. J. Gen. Virol. 2005;86:1509–1513. doi: 10.1099/vir.0.80572-0. [DOI] [PubMed] [Google Scholar]
- 59.Fang J., Kubota S., Pomerantz R.J. A trans-dominant negative HIV type 1 Rev with intact domains of NLS/NOS and NES. AIDS Res. Hum. Retroviruses. 2002;18:705–709. doi: 10.1089/088922202760072320. [DOI] [PubMed] [Google Scholar]
- 60.Daugherty M.D., Liu B., Frankel A.D. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct. Mol. Biol. 2010;17:1337–1342. doi: 10.1038/nsmb.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DiMattia M.A., Watts N.R., Stahl S.J., Rader C., Wingfield P.T., Stuart D.I. Implications of the HIV-1 Rev dimer structure at 3.2 Å resolution for multimeric binding to the Rev response element. Proc. Natl Acad. Sci. USA. 2010;107:5810–5814. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsu C.A., Uhlenbeck O.C. Kinetic analysis of the RNA-dependent adenosinetriphosphatase activity of DbpA, an Escherichia coli DEAD protein specific for 23S ribosomal RNA. Biochemistry. 1998;37:16989–16996. doi: 10.1021/bi981837y. [DOI] [PubMed] [Google Scholar]
- 63.Vanzo N.F., Li Y.S., Py B., Blum E., Higgins C.F., Raynal L.C. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 1998;12:2770–2781. doi: 10.1101/gad.12.17.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.von Moeller H., Basquin C., Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat. Struct. Mol. Biol. 2009;16:247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 65.Andersen C.B.F., Ballut L., Johansen J.S., Chamieh H., Nielsen K.H., Oliveira C.L.P. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 66.Bono F., Ebert J., Lorentzen E., Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 67.Henn A., Cao W., Hackney D.D., De La Cruz E.M. The ATPase cycle mechanism of the DEAD-box rRNA helicase, DbpA. J. Mol. Biol. 2008;377:193–205. doi: 10.1016/j.jmb.2007.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]