Abstract
Ancient DNA methodologies can be applied in the investigation of the genetics of extinct populations. A search for beta thalassemia mutations was performed on 49 Minoan individuals from the Bronze Age who were living in the island of Crete approximately 4,000 Years Before Present (YBP). Standard precautionary measures were employed in the laboratory to ensure authenticity of the DNA extracted from the ancient bones, resulting in the successful analysis of DNA of 24 Minoans. DNA sequencing focused on the Intervening Sequence 1 (IVS-1) of the beta globin gene and its splicing junctions. 63% of the thalassemia mutations observed among modern Cretans reside in beta IVS -1. None of the Minoan individuals carried one of the IVS-1 mutations known to cause beta thalassemia; however, only one was expected to be observed if the average frequency of beta thalassemia heterozygotes in the Minoan population was the same with that of modern day Cretans (7.6 %). One individual contained a C to G substitution in position 91 of the IVS-1, located 40 bp 5′ to the intron 1/exon 2 junction. Functional studies indicated that the mutation did not affect mRNA splicing or stability, and most likely represented an innocent single nucleotide polymorphism.
Keywords: ancient DNA, beta globin, IVS-1, Minoan, SNP
Introduction
Since its inception in 1984 the use of ancient DNA in addressing biological questions has led to the genetic characterization of species and populations from all major biological lineages [1,2]. The discipline, molecular archaeology, did not however immediately flourish without debate. Its inauguration was led by a series of sensational claims, many of which were invalidated [3]. Despite its dubious beginning, the discipline now operates under strict guidelines that reduce false claims [4]. The field has since triumphed with the publication of the Neandertal genome [5], and in recent years has explored the use of ancient DNA to address historically significant diseases, such as the plague [6], tuberculosis [7], leprosy [8], and the pathology of King Tutankhamun’s family [9]. The earliest paper on the subject of ancient DNA and blood, targeted various regions of the β-Globin gene [10]. In it, the analysis of nuclear polymorphisms from bone was shown to be feasible in materials as old as 14,000 Years Before the Present (YBP). The latest paper to demonstrate the use of ancient DNA in the field of hematology focused on ABO typing in 2,000–3,000 year old specimens from northern Japan [11].
The thalassemias are mutations of the alpha or the beta hemoglobin genes that result in absence or decreased production of the normal globin polypeptide chains [12,13]. Thalassemia mutations are the most common genetic defects in humans, their high frequency resulting from the positive selection of heterozygotes in an environment of endemic malaria. Geographic distribution considerations and population frequencies of thalassemia mutants suggest that the most common thalassemia mutations most likely originated in neolithic or late paleolithic human populations. Characteristic of severe homozygous β thalassemia are bony abnormalities such as the “hair on brush” appearance seen on X-ray examinations of the skull of patients [13]. Hence, several attempts have been made in the past to identify severe thalassemia in ancient skeletal remains by examining bone morphology. Examination of skeletal material from a number of ancient Greek tombs and ossuaries has identified bones showing osseous abnormalities suggestive of severe thalassemia disease [14]. However these conclusions remain tentative [15] because severe iron deficiency and nutritional anemia [16] and pathologies that are infectious in origin [17] can also produce osseous abnormalities resembling those of severe β thalassemia.
The purpose of this study was to use technologies for molecular analysis of ancient DNA in order to search for β thalassemia mutations in an ancient Cretan population, the Minoans. The Minoans flourished on the island of Crete, Greece, during the early- to mid-Bronze Age (5,000 to 3,500 YBP) and established one of the oldest civilizations of Europe [18]. Since DNA degradation is characteristic of ancient DNA, we analyzed a small target, the IVS 1 region of the β-globin gene. This intron is 130 nucleotides in length [19], and it and its junctions account for approximately 63% of all the β-thalassemia mutations in the present Cretan population [20, 21].
Materials and Methods
Tooth samples
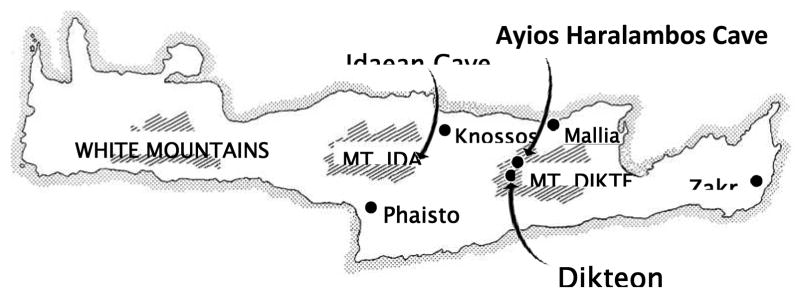
Sixty-nine specimens, representing 49 individuals were excavated from the Ayios Haralambos cave, a secondary ossuary located in central eastern Crete on the Lasithi Plateau (Figure 1). The cave was completely sealed and was accidentally opened during road construction in 1976. The samples come from a deposit which dates from the Neolithic to the Middle Minoan IIB and were preserved in excellent condition.
Figure 1.
Map of the island of Crete showing the location of the Ayios Haralambos cave where the Minoan ossuary was found. The cave is located in the plateau of Mount Dikte (Lassithi plateau). The Ayios Haralambos cave is close to the Dikteon Cave where, according to the ancient Greek Mythology, Zeus was born.
DNA extraction, PCR, cloning, and sequencing
To reduce the probability of analyzing individuals multiple times, teeth were extracted directly from jaws. In the laboratory they were decontaminated by removing the outer layer with sand paper, soaking in 100% bleach for 15 sec, rinsing 8X with DNA free water, and UV treating on all sides for 3 hours. They were then pulverized with a Spex CertiPrep 6750 Freezer/Mill for 2 min at a setting of four. 400 mg of the resulting powder was decalcified and digested following Krings et al. [22] using Ultra reagents (Fluka BioChemika). Samples were centrifuged for 1 min at 4,000 × g and the supernatant removed and extracted with an equal volume of UltraPure phenol, chloroform, isoamyl alcohol (25:24:1) (Invitrogen). Supernatant was concentrated to 100 μL using Microcon MW-30 columns (Millipore). DNA from concentrate was isolated using the MinElute Qiagen PCR Purification Kit [23] and eluted with 70 μL of DNA-Free Elution Solution (QBIOgene). 6 μL of the DNA extract was added to each 25 μL reaction containing HotStart Taq DNA Polymerase (Qiagen) following the manufacturers protocol. Three primer pairs were used to amplify a 267 bp target that included the 130 bp of beta globin IVS-1. The first pair flanked intron 1 and the other two (inner 54 forward and reverse) primers were used to amplify the intron in two amplicons (Supplementary Table 1, Primers A and B). Reactions were cycled in a PTC-150HB PCR MiniCycler (MJ Research) using the parameters: 95°C for 15 mins, 42 cycles of 94°C for 30 s, 55°C for 60 s, 72°C for 60 s, and 72°C for 7 min. PCR products were cloned using the 2.1-TOPO® TA Cloning Kit (Invitrogen).
Authentication of ancient DNA
Authentication of molecular archaeological results necessitates compliance with ancient DNA criteria [4]. We validated our results through the following: (i) all DNA extractions and amplification preparations were carried out in a physically isolated work area in a flow hood exclusively dedicated to the study of ancient DNA; (ii) DNA extracts were maintained in a dedicated, bleach treated freezer in a separate wing of the building from the PCR machine; (iii) multiple blank extractions were processed in parallel and negative controls were included in all reactions; (iv) positive controls were excluded from extractions and amplifications; (v) DNA samples were tested for appropriate molecular behavior [24]; (vi) amino acid racemization [25] and concentration ratios [26] were determined by MicroAnalytica LLC on 15 of the 49 samples to test biomolecular preservation; (vii) PCR copy number was estimated using mtDNA primers L16055-H16155 [22] and SYBR Green (Qiagen) dye on a DNA Engine Opticon 2 Real-Time PCR Detection System (MJ Research); (viii) protective surgical clothing and mask were worn during the handling and extraction of materials; (ix) equipment, sand paper, and tubes were illuminated with UV for 3 hours prior to each use; and (x) all commercial reagents (Taq Polymerase, primers, water, and buffers) were screened for modern DNA prior to use.
Functional studies
Construction of β-globin gene expression plasmids
The β-globin expression plasmid consisted of the CMV promoter, a 1.7 kb human β-globin gene (from −71 to +1616), and the SV40 polyA signal sequence at the 3′ end. The 1.7 kb β-globin gene was generated by PCR using Pfu DNA polymerase (Stratagene) on the template containing a 5.5 kb BglII β-globin gene in pUC18. The pair of primers used in the PCR (Primer C, Supplementary Table 1) contained a 5′ extension of a BglII site in the forward one and a PstI site in the reverse one. The amplified PCR product was purified by QIAgen Gel Extraction Kit (Qiagen), digested with BglII and PstI, and then sub-cloned into a eukaryotic expression vector N1-EGFP (Clonetech). The integrity of the β globin gene in the expression vector was verified by DNA sequencing. The EGFP gene in the original vector was deleted by double digestion with SalI and NotI to avoid transcriptional interference. The resulting construct, pCMVhβ, is shown in Supplementary Fig. 1. The C→G mutation in intron 1 was introduced into pCMVhβ using QuikChange® Lightning Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. The oligo sequences used in this mutagenesis were Primers D (Supplementary Table 1). The resulting plasmid was named pCMVhβ(mut) and its mutation was verified by DNA sequencing. Plasmid DNA of pCMVhβ and pCMVhβ(mut) were prepared and purified using QIAprep Miniprep kit (Qiagen).
Cell culture and transfection
HeLa cells were maintained in RPMI-1640 medium supplemented with 10% FBS, Penicillin (100 unit/ml), and Streptomycin (100 μg/ml). Twenty-four hours prior to transfection, cells were trypsinized and counted. Approximately 2×105 cells, which were in 50–80% confluent, were plated into each well containing 2 ml of antibiotic-free medium in a 6-well plate, and transfected with 1.25 μg of plasmid DNA (pCMVhβ or pCMVhβ(mut)) in a serum-free medium using Lipofectamine™ LTX and PLUS™ Reagent (Invitrogen). On the second day (~ 24 hours) after transfection the cells were transferred to complete medium, and harvested after 60–72 hour culture. Duplication was done for each sample in a transfection experiment and the experiment was repeated.
Analysis of transcription products
Total RNA was isolated from cells using PureLink™ RNA Mini Kit (Invitrogen). The cDNA was synthesized using SuperScript™ II First-Strand Synthesis System for the RT-PCR Kit (Invitrogen). β-globin specific cDNA was amplified via PCR using primers spanning exon1 to exon 3 of the gene (Supplementary Table 1, Primers G) and Pfu DNA polymerase (Stratagene). The PCR-amplified wild type and mutated cDNA product was cloned into T-vector (Promega). Individual colonies were selected and plasmid DNA was purified using QIAprep Miniprep kit (Qiagen), and then sequenced. Sequence comparison was performed using Clustalw 2.0.12 multiple alignment software.
Evaluation of β-globin gene expression
β-globin gene expression was evaluated by quantitative real-time PCR using two pairs of primers, which amplified the regions between exon 1 and exon 2 and between exon 2 and exon of the gene respectively (Primers E and F, Supplementary Table 1). PCR was performed using SYBR Green (Qiagen) on the LightCycler (Roche). The GAPDH gene (Primers H) was used as endogenous control to correct cDNA input. The measurements were repeated at least three times for each sample.
Results and Discussion
The Minoan bones were obtained from a cave located near the village Ayios Haralambos in the mountainous plateau of Lassithi in Crete, where these bones have been kept undisturbed and in relatively low temperatures. Amino acid analysis on the materials showed that the Ayios Haralambos cave contains skeletal materials with outstanding biomolecular preservation. Dextrorotatory/Levorotatory (D/L) racemization ratios for aspartic acid (Asp) and alanine (Ala) were calculated and found to fall comfortably below material from which authentic ancient DNA has been obtained. Generally, D/L Asp ratios lower than 0.12 indicate high biomolecular preservation and correlate with DNA recovery [25], while D/L Ala ratios higher than Asp ratios suggest contamination from exogenous modern sources. The latter was not observed in our samples, indicating high quality skeletal materials and that proper cleaning procedures were implemented. Concentration ratios were also estimated to determine if the Minoan bones have similar proportions of amino acids compared to a modern reference standard. Ratios here were also found to fit within the specifications of material from which authentic ancient DNA has been obtained [26].
DNA from twenty-four of 49 individuals was successfully amplified. The DNA extractions were estimated to contain high copy numbers, ranging from 6,250–13,125 copies (aver.= 10,500) per PCR reaction. Of the 24 samples, none amplified when screened for evidence of modern contamination with the L16055-H16379 mtDNA primer pair. Racemization results on teeth for aspartic acid ranged from 0.057–0.103 (aver.= 0.08) and for alanine from 0.004–0.011 (aver.= 0.0076). Concentration ratios for Asp/Glu ranged from 0.65–0.79 (aver.= 0.71), Ser/Glu 0.44–0.47 (aver.= 0.45), and Ala/Glu 1.56–1.71 (aver.= 1.63).
In 13 individuals the entire IVS 1 target was amplified. In 11 individuals the IVS-1 was amplified in two parts: in 5 only the front half of the IVS 1 was amplified; in 3 the back half of IVS 1; and in 3 both halves were amplified. A total of 304 clones were sequenced. 296 of these contained the β-globin IVS-1 sequences. 8 were non-specific amplifications (when the forward primer was used in conjunction with the internal reverse primer) of the delta globin IVS-1.
Examination of the 296 cloned sequences found that many showed evidence of DNA lesions (Supplementary Table 2) which is a common finding in the analysis of ancient DNA. Several clones had singleton substitutions (Supplementary Table 2). Individuals 3, 4, 8, 22, and 24 of the Supplementary Table 2 contained consistent transitions in several clones. Thus, all 10 sequenced clones of individual 3 contained consistent C to T transitions in two nucleotides (positions IVS-1-22 and IVS-1-99). In individual 4, half of the clones sequenced contained a G to A substitution (IVS-1-77). In individual 8, nine of 47 clones contained consistent G to A transitions in five nucleotides (positions IVS-1-12, IVS-1-13, IVS-1-20, IVS-1-63, and IVS-1-110). In individual 22, seven of the 16 clones sequenced contained consistent G to A transition in three nucleotides (positions IVS-1-24, IVS-1-65, and IVS-1-76). In individual 24, eight of the 33 sequenced clones had a G to A transition in one nucleotide (position IVS-1-111). It is well established that C to T and G to A nucleotide misincorporations are frequently seen in ancient DNA [27]. Therefore the transition mutations of these individuals reflect the damage the ancient DNA had suffered through the millennia. That several clones of an individual contained the same transition of the same nucleotide is due to the amplification of the same parental damaged ancient DNA fragment.
One individual Minoan 7, contained a transversion, a C to G nucleotide substitution in position 91 of the β-globin IVS-1, 40 bp 5′ to the intron 1/exon 2 junction (Supplementary Figure 2). Of twelve clones analyzed 7 carried this C to G transversion, indicating that Minoan 7 was a C/G heterozygote at position 91 of the β-globin IVS-1.
Nucleotide substitutions in IVS-1 of the β-globin gene are frequent causes of β thalassemia. To test whether the IVS-1 substitution detected in Minoan 7 was a thalassemia mutation or an innocent SNP, functional studies were performed to determine whether the mutation affected mRNA splicing or expression of the β-globin gene. As described in the Materials and Methods, the β-globin expression plasmid, pCMVhβ, contained a 1.7 kb DNA fragment encompassing the entire human β-globin gene driven by the CMV promoter. The G to C mutation at IVS-1 position 91 was introduced into the expression plasmid, resulting in plasmid pCMVhβ(mut). The normal and mutant β-globin expression plasmids were transfected into HeLa cells. Total RNA was prepared from the transfected cells, β-globin specific cDNA was amplified by PCR and then cloned into T-vector. The positive clones were sequenced.
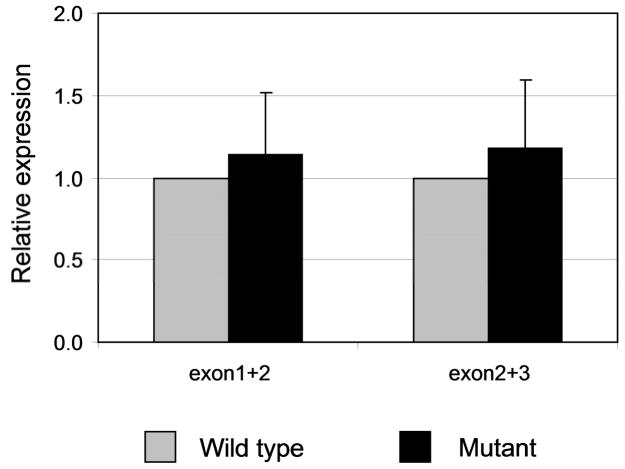
We sequenced three plasmid DNAs prepared from independent clones containing the wild type β-globin construct and four DNAs from mutated β-globin clones (Supplementary Fig. 3). The DNA sequences of all four mutant clones were identical to the sequences of the wild type control, indicating that the exon 1/exon 2 and the exon 2/exon3 junctions were normal in the mRNA transcribed from the mutated β-globin gene. The yield of globin mRNA was estimated in transient transfected HeLa cells by quantitative PCR. The results showed that the yield of mRNA from the mutated β-globin gene was 114% to 118% of the wild type (Figure 2), indicating that the mutant does not affect the efficiency of β-globin gene transcription. We conclude that the observed mutation represents an innocent single nucleotide polymorphism (SNP). This polymorphism is not recorded among the SNPs of the HapMap; however, another SNP mapping at position 91 of the β IVS-I (a C to T substitution) has been found in the analysis of DNA of 3,796 thalassemic heterozygotes in Greece [28].
Figure 2.
Comparison of the yield of β-globin mRNA in HeLa cells transfected with the wild and the mutated β-globin genes, respectively. Quantification of cDNA was performed by real time PCR using SYBR green mix on the Roche LightCycler. Two portions of globin cDNA were measured which covered the regions between exon 1 and exon 2 and between exon 2 and exon 3, respectively. The input of cDNA templates was normalized by the endogenous gene GAPDH. The level of the wild type β-globin gene expression set as 1. The values represent means ± SD.
Our results do not allow us to draw any definite conclusions about the frequency of β thalassemias in the Minoans. Thalassemias due to IVS-1 mutations represent the majority of thalassemia mutants in the Mediterranean populations. On the average, 64% of the β thalassemias in the Greek population are due to mutations at IVS-1 [20]. In the modern Cretan population, four mutations at IVS-1, IVS-6, IVS-110, and IVS-116 account for 63% of all β thalassemia mutations [21]. Presumably high frequencies of IVS-1 mutations were characteristic of the ancient Mediterranean and Anatolian populations. The average frequency of heterozygous β thalassemia in the modern Cretan population is 7.6% [29]. If that were also the frequency among the Minoans, we would expect to find one IVS-1 mutation among the 24 Minoans we sequenced, and we found none. It is therefore likely that the frequency of thalassemia mutations among the Minoans was relatively low.
Supplementary Material
Acknowledgments
We are grateful to Dr. Photini P.J. McGeorge for generously supplying us with the Ayios Haralambos materials. This work was supported by NIH Grants DK 045365 and GM 007454.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Higuchi R, Bowman B, Freiberger M, et al. DNA sequences from the quagga, an extinct member of the horse family. Nature. 1984;312:282–284. doi: 10.1038/312282a0. [DOI] [PubMed] [Google Scholar]
- 2.Hofreiter M, Serre D, Poinar HN, et al. Ancient DNA. Nat Rev Genet. 2001;2:353–359. doi: 10.1038/35072071. [DOI] [PubMed] [Google Scholar]
- 3.Wayne RK, Leonard JA, Cooper A. Full of sound and fury: the recent history of ancient DNA. Annu Rev Ecol Syst. 1999;30:457–477. [Google Scholar]
- 4.Cooper A, Poinar HN. Ancient DNA: do it right or not at all. Science. 2000;289:1139. doi: 10.1126/science.289.5482.1139b. [DOI] [PubMed] [Google Scholar]
- 5.Green RE, Krause J, Briggs AW, et al. A Draft Sequence of the Neandertal Genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drancourt M, Aboudharam G, Signoli M, et al. Detection of 400-year-old Yersinia pestis DNA in human dental pulp: An approach to the diagnosis of ancient septicemia. Proc Natl Acad Sci USA. 1998;95:12637–12640. doi: 10.1073/pnas.95.21.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mays S, Fysh E, Taylor G. Investigation of the link between visceral surface rib lesions and tuberculosis in a Medieval skeletal series from England using ancient DNA. Am J Phys Anthropol. 2002;119:27–36. doi: 10.1002/ajpa.10099. [DOI] [PubMed] [Google Scholar]
- 8.Taylor GM, Donoghue HD. Multiple loci variable number tandem repeat (VNTR) analysis (MLVA) of Mycobacterium leprae isolates amplified from European archaeological human remains with lepromatous leprosy. Microbes Infect. doi: 10.1016/j.micinf.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Hawass Z, Gad YZ, Ismail S, et al. Ancestry and pathology in King Tutankhamun’s family. JAMA. 2010;303:638–647. doi: 10.1001/jama.2010.121. [DOI] [PubMed] [Google Scholar]
- 10.Béraud-Colomb E, Roubin R, Martin J, et al. Human beta-globin gene polymorphisms characterized in DNA extracted from ancient bones 12,000 years old. Am J Hum Genet. 1995;57:1267–1274. [PMC free article] [PubMed] [Google Scholar]
- 11.Sato T, Kazuta H, Amano T, et al. Polymorphisms and allele frequencies of the ABO blood group gene among the Jomon, Epi-Jomon and Okhotsk people in Hokkaido, northern Japan, revealed by ancient DNA analysis. J Hum Genet. 2010;55:691–696. doi: 10.1038/jhg.2010.90. [DOI] [PubMed] [Google Scholar]
- 12.Stamatoyannopoulos G, Majerus PW, Perlmutter RM, Varmus H. Molecular basis of blood diseases. 3. The W.B. Saunders Publishing Co; Philadelphia: 2001. [Google Scholar]
- 13.Weatherall DJ, Clegg JB. The Thalassaemia Syndromes. Blackwell Scientific Publications; Oxford: 1981. [Google Scholar]
- 14.Chini V, Maloaguzzi Valeri C. Mediterranean Hemopathic Syndromes. Blood. 1949;4:989–1013. [PubMed] [Google Scholar]
- 15.Angel JL. Osteoporosis: Thalassemia? Am J Phys Anthropol. 1964;22:369–373. [Google Scholar]
- 16.Stuart-Macadam P. Porotic hyperostosis: A new perspective. Am J Phys Anthropol. 1992;87:39–47. doi: 10.1002/ajpa.1330870105. [DOI] [PubMed] [Google Scholar]
- 17.Walker PL. Porotic hyperostosis in a marine-dependent California Indian population. Am J Phys Anthropol. 1986;69:345–354. doi: 10.1002/ajpa.1330690307. [DOI] [PubMed] [Google Scholar]
- 18.Evans A. The Palace of Minos; a comparative account of the successive stages of the early Cretan civilization as illustrated by the discoveries at Knossos. Macmillan; London: 1921–1935. [Google Scholar]
- 19.Forget BG. Molecular mechanisms of b-thalassemia. In: Steinberg MH, Forget BG, Higgs DR, Nagel RL, editors. Disorders of hemoglobin: genetics, pathophysiology, and clinical management. Cambridge University Press; Cambridge: 2001. pp. 252–276. [Google Scholar]
- 20.Georgiou I, Makis A, Chaidos A, et al. Distribution and frequency of β-thalassemia mutations in northwestern and central Greece. Eur J Haematol. 2003;70:75–78. doi: 10.1034/j.1600-0609.2003.00017.x. [DOI] [PubMed] [Google Scholar]
- 21.Loutradi A. personal communication.
- 22.Krings M, Stone A, Schmitz RW, et al. Neandertal DNA sequences and the origin of modern humans. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 23.Yang DY, Eng B, Waye JS, et al. Technical note: improved DNA extraction from ancient bones using silica-based spin columns. Am J Phys Anthropol. 1998;105:539–543. doi: 10.1002/(SICI)1096-8644(199804)105:4<539::AID-AJPA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 24.Malmstrom H, Svensson EM, Gilbert MTP, et al. More on contamination: the use of asymmetric molecular behavior to identify authentic ancient human DNA. Mol Biol Evol. 2007;24:998–1004. doi: 10.1093/molbev/msm015. [DOI] [PubMed] [Google Scholar]
- 25.Poinar HN, Hoss M, Bada JL, Pääbo S. Amino acid racemization and the preservation of ancient DNA. Science. 1996;272:864–866. doi: 10.1126/science.272.5263.864. [DOI] [PubMed] [Google Scholar]
- 26.Klinken GJ, Mook WG. Preparative high-performance liquid chromatographic separation of individual amino acids derived from fossil bone collagen. Radiocarbon. 1990;32:155–164. [Google Scholar]
- 27.Hofreiter M, Jaenicke V, Serre D, et al. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 2001;29:4793–4799. doi: 10.1093/nar/29.23.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boussiou M, Karababa P, Sinopoulou K, et al. The molecular heterogeneity of beta-thalassemia in Greece. Blood Cells Mol Dis. 2008;40:317–319. doi: 10.1016/j.bcmd.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Malamos B, Fessas P, Stamatoyannopoulos G. Types of thalassaemia-trait carriers as revealed by a study of their incidence in Greece. Br J Haematol. 1962;8:5–14. doi: 10.1111/j.1365-2141.1962.tb06489.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.