Abstract
The interferon (IFN)-inducible p200-protein family includes structurally-related murine (for example, p202a, p202b, p204, and Aim2) and human (for example, AIM2 and IFI16) proteins. All proteins in the family share a partially-conserved repeat of 200-amino acid residues (also called HIN-200 domain) in the C-terminus. Additionally, most proteins (except the p202a and p202b proteins) also share a protein-protein interaction pyrin domain (PYD) in the N-terminus. The HIN-200 domain contains two consecutive oligosaccharide/oligonucleotide binding folds (OB-folds) to bind double stranded DNA (dsDNA). The PYD domain in proteins allows interactions with the family members and an adaptor protein ASC. Upon sensing cytosolic dsDNA, Aim2, p204, and AIM2 proteins recruit ASC protein to form an inflammasome, resulting in increased production of proinflammatory cytokines. However, IFI16 protein can sense cytosolic as well as nuclear dsDNA. Interestingly, the IFI16 protein contains a nuclear localization signal (NLS). Accordingly, the initial studies had indicated that the endogenous IFI16 protein is detected in the nucleus and within the nucleus in the nucleolus. However, several recent reports suggest that subcellular localization of IFI16 protein in nuclear versus cytoplasmic (or both) compartment depends on cell type. Given that the IFI16 protein can sense cytosolic as well as nuclear dsDNA and can initiate different innate immune responses (production of IFN-β versus proinflammatory cytokines), here we evaluate the experimental evidence for the regulation of subcellular localization of IFI16 protein in various cell types. We conclude that further studies are needed to understand the molecular mechanisms that regulate the subcellular localization of IFI16 protein.
Keywords: Interferon, p200-family proteins, IFI16, inflammasome, IFN-β production
1. The p200-family protein IFI16
Interferon (IFN)-inducible p200 family proteins in humans include IFI16, MNDA, IFIX, and AIM2 (encoded by the IFI16, MNDA, IFIX, and AIM2 genes) (Asefa et al., 2004; Choubey et al., 2008; Johnstone and Trapani, 1999; Ludlow et al., 2005; Mondini et al., 2010; Ouchi and Ouchi, 2008). These proteins share a partially conserved repeat of 200-amino acid residues (the HIN-200 domain) towards the C-terminus, which allows these proteins to bind dsDNA (Dawson and Trapani, 1995b; Yan et al., 2008). Most p200-family proteins (except the murine p202a and p202b proteins) also contain a homotypic protein-protein interaction PYRIN domain (PYD) in the N-terminus.
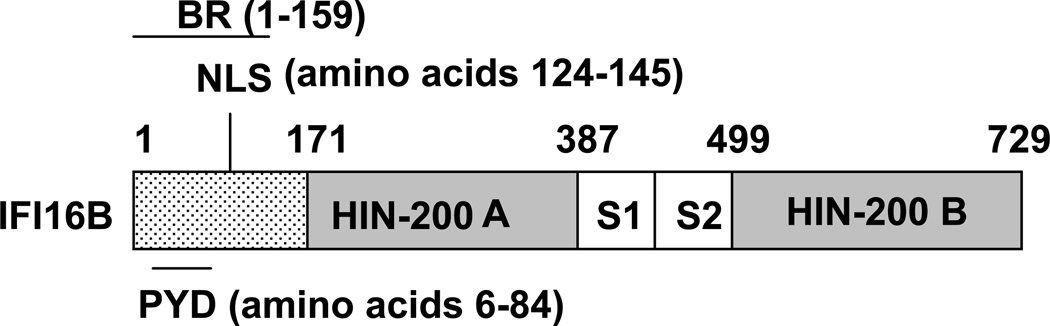
The IFI16 gene encodes three isoforms (A, B, and C) of the IFI16 protein through an alternative splicing of mRNA (Johnstone and Trapani, 1999; Choubey et al., 2008). The B form of IFI16 protein is the predominant form that is detected in human normal prostate epithelial cells and fibroblasts. The IFI16 protein contains two repeats (the repeat A and B) of 200-amino acid residues (or HIN-200 domain) and a serine-threonine-proline (S/T/P)-rich spacer region separates the two repeats (Fig. 1). The size of the spacer region in the IFI16 protein is regulated by mRNA splicing and can contain one, two, or three copies of highly conserved 56-amino acids S/T/P domain encoded by distinct axons. The N-terminus of IFI 16 protein contains a PYD (Choubey et al., 2010).
FIG. 1.
Schematic representation of structural and functional domains in IFI16 protein. Light dotted area in the amino terminus includes the basic region (BR; amino acid residues 1–159), which is sufficient to bind dsDNA in vitro, a PYD domain (amino acid residues 6–84), and a nuclear localization signal (NLS; amino acid residues 124–145). The dark gray boxes in the IFI16 protein denote a type-A and a type-B 200-AA repeat (or HIN-200 domain), respectively (Ludlow et al., 2005). White boxes (the S1 and S2) between the two repeats denote two spacer regions in the IFI16B protein.
Accumulated experimental evidence has attributed diverse functions to the p200-family proteins ranging from transcriptional regulation, apoptosis, cell growth regulation, autoimmunity, viral resistance, inflammasome assembly in response to cytosolic dsDNA, and cell differentiation (Johnstone and Trapani, 1999; Ludlow et al., 2005; Choubey et al., 2008; Ouchi and Ouchi, 2008; Mondini et al., 2010). Interestingly, the PYD domain of the AIM2 protein has been shown to heterodimerize with an adaptor protein ASC in response to cytoplasmic dsDNA to form the AIM2 inflammasome (Choubey et al., 2010; Fernandes-Alnemri et al., 2009). Moreover, upon sensing cytosolic dsDNA, the IFI16 protein has been reported to recruit the stimulator of interferon genes (STING) protein to stimulate the expression of IFN-β through the activation of the transcriptional activity of IRF3 and NF-κB (Unterholzner et al., 2010). Accordingly, a recent study noted that IFI16 protein may play a role in human dendritic cell activation by dsDNA and in the subsequent activation of the adaptive immune system (Kis-Toth et al., 2011). However, during the Kaposi Sarcoma-associated herpesvirus (KSHV) infection of human endothelial cells, IFI16 protein is shown to interact with the ASC protein through the PYD to form a functional IFI16-ASC inflammasome (Kerur et al., 2011). This protein complex containing the IFI16 protein was initially detected in the nucleus and then in the perinuclear area. Given that ASC adaptor protein is detected primarily in the nucleus in resting human monocytes/ macrophages (Bryan et al., 2009) and, upon infection, it redistributes to the cytosol (Bryan et al., 2009), it is likely that interactions between ASC and IFI16 proteins through the PYD in the nucleus contribute to the translocation of IFI16 protein to the cytoplasm.
The HIN-200 domain in the p200-family proteins consists of two oligonucleotide/oligosaccharide binding folds (OB-folds), which recognizes nucleic acids (Albrecht et al., 2005). Accordingly, the AIM2 protein requires this domain to sense cytosolic dsDNA and to assemble an inflammasome (Choubey et al., 2010). Similarly, the IFI16 protein can bind both single and double-stranded DNA in vitro (Dawson and Trapani, 1995b; Yan et al., 2008).
Expression of IFI16 protein is detectable in epithelial cells, fibroblasts, endothelial cells, and cells of the hematopoietic origin (Choubey et al., 2008; Johnstone and Trapani, 1999; Ouchi and Ouchi, 2008; Kis-Toth et al., 2011). Subcellular localization of IFI16 protein has been examined in different cell types (primary cells and cancer cell lines) and the primary tissues (Johnstone and Trapani, 1999). In contrast to the exclusive nuclear or cytoplasmic localization pattern of the other 200-family proteins, the IFI16 protein has been detected in the nucleus (within nucleus, both in the nucleolus and nucleoplasm), cytoplasm, or both (Table 1). As the subcellular localization of IFI16 protein is likely to determine the nature of an innate immune response (production of IFN-β versus activation of an inflammasome) following sensing of dsDNA, we decided to review the experimental evidence for the regulation of subcellular localization of IFI16 protein. Here we evaluate the experimental evidence for the regulation of subcellular localization of IFI16 protein. Additionally, we discuss various factors that are likely to regulate the subcellular localization of the IFI16 protein, thus, its functions.
Table 1.
Subcellular localization of IFI16 protein in various cell types
| Localization | Approach | Cell line (s)/tissue | Protein detected | Reference |
|---|---|---|---|---|
| Nuclear | CF* | HL-60 (a leukemia line) treated with IFN-γ | Endogenous | (Dawson and Trapani, 1995a) |
| Nuclear | IHC | Peripheral blood leukocytes | Endogenous | (Dawson and Trapani, 1995a) |
| Nuclear (nucleolar) | IIF | Peripheral blood mononuclear cells | Endogenous | (Dawson and Trapani, 1995b) |
| Nuclear | IIF | HL-60 (a leukemia line) treated with IFN-γ | Endogenous | (Dawson and Trapani, 1995b) |
| Nucleolus | CF | Daudi (a lymphoma line) | Endogenous | (Dawson and Trapani, 1995b) |
| Cytoplasmic and nuclear | IIF | HTC rat hepatoma tissue-culture line | Overexpressed fusion protein | (Briggs et al., 2001) |
| Nuclear (nucleolar) | IIF | HCC1937 (a breast cancer line with BRCA1 mutation) | Endogenous | (Aglipay et al., 2003) |
| Cytoplasmic and nuclear | IIF | PC-3 (a prostate cancer line) and PrECs | Endogenous | (Xin et al., 2003) |
| Cytoplasmic and nuclear | IIF | A431 (a skin carcinoma line) | Endogenous | (Barbe et al., 2008) |
| Nuclear and cytoplasmic | IIF | U2OS (an osteosarcoma line) | Endogenous | (Barbe et al., 2008) |
| Nuclear and nucleolus | IIF | U-251 MG (a glioma cell line) | Endogenous | (Barbe et al., 2008) |
| Nuclear | IIF | 293T (a human kidney transformed cell line) | Overexpressed fusion protein | (Hornung et al., 2009) |
| Nuclear | IIF | HeLa (a cervical cancer line with HPV-18) | Overexpressed fusion protein | (Burckstummer et al., 2009) |
| Nuclear and nucleolus | IIF | Primary human foreskin fibroblasts | Endogenous | (Cristea et al., 2010) |
| Cytoplasmic and nuclear | IIF | HeLa | Endogenous and overexpressed | (Berry et al., 2010) |
| Cytoplasmic and nuclear | IHC | Lung tissue sections | Endogenous | (Berry et al., 2010) |
| Cytoplasmic and nuclear | IIF | Primary skin keratinocytes | Endogenous | (Costa et al., 2011) |
| Cytoplasmic and nuclear | CF | Primary lung fibroblasts (WI-38) | Endogenous | (Duan et al., 2011) |
| Cytoplasmic | CF | THP-1 | Endogenous | (Veeranki et al., 2011) |
CF, Cell fractionation; IHC, Immunohistochemistry; IIF, Indirect immunofluorescence; PrECs, Prostate epithelial cells;
2. Subcellular localization of IFI16 protein
As noted above, the amino terminus of IFI16 protein contains a bi-partite nuclear localization signal (NLS; Briggs et al., 2001). Accordingly, a study noted that nuclear localization signal in IFI16 protein is sufficient to drive the nuclear localization of the β-Gal fusion protein (Briggs et al., 2001). However, the study noted the following deviations from the conventional nuclear import mechanisms: (i) the lack of strong binding of IFI16 NLS-fusion proteins with the importin heterodimers; (ii) the requirement for ATP, but not the cytosolic factors, for the nuclear import; and (iii) the Ran-independent import of the IFI16NLS-fusion proteins in the nucleus. Additionally, the study noted that the IFI16NLS fusion protein interacted with the Casein kinase 2 (CK2) and the CK2 phosphorylation site in the IFI16 protein also regulated the extent of the nuclear accumulation as well as the nuclear retention of the IFI16NLS-fusion protein. Moreover, several previous studies, using approaches involving indirect immunofluorescence and/or cell fractionation, reported different subcellular localization of IFI16 protein in a variety of cell types (Table 1). Notably, within the nuclear compartment, IFI16 protein is also detected in the nucleolus (Dawson and Trapani 1995b; Aglipay et al., 2003; Barbe et al., 2008; Cristea et al., 2010). Moreover, the murine ortholog of the IFI16 protein, the p204 protein, is also detected in the nucleolus (Choubey and Lengyel 1992). Although, the motifs that are involved in regulating nucleolar localization of proteins are not well defined (Emmott and Hiscox, 2009), and it has been proposed that the nucleolar localization of a protein results from interactions with rDNA and/or a protein that is located within the nucleolus. Given that IFI16 protein can bind to dsDNA (Dawson and Trapani, 1995b; Yan et al., 2008), and can associate with proteins, such as p53 (Johnstone et al., 2000; Liao et al., 2011), which are detected in the nucleolus (Rubbi and Milner, 2000), it is likely that interactions of IFI16 protein with rDNA and/or other proteins might contribute to its nucleolar localization in certain cell types.
Interestingly, upon exposure of keratinocytes to UV-B light, IFI16 protein was redistributed from nucleus to cytoplasm (Costa et al., 2011). This redistribution of the IFI16 protein was also associated with apoptosis of cells. However, the molecular mechanisms that regulate IFI16 protein redistribution between the nuclear and cytoplasmic compartment remain unknown. In this regard, studies indicate that several factors may regulate the subcellular localization of IFI16 protein (see below).
2.1 Polymorphisms in the IFI16 gene
We first reported cytoplasmic localization of IFI16 protein in PC-3 human prostate cancer cell line (Xin et al., 2003). Based on sequencing of the IFI16 cDNA (the sequence accession # AAM96005) isolated from the PC-3 cell line, we noted at least six non-conservative amino acid substitutions in the IFI16 protein. Consequently, we proposed that polymorphisms in the IFI16 gene that cause amino acid substitutions could affect the subcellular localization of IFI16 protein (Xin et al., 2003). Moreover, it has been reported that the SNP rs866484, which causes a Thr to Ser amino acid substitution at amino acid 178 in the IFI16 protein, is significantly associated with increased incidences of systemic lupus erythematosus (Kimkong et al., 2010). However, the functional significance of this amino acid substitution in the IFI16 protein remains unknown. These observations make it conceivable that amino acid substitutions in the IFI16 protein alter its interactions with other proteins and/or its sub-cellular localization.
2.2 Protein-protein interactions
Protein-protein interactions are known to affect the sub-cellular localization of proteins. Given that the IFI16 protein can form homo- and heterodimers with other proteins (for example, p53, Rb, AR, GR, BRCA1, and ASC) (Choubey et al., 2008; see Table 2), it is likely that the subcellular localization of the endogenous IFI16 protein in part depends on the cellular levels of these interacting proteins. The protein-protein interaction-dependent subcellular localization of IFI16 protein in certain cell types may also dictate the cell type-dependent functions. For example, though p53 protein has been mainly considered to be a nuclear protein, under certain circumstances (Shaulsky et al., 1990), it is also localized in the cytoplasm (Mihara et al., 2003). Therefore, to what extent interactions between the IFI16 and p53 proteins affects the sub-cellular localization of IFI16 protein (or possibly of the p53 protein) requires further investigation. Furthermore, the oxidative stress has been shown to stabilize the IFI16 protein (Gugliesi et al., 2005). Therefore, it is conceivable that, in response to oxidative stress, protein-protein interaction-dependent sequestration of IFI16 protein in a particular subcellular compartment may make the IFI16 protein inaccessible for protein degradation.
Table 2.
IFI16 interacts with other proteins through the HIN-200 or PYD domain.
| IFI16 interacting protein | Interacting IFI16 domain | Source of interacting protein | Reference |
|---|---|---|---|
| p53 | HIN-200A | Nuclear extracts | (Johnstone et al., 2000) |
| Absent in melanoma 2 (AIM2) | Not known | Cytosolic and nuclear extracts | (Veeranki et al., 2011) |
| Retinoblastoma protein (Rb) | HIN-200 | Total cell lysates | (Xin et al., 2003) |
| E2F1 | HIN-200 | Total cell lysates | (Xin et al., 2003) |
| Androgen receptor (AR) | Not known | Total cell lysates | (Alimirah et al., 2006) |
| Glucocorticoid receptor (GR) | Not known | Total cell lysates | (Berry et al., 2010) |
| ASC | Not known | Total cell lysates | (Kerur et al., 2011) |
| BRCA1 | PYD domain | Total cell lysates | (Aglipay et al., 2003) |
| STING | Not known | Total cell lysates | (Unterholzner et al., 2010) |
The IFI16 isoforms can form homo and heterodimers and the interaction domain was mapped in the amino terminal region (1–159 amino acid residues), which also contains PYD (amino acids 6–84) and NLS (amino acids 124–145) (Fig. 1) (Dawson and Trapani, 1995b; Briggs et al., 2001). Therefore, it remains to be determined whether the heterodimerization of IFI16 protein affects the accessibility of the NLS to the nuclear import machinery. Furthermore, IFI16 protein can also form heterodimers with the other family members, such as MNDA and AIM2 (Dawson and Trapani, 1995a). Because AIM2 and MADA proteins exhibit a contrasting subcellular localization patterns: the former being localized in primarily the cytoplasm and the later being in the nucleus, the relative levels of these two proteins within a cell type may regulate the subcellular localization of the endogenous IFI16 protein. Additionally, the IFN-treatment of IFN-responsive cells induces the expression of p200-family proteins. However, it is not clear how IFN-induced increased levels of these proteins affect the subcellular localization.
It has been reported that the IFI16 protein binds to DNA in vitro (Dawson and Trapani, 1995a; Yan et al., 2008) and in vivo (Dawson and Trapani, 1995b). Moreover, the amino terminal region (1–159 AA) of the IFI16 protein, which is rich in the basic amino acid residues and also contains the nuclear localization signal (Briggs et al., 2001), was found to be sufficient to mediate DNA-binding (Dawson and Trapani, 1995b). Consistent with the DNA-binding ability of IFI16 protein, it has been reported that IFI16 can act as cytosolic DNA sensor to viral DNA and play a critical role in modulation of immune responses by enhancing the IFN-β production (Unterholzner et al., 2010). Furthermore, it has been shown that presence of cytosolic viral DNA enhances interaction of IFI16 with STING and this interaction is necessary for cell’s ability to eliminate viral infections. The mouse ortholog of the human IFI16 protein, p204 protein, was also shown to orchestrate viral DNA-induced gene transcription by activating IRF3 and NF-κB transcription factors (Unterholzner et al., 2010). Studies also showed that IFI16 can assemble inflammasome in response to KSHV infection in the nucleus (Kerur et al., 2011). Moreover, our recent study revealed that interactions of IFI16 protein with AIM2 protein in the cytoplasmic fractions can attenuate caspase-1 activation by inflammasomes (Veeranki et al., 2011). These observations suggest that protein-protein interactions as well as binding of IFI16 protein to DNA (derived from infectious agents or self) may contribute to the regulation of its subcellular localization.
2.3 Hormonal regulation
IFI16 protein has been shown to bind the androgen receptor (AR) within the DNA-binding domain (Alimirah et al., 2006). Because in the absence of the male hormone (androgen), the majority of the AR is present in the cytoplasm (Simental et al., 1991), the interaction between IFI16 and AR in the absence of the AR ligand suggests a cytoplasmic localization of IFI16 protein. Furthermore, co-localization of both endogenous and overexpressed IFI16 protein along with the glucocorticoid receptor (GR) was observed in the HeLa cells as well as in human lung sections (Berry et al., 2010). Interestingly, the IFI16 protein was re-localized into the nucleus upon treatment of cells with the corticosteroids along with GR. However, only the IFI16B isoform was involved in the interaction with GR, but not the other two isoforms (the isoforms A and C). This is the first report that suggests the IFI16 isofrom-specific protein-protein interactions. These observations suggest sex hormone-dependent regulation of subcellular localization of IFI16 protein in certain cell types.
2.4 Posttranslational modifications
Though IFI16 protein is phosphorylated on Serine/Threonine residues, neither the upstream protein kinases nor the significance of such phosphorylation of IFI16 protein is known (Briggs et al., 2001). The isoform IFI16C was suggested to be modified by glycosylation (Johnstone et al., 1998). Given that the glycosylation also affect the nuclear localization of NLS containing proteins (Guinez et al., 2005), it is not clear to what extent, if any, the glycosylation of IFI16 protein plays a role in the regulation of its subcellular localization.
2.5 Expression levels
The onset of cellular senescence in human diploid fibroblasts (HDFs) increases levels of IFI16 protein in cells (Xin et al., 2004). Interestingly, our recent study (Duan et al., 2011) revealed that increased levels of IFI16 protein in old (versus young) HDFs were primarily detected in the cytoplasm and this subcellular localization of IFI16 protein was associated with increased levels of STING, IRF3, activated IRF3, and increased production of IFN-β. However, in the senescent cells (versus young or old cells), the IFI16 protein levels were lower. Moreover, the ratio between IFI16 and AIM2 protein decreased significantly in senescent HDFs, thus, favoring formation of the AIM2 inflammasome and production of proinflammatory cytokine IL-1β (Duan et al., 2011). These observations raise an interesting possibility that reduced cytosolic levels of IFI16 protein in senescent cells contribute to the activation of AIM2 inflammasome and cellular senescence-associated secretory phenotype (SASP).
3. Conclusions and future directions
It is likely that the subcellular localization of the endogenous IFI16 protein depends on the cell type and several factors described above. Given that the IFI16 protein can modulate diverse cellular functions, including inhibition of cell growth, modulation of apoptosis, inflammatory responses, DNA surveillance, and cellular senescence, it is of significance to correlate these various functions of IFI16 protein with its subcellular localization (cytoplasmic versus nuclear or both). It is likely that cells use both basal and the induced IFI16 protein levels to fine tune certain cellular responses. Therefore, a complete understanding of the molecular mechanisms that regulate the subcellular localization of the endogenous IFI16 protein in various cell types is needed to improve our understanding of the role of IFI16 protein in cell growth regulation and as an innate immune sensor.
Highlights.
-
➢
The p200-family protein IFI16 is a newly identified DNA sensor.
-
➢
The IFI16 protein can sense cytosolic as well as nuclear DNA to initiate different innate immune responses.
-
➢
Increased levels of the IFI16 protein are associated with lupus susceptibility.
-
➢
Subcellular localization of IFI16 protein is cell type-dependent.
-
➢
Several factors may regulate subcellular localization of IFI16 protein.
Acknowledgments
The work in the laboratory has been supported by grant awards from the National Institutes of Health (AI066261 and AG025036) and Merit Awards from the Department of Veterans Affairs to D. C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement
No competing financial interests exist.
References
- Aglipay JA, Lee SW, Okada S, Fujiuchi N, Ohtsuka T, Kwak JC, Wang Y, Johnstone RW, Deng C, Qin J, Ouchi T. A member of the Pyrin family, IFI16, is a novel BRCA1-associated protein involved in the p53-mediated apoptosis pathway. Oncogene. 2003;22:8931–8938. doi: 10.1038/sj.onc.1207057. [DOI] [PubMed] [Google Scholar]
- Albrecht M, Choubey D, Lengauer T. The HIN domain of IFI-200 proteins consists of two OB folds. Biochem Biophys Res Commun. 2005;327:679–687. doi: 10.1016/j.bbrc.2004.12.056. [DOI] [PubMed] [Google Scholar]
- Alimirah F, Chen J, Xin H, Choubey D. Androgen receptor auto-regulates its expression by a negative feedback loop through upregulation of IFI16 protein. FEBS Lett. 2006;580:1659–1664. doi: 10.1016/j.febslet.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Asefa B, Klarmann KD, Copeland NG, Gilbert DJ, Jenkins NA, Keller JR. The interferon-inducible p200 family of proteins: a perspective on their roles in cell cycle regulation and differentiation. Blood Cells Mol Dis. 2004;32:155–167. doi: 10.1016/j.bcmd.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Barbe L, Lundberg E, Oksvold P, Stenius A, Lewin E, Bjorling E, Asplund A, Ponten F, Brismar H, Uhlen M, Andersson-Svahn H. Toward a confocal subcellular atlas of the human proteome. Mol Cell Proteomics. 2008;7:499–508. doi: 10.1074/mcp.M700325-MCP200. [DOI] [PubMed] [Google Scholar]
- Berry A, Matthews L, Jangani M, Plumb J, Farrow S, Buchan N, Wilson PA, Singh D, Ray DW, Donn RP. Interferon-inducible factor 16 is a novel modulator of glucocorticoid action. FASEB J. 2010;24:1700–1713. doi: 10.1096/fj.09-139998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs LJ, Johnstone RW, Elliot RM, Xiao CY, Dawson M, Trapani JA, Jans DA. Novel properties of the protein kinase CK2-site-regulated nuclear- localization sequence of the interferon-induced nuclear factor IFI 16. Biochem J. 2001;353:69–77. [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Choubey D, Deka R, Ho SM. Interferon-inducible IFI16 protein in human cancers and autoimmune diseases. Front Biosci. 2008;13:598–608. doi: 10.2741/2705. [DOI] [PubMed] [Google Scholar]
- Choubey D, Duan X, Dickerson E, Ponomareva L, Panchanathan R, Shen H, Srivastava R. Interferon-inducible p200-family proteins as novel sensors of cytoplasmic DNA: role in inflammation and autoimmunity. J Interferon Cytokine Res. 2010;30:371–380. doi: 10.1089/jir.2009.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa S, Borgogna C, Mondini M, De Andrea M, Meroni PL, Berti E, Gariglio M, Landolfo S. Redistribution of the nuclear protein IFI16 into the cytoplasm of ultraviolet B-exposed keratinocytes as a mechanism of autoantigen processing. Br J Dermatol. 2011;164:282–290. doi: 10.1111/j.1365-2133.2010.10097.x. [DOI] [PubMed] [Google Scholar]
- Cristea IM, Moorman NJ, Terhune SS, Cuevas CD, O'Keefe ES, Rout MP, Chait BT, Shenk T. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J Virol. 2010;84:7803–7814. doi: 10.1128/JVI.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MJ, Trapani JA. IFI 16 gene encodes a nuclear protein whose expression is induced by interferons in human myeloid leukaemia cell lines. J Cell Biochem. 1995a;57:39–51. doi: 10.1002/jcb.240570106. [DOI] [PubMed] [Google Scholar]
- Dawson MJ, Trapani JA. The interferon-inducible autoantigen, IFI 16: localization to the nucleolus and identification of a DNA-binding domain. Biochem Biophys Res Commun. 1995b;214:152–162. doi: 10.1006/bbrc.1995.2269. [DOI] [PubMed] [Google Scholar]
- Duan X, Ponomareva L, Veeranki S, Panchanathan R, Dickerson E, Choubey D. Differential roles for the interferon-inducible IFI16 and AIM2 innate immune sensors for cytosolic DNA in cellular senescence of human fibroblasts. Mol Cancer Res. 2011;9:589–602. doi: 10.1158/1541-7786.MCR-10-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugliesi F, Mondini M, Ravera R, Robotti A, de Andrea M, Gribaudo G, Gariglio M, Landolfo S. Up-regulation of the interferon-inducible IFI16 gene by oxidative stress triggers p53 transcriptional activity in endothelial cells. J Leukoc Biol. 2005;77:820–829. doi: 10.1189/jlb.0904507. [DOI] [PubMed] [Google Scholar]
- Guinez C, Morelle W, Michalski JC, Lefebvre T. O-GlcNAc glycosylation: a signal for the nuclear transport of cytosolic proteins? Int J Biochem Cell Biol. 2005;37:765–774. doi: 10.1016/j.biocel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RW, Kershaw MH, Trapani JA. Isotypic variants of the interferon-inducible transcriptional repressor IFI 16 arise through differential mRNA splicing. Biochemistry. 1998;37:11924–11931. doi: 10.1021/bi981069a. [DOI] [PubMed] [Google Scholar]
- Johnstone RW, Trapani JA. Transcription and growth regulatory functions of the HIN-200 family of proteins. Mol Cell Biol. 1999;19:5833–5838. doi: 10.1128/mcb.19.9.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RW, Wei W, Greenway A, Trapani JA. Functional interaction between p53 and the interferon-inducible nucleoprotein IFI 16. Oncogene. 2000;19:6033–6042. doi: 10.1038/sj.onc.1204005. [DOI] [PubMed] [Google Scholar]
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimkong I, Avihingsanon Y, Hirankarn N. Association of IFI200 gene polymorphisms with susceptibility to systemic lupus erythematosus. J Rheumatol. 2010;37:1544–1547. doi: 10.3899/jrheum.091255. [DOI] [PubMed] [Google Scholar]
- Kis-Toth K, Szanto A, Thai TH, Tsokos GC. Cytosolic DNA-activated human dendritic cells are potent activators of the adaptive immune response. J Immunol. 2011;187:1222–1234. doi: 10.4049/jimmunol.1100469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow LE, Johnstone RW, Clarke CJ. The HIN-200 family: more than interferon-inducible genes? Exp Cell Res. 2005;308:1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Mondini M, Costa S, Sponza S, Gugliesi F, Gariglio M, Landolfo S. The interferon-inducible HIN-200 gene family in apoptosis and inflammation: implication for autoimmunity. Autoimmunity. 2010;43:226–231. doi: 10.3109/08916930903510922. [DOI] [PubMed] [Google Scholar]
- Ouchi M, Ouchi T. Role of IFI16 in DNA damage and checkpoint. Front Biosci. 2008;13:236–239. doi: 10.2741/2673. [DOI] [PubMed] [Google Scholar]
- Shaulsky G, Goldfinger N, Ben-Ze'ev A, Rotter V. Nuclear accumulation of p53 protein is mediated by several nuclear localization signals and plays a role in tumorigenesis. Mol Cell Biol. 1990;10:6565–6577. doi: 10.1128/mcb.10.12.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simental JA, Sar M, Lane MV, French FS, Wilson EM. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J Biol Chem. 1991;266:510–518. [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeranki S, Duan X, Panchanathan R, Liu H, Choubey D. IFI16 protein mediates the antiinflammatory actions of the type-I interferons through suppression of activation of caspase-1 by inflammasomes. PLoS ONE. 2011;6:e27040. doi: 10.1371/journal.pone.0027040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, Curry J, Johnstone RW, Nickoloff BJ, Choubey D. Role of IFI 16, a member of the interferon-inducible p200-protein family, in prostate epithelial cellular senescence. Oncogene. 2003;22:4831–4840. doi: 10.1038/sj.onc.1206754. [DOI] [PubMed] [Google Scholar]
- Yan H, Dalal K, Hon BK, Youkharibache P, Lau D, Pio F. RPA nucleic acid-binding properties of IFI16-HIN200. Biochim Biophys Acta. 2008;1784:1087–1097. doi: 10.1016/j.bbapap.2008.04.004. [DOI] [PubMed] [Google Scholar]