Abstract
Cystic fibrosis (CF), the most common lethal genetic disease among Caucasians, is caused by mutations in cystic fibrosis transmembrane conductance regulator (CFTR). CFTR’s main role is to transport chloride ions across epithelial cell membranes. It also regulates many cell functions. However, the exact role of CFTR in cellular processes is not yet fully understood. It is recognized that a key factor in CFTR-related regulation is its phosphorylation state. The important kinases regulating CFTR are cAMP-dependent protein kinase A (PKA) and 5′-AMP-activated protein kinase (AMPK). PKA and AMPK have opposite effects on CFTR activity despite their highly similar structures and recognition motifs. Utilizing homology modeling, in silico mutagenesis and literature mining, we supplement available information regarding the atomic-resolution structures of PKA, AMPK and CFTR, and the complexes CFTR–PKA and CFTR–AMPK. The atomic-resolution structural predictions reveal an unexpected availability of CFTR Ser813 for phosphorylation by both PKA and AMPK. These results indicate the key role of the structural flexibility of the serine-rich R-domain in CFTR regulation by phosphorylation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00894-011-1029-0) contains supplementary material, which is available to authorized users.
Keywords: CFTR, AMPK, PKA, Interaction, Molecular modeling, Docking
Introduction
Cystic fibrosis (CF) is caused by mutations in cystic fibrosis transmembrane conductance regulator (CFTR) [1]. The main function of CFTR is its role as a protein kinase-regulated chloride channel, but it is a multifunctional protein. CFTR, located in the apical membranes of secretory epithelial cells, also anchors a dynamic protein macro complex involved in multiple cell functions [2].
Clinical observations have identified a large spectrum of phenotypes of CF, even for patients bearing the same mutation. These observations cannot be solely explained by mutations in CFTR. Modifier genes and/or epigenetic factors may be potential severity modulating factors. Studies suggest that dynamic interactions within the CFTR interactome—important players in the regulation of cell and epithelial functions—can serve as severity modulation factors as well [3].
CFTR belongs to the ATP-binding cassette transporter (ABC) superfamily of proteins. It is a 1480-amino acid transmembrane protein with a symmetrical structure: a repeat composed of a transmembrane region (TMD) and a nucleotide binding domain (NBD) separated by an intrinsically disordered [4, 5] hydrophilic regulatory domain (R domain). The R domain is a unique feature among ABC transporters. A molecular model of the CFTR structure model was prepared by Riordan’s group [6], but it is not complete. During this study we successfully extended this model.
Several regulatory factors affecting CFTR chloride channel activity have been reported, but CFTR chloride channel activity is regulated mostly by protein kinases [7]. The following kinases have been reported to be involved in this regulation: cAMP-dependent protein kinase A (PKA), protein kinase C, Ca2+/calmodulin-dependent kinase, cGMP-dependent kinase, CK2 protein kinase [8], and 5′-AMP-activated protein kinase (AMPK) [9]. This list is likely still incomplete.
The best described is the process of activating CFTR channels via phosphorylation by PKA in the presence of ATP. There are 15 serines that need to be mutated into alanines before CFTR loses its PKA activation sensitivity; none of these serines alone is necessary or sufficient for full PKA response [10]. PKA phosphorylation sites are found primarily in the R region, but are also found within the NBD1 sequence [11].
Understanding the mechanisms underlying the regulation of CFTR by PKA also requires an understanding of the mechanisms underlying the opposite effect of another kinase, AMPK, which inhibits the PKA-dependent activation of CFTR and leads to the inhibition of Cl− secretion in vitro and in vivo [12].
AMPK forms a heterotrimer consisting of subunits α, β and γ. Subunits β, γ and the C-terminal domain of subunit α form the regulatory part of the protein. The N-terminal catalytic domain of subunit α possesses the kinase functionality. Catalytic and regulatory parts are connected with a flexible linker. It has been shown that the AMPK regulatory part binds to the NBD domains of CFTR [13–16]. The R-domain sequence carries two sites recognized by AMPK: “inhibitory” serines S737 and S768; however, CFTR phosphorylation by AMPK is decreased by only ∼80% when those two serines are replaced by alanines [7].
In order to gain insight into the interaction between CFTR and its regulatory kinases, the aim of the present work was to create atomic-resolution models of the complexes of CFTR with PKA and AMPK.
Our results allowed us to draw conclusions about the AMPK–CFTR and PKA–CFTR binding interfaces and to prove at the structural level a hypothesis regarding the role of the flexibility of the R domain in CFTR phosphorylation.
Materials and methods
Sequence analysis
The SwissProt/Trembl [17] database was used as the source of sequences of human 5′-AMP-activated protein kinase (AMPK) subunits α1 (id: Q13131), β1 (id: Q9Y478), γ2 (id: Q9UGJ0), human PKA (id: P17612) and human cystic fibrosis transmembrane conductance regulator (CFTR) (id: P13569).
Templates for structural modeling were identified as sequence homologs among the proteins deposited in the Protein Data Bank [18] using the BLAST algorithm [19] and the HHPRED program [20]. Pairwise alignments of sequences of the AMPK subunits with the chosen templates were prepared with the Needleman–Wunsch algorithm [21], and are included in the “Electronic supplementary material.”
Identified AMPK α1 catalytic domain templates fulfilled the following conditions: (i) the structure has bound ATP/ADP and a substrate peptide; (ii) the structure is not altered by interactions; (iii) sequence similarity enables homology modeling; (iv) the template structure should have the least number of gaps in the structure.
Using these conditions, the α-catalytic subunit of mouse cAMP-dependent protein kinase (pdb: 1JBP:E [22]) was identified as the best template candidate.
For AMPK β1, the structures of the yeast proteins SPCC1919.03c and SIP2 (pdb ids: 2OOX:B [23] and 2QLV:B [24] respectively) were used as templates. The best templates to model AMPK γ2 were found to be rat 5′-AMP-activated protein kinase subunit γ-1, yeast hypothetical protein C1556.08c, and nuclear protein SNF4 (pdb ids: 2V8Q:E, 2OOX:G and 2QLV:C, respectively). In cases where more than one template was chosen to model a given subunit, the structures of the templates were aligned using CE [25] and FATCAT [26].
The sequence of the α-catalytic subunit of mouse cAMP-dependent protein kinase (pdb: 2GU8 [27]) exhibits 97.6% identity to the sequence of human PKA in the covered fragment 15–351.
Modeling tools
Loop modeling
Loops were modeled by searching the PRODAT database with SYBYL 8.0.
Minimization
Models were subjected to energy minimization using the AMBER7 99 force field [28], Gasteiger–Hückel charges, a dielectric constant of 80, and simplex initial optimization followed by Powell minimization terminated at a gradient change of 0.1 kcal/(mol Å), as implemented in SYBYL 8.0.
In silico mutagenesis
Side chains of mutated residues were replaced using SYBYL 8.0.
Visualization
Figures representing protein structures were prepared using PyMol [29].
CFTR modeling
To model CFTR fragment 1156–1208, a secondary structure prediction was obtained using a Jpred [30] server. We decided to rely on Jpred because other (transmembrane-oriented) tools predicted that the sixth helix ends at residue 1148, which is not likely. The predicted helical fragment (CFTR 1156–1180) was modeled using the sixth helix of the TMD1 domain as a template, and connected with NBD2 by modeled loop (CFTR 1181–1208).
CFTR fragment 807–817 was remodeled using peptide bound to the α-catalytic subunit of mouse cAMP-dependent protein kinase as template substrate. The template peptide’s residues RRASI were replaced in silico by the corresponding CFTR residues RRLSQ; the residues flanking this sequence were removed. Rotamers of sidechains had to have: (i) minimal structural overlap with the existing structure; (ii) minimal full atom RMSD to the old residues. As a next step, the structure of the prepared pentapeptide was superimposed on the original CFTR model 810–814 fragment. The residues 807–817 were removed from the R domain, and the loops CFTR 807–809 and CFTR 815–817 that link the prepared 810–814 pentapeptide with the R domain were modeled.
AMPK modeling
Catalytic domain: SWISS-MODEL was used to model AMPK α1 1–274. The modeling of fragments 289–293 and 302–307 using 1JBP:E as a template and linking all fragments with loops was carried out in SYBYL.
Regulatory part: SWISS-MODEL [31] was used to model the C-terminal regulatory domain of AMPK α1, while SYBYL 8.0 was used to model subunits β1 and γ2. Two AMPK α1 models with two templates were prepared: (i) AMPK α1 358–478, using carbon catabolite derepressing protein kinase (pdb: 2QLV:A); (ii) AMPKα1 398–550, using rat 5′-AMP-activated protein kinase subunit α1 (pdb: 2V8Q:A). Overlapping fragments were structurally aligned (RMSD 3.06 Å) then removed. Models were merged, with the missing fragment 469–471 modeled as a loop.
There are more gaps in the C-terminal part of the alignment of AMPK β1 fragment 73–270 with the SIP2 protein β subunit (pdb: 2QLV:B) than in the corresponding part of the alignment with protein SPCC1919.03c (pdb: 2OOX:B). In the modeling of AMPK β1 73–157, 2QLV:B was used as a template; in the modeling of fragment 167–270, 2OOX:B was used. The models of fragments 73–157 and 167–270 were then structurally aligned with structure 2QLV:B to obtain their position in the AMPK trimer (as we used 2QLV:A and C to model other subunits of the trimer). The missing linker AMPK β1 158–166 was modeled as a loop.
AMPK γ2 was modeled using nuclear protein SNF4 (pdb: 2QLV:C) as template.
Human PKA modeling
The structure of the mouse PKA catalytic [27] subunit was used as the template. Eight residues differentiating mouse PKA from human PKA (T19, S21, Q26, D31, S52, A111, E163, and T335) were replaced in silico in SYBYL 8.0 with the corresponding human PKA residues S, A, H, E, T, P, Q, and S, respectively. The structure obtained was subjected to energy minimization.
Modeling of the CFTR–AMPK complex
The interface region of the regulatory domain CFTR was defined on the basis of experimentally determined interaction sites described in the literature. The surface and structure of the interface region was visually scanned for spacial and hydrophobic complementarity. The identified area was used to manually construct the complex subjected to minimization. The catalytic domain was docked to the CFTR R domain using the previously docked PKA structure as a template. Complex analysis was performed using the Solvent Accessible Surface calculator of Michel Sanner’s Molecular Surface algorithm [32] and the Protein Interactions Calculator [33]
Results
AMPK model extension
A database search revealed that the available AMPK model [34] could be extended. Most of the potential templates are fragmentary and contain different fragments that are homologous to AMPK. The protein of the sequence most similar to the catalytic domain of AMPK α1—protein kinase Chk2—is crystallized as a homodimer, with inhibitory T-loops projecting from the bodies of the subunits involved in the formation of the interface [35]. This dimerization-enabling feature, specific to Chk2, disqualified its structure. The 1JBP:E structure we used has a RMSD from the Chk2 structure (with T-loop excluded) of below 2 Å.
When modeling the regulatory part, the 2V8Q structure—which was the most sequentially similar—seemed to be the best template for trimer modeling. However, the 2V8Q:E chain was truncated prior to crystallization, resulting in the deformation of the 190–198 fragment. The 2V8Q structure was excluded from further modeling of the regulatory part of AMPK. Structural alignments demonstrated that the structures 2QLV and 2OOX are almost identical to 2V8Q (their similarities in terms of the RMSDs of the protein backbone atom positions are presented in Table 1 in the “Electronic supplementary material”).
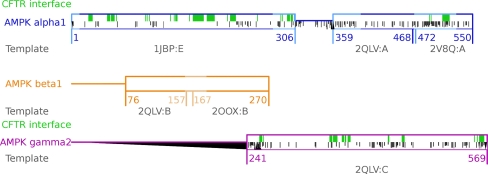
For structure-based interaction predictions, it is of the upmost importance to use continuous structural models of the proteins involved, as any discontinuity may contain potential obstacles or introduce clues for complex formation. All of our models were extended to meet the continuity criterion in the interface areas. The range of extension of the AMPK model is presented in Fig. 1.
Fig. 1.
In-scale visualization of the AMPK modeling effort. Horizontal boxes depict modeled fragments. Horizontal lines depict fragments of unknown homologs with known structures, probably unstructured fragments. Lighter box lines depict fragments of the AMPK model created in this work; darker lines depict fragments that were previously modeled. Green fill depicts the predicted interface of AMPK with CFTR. Grayscaling depicts differences in sequence between isoforms that interact and do not interact with CFTR (position-specific score of Needleman–Wunsch alignment of AMPK α1 vs. AMPK α2 and AMPK γ2 vs. γ1). Black triangles depict gaps: those pointing up depict gaps in the presented isoform; those pointing down depict gaps in isoforms that do not interact with CFTR. Shorter and darker lines depict different residues. Numbers below the lines describe the range of the model: the pdb id of the template structure used to model these fragments is shown in gray
CFTR–PKA catalytic domain interaction
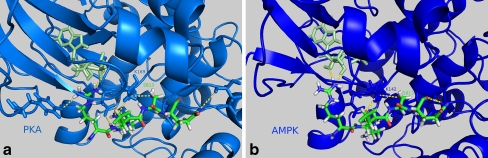
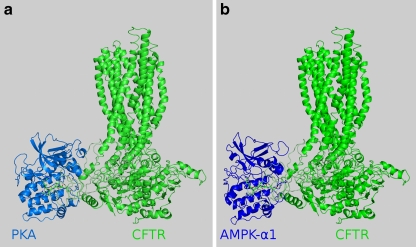
Ten residues of CFTR are known to be phosphorylated by protein kinase A (PKA). However, even mutations of all ten of these residues do not prevent the activation of CFTR by PKA [36]. In our model, S813—which is known to have the strongest activating effect among all serines phosphorylated in CFTR—is the most exposed. We were aware that the R domain is highly unstable, and that the conformation under study was just one of many possible. We confirmed that this conformation allowed the binding of PKA to S813. After docking human PKA to CFTR, we observed that the following H-bonds, responsible for substrate peptide recognition, formed between the recognized pentapeptide of CFTR (810–814) and both PKA and PKA-bound ADP: CFTR Arg810–ADP; CFTR Arg810–PKA Tyr331; CFTR Arg811–PKA Lys169, Glu171; CFTR Ser813–PKA Lys169; CFTR Glu814–PKA Leu199. These bonds are presented in Fig. 2a. Lys169 is a residue in the active site associated with the kinase activity of PKA. The surface position of S813, on flexible linkers, agrees with data showing an immediate CFTR response to PKA activation. S813 is accessible, and its environment allows the rapid binding of PKA. The overall structure of the CFTR–PKA complex is presented in Fig. 3a.
Fig. 2.
Interactions of CFTR pentapeptide 810–814 with PKA (left) and AMPK (right). Kinase structures are simplified, and only residues that form H-bonds with the pentapeptide are drawn in detail (yellow dashed lines). CFTR S813 and the lysines at the active site of the kinases are labeled
Fig. 3.
Overall structures of the CFTR–PKA catalytic domain (left) and the CFTR–AMPK (right) complexes. Kinases and CFTR are represented in cartoon form, while the CFTR pentapeptide 810–814 is presented in both pictures in stick form
CFTR–AMPK catalytic domain complex formation
Riordan’s de novo modeling [4] of the R domain provides many clusters with different conformations. The conformation presented in the template model used is considered to be one of many that occur naturally in a cell. The R domain is connected to other domains via linkers: 658–679 and 843–847. NMR experiments have shown that the flexibilities of some R-domain fragments decrease after NBD1 binding [5]. In the conformation used, these parts are internally stabilized, so they are not predicted to interact directly with NBD1. There is experimental evidence showing large movements of the R domain vs. the NBDs and TMDs [37]. As the relative position of the R domain with respect to channel-forming domains has not been specified, we treated the linkers as flexible strings, and the R-domain position was not fixed during the docking of the catalytic domains of AMPK α1 and PKA. The R domain extracted from the whole CFTR model was scanned for the AMPK recognition motif. Two serines—S737 and S768—were identified. Experimental evidence of the inhibitory role of AMPK after the phosphorylation of S737 and S768 was provided in previous research [37–39]. However, experiments [7] have shown an 80% decrease in AMPK phosphorylation for the CFTR double mutant S737A-S768A. We decided to use relaxed AMPK recognized motif, RRXS/TY, to identify other potential target serines. S813 was identified. We decided to use PKA previously bound to S813 as a template for AMPK α1 catalytic domain docking. After preparing the R domain–AMPK catalytic domain complex, the R domain was re-attached to CFTR.
The R domain with the docked AMPK α1 catalytic subunit was minimized. The bonds required for substrate peptide binding by protein kinase [22] were satisfied.
In our model, we observed the following H-bonds responsible for substrate peptide recognition between the recognized pentapeptide of CFTR (810–814) and AMPK, and between ADP and AMPK: CFTR Arg810–ADP; CFTR Arg811–AMPK Glu145, Tyr181; CFTR Ser813–AMPK Lys142, Ser178; CFTR Gln814–AMPK Gly177. These bonds are presented in Fig. 2b. Lys142 is a residue in the active site associated with the kinase activity of AMPK. The overall structure of CFTR in complex with the AMPK catalytic domain is presented in Fig. 3b.
Comparison of the CFTR–AMPK catalytic domain and CFTR–PKA complexes
Our analysis revealed high probabilities of both complexes. The interface areas of both complexes measured with MSMS [32] differed slightly: 2548 Å2 for CFTR–PKA compared to 2149 Å2 for CFTR-AMPK, which suggests that the PKA–CFTR complex is more stable. However, both interface areas are typical of heterodimers. More detailed analysis with PIC [33] showed that although the AMPK–CFTR interface has only six hydrophobic interactions (in contrast to the nine present at the PKA–CFTR interface), and it does not form (main chain)–(main chain) hydrophobic bonds (in contrast to the three bonds formed in the PKA–CFTR case), its higher number of (main chain)–(side chain) hydrogen bonds (30 compared to 22) and (side chain)–(side chain) hydrogen bonds (29 compared to 20) suggest that both complexes have similar stabilities.
CFTR–AMPK regulatory domain complex
Experimental data for the CFTR–AMPK complex indicate that (i) the CFTR region 1420–1457 interacts with the 407–550 region of AMPK α1 [16], and (ii) the R domain is phosphorylated by the catalytic domain of AMPK α1 [12]. Prepared models of AMPK and CFTR lack unstructured tails: the final complex was defined as exposing terminal residues of the truncated subunits to the solvent in a way that enables chain extension. The AMPK regulatory domain was manually docked to CFTR in the experimentally confirmed interface area in a manner that conforms with established restrictions.
The CFTR interface region consists of two parts: fragment 1420–1428 is part of the NBD, while 1429–1450 is an unstructured CFTR tail. In our work, we modeled the interaction with a structured part (1420–1428), ensuring that the model provides the space required for the C-terminal tail. The complex was minimized. Interfaces did not introduce any energetically unfavorable conformations for any of the proteins; nor were any of the predicted H-bonds broken due to an unfavorable environment.
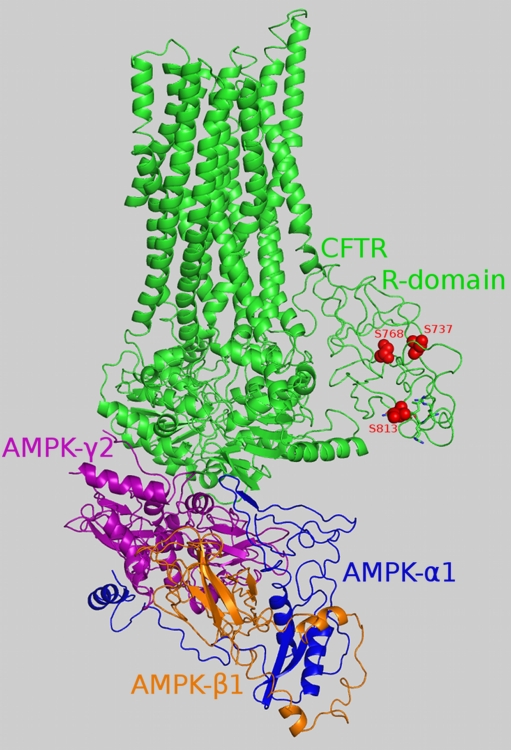
The structure of the CFTR–AMPK regulatory domain complex is shown in Fig. 4. In the interface created, 52 and 49 residues are involved on the CFTR and AMPK sides respectively, with a total area of 1856 Å2. This is an average surface area for a nonobligatory complex interface. As further support for our model, we used information on mutations disrupting the interaction between AMPK α1 and CFTR [16].
Fig. 4.
Overall structure of the CFTR–AMPK regulatory domain complex. The main chains of the proteins are colored by chain, with the R domain highlighted in a lighter green color compared to the rest of the CFTR protein. Atoms from S737, S768 and S813 in the R domain are presented as red spheres. CFTR pentapeptide 810–814 is represented in stick form (except for S813, which is presented in sphere form)
Discussion
Our models show that two counteracting kinases, PKA and AMPK, can phosphorylate CFTR S813, suggesting that the different effects of AMPK and PKA on CFTR Cl− channel activity result from interactions with other proteins and/or S813 availability.
AMPK binding to CFTR
Our model shows that AMPK regulatory binding to CFTR is not required for CFTR phosphorylation. However, the R domain undergoes constant structural changes, hampering the formation of an appropriate interface. Furthermore, the R domain has only two phospho sites in the precise AMPK-recognition motif (S737 and S768), which are inaccessible in the conformation used. The binding of AMPK to CFTR with the regulatory domain may provide the time required for the R domain to change conformation, thus allowing access to the “inhibitory” serines.
S813 phosphorylation by PKA and AMPK
The complexes of CFTR with the catalytic subunit of PKA and the catalytic domain of AMPK α1 are bound at the S813 site within the R domain.
Our model of S813 binding by PKA is consistent with experimental evidence: the active-site lysine (K169) is able to react with S813 and bind adenosine phosphate. AMPK bound to S813 in the same conformation as PKA, and did not differ from it in terms of the H-bonds formed and interface stability. This result suggests that AMPK may phosphorylate S813. This shows that, at the molecular level, it is possible that 20% of the currently unexplained phosphorylation signal observed in the CFTR double mutant S737A-S768A incubated with AMPK may come from “activator” serines such as S813.
Role of CFTR R-domain flexibility in phosphorylation
CFTR activity depends on R-domain phosphorylation [10, 40]. In the process of CFTR regulation, the R domain acts as an inhibitor that needs to be phosphorylated to enable CFTR channel activity [41]. Based on the results of NMR experiments, Baker et al. described the R domain as “sampling multiple, heterogeneous conformations, […] with rapid interconversion between conformers [5].” In our work, we decided to use one conformation among the many derived by Riordan et al. by de novo modeling [4]. In our model, the R domain’s structure prohibits the phosphorylation of the PKA and PKC phospho sites S686, S712, S768, and S790, which are known to be phosphorylated in vitro under controlled conditions. The latest experimental results show that the “inhibitory” serines S737 and S768 introduce an insensitive CFTR mutant into PKA (in which 15 potential phosphoserines are mutated to alanines) and restore PKA sensitivity [10]. Considering the diminishing differences between “activating” and “inhibiting” serines as well as “serines phosphorylated by activating and inhibiting kinases,” further structural analyses of the R domain and the nature of its interaction with suitable kinases appear to be crucial elements for elucidating CFTR regulation.
Our results present a molecular explanation for the phenomenon of the dependence of CFTR phosphorylation on the flexibility of the R domain. A new question can thus be formulated: what are the factors that influence the structural state of the R domain prior to phosphorylation?
Electronic supplementary material
Below is the link to the electronic supplementary material.
Structural similarities of AMPK subunit homologs (DOC 56 kb)
Acknowledgments
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- CF
Cystic fibrosis
- CFTR
Cystic fibrosis transmembrane conductance regulator
- ABC
ATP-binding cassette
- TMD
Transmembrane domain
- NBD
Nucleotide binding domain
- PKA
cAMP-dependent protein kinase A
- AMPK
5′-AMP-activated protein kinase
- RMSD
Root mean square deviation
References
- 1.Wine JJ. Cystic fibrosis: how do CFTR mutations cause cystic fibrosis? Curr Biol. 1995;5:1357–1359. doi: 10.1016/S0960-9822(95)00269-7. [DOI] [PubMed] [Google Scholar]
- 2.Kirk KL. New paradigms of CFTR chloride channel regulation. Cell Mol Life Sci. 2000;57:623–634. doi: 10.1007/PL00000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ollero M, Brouillard F, Edelman A (2006) Cystic fibrosis enters the proteomics scene: New answers to old questions. Proteomics 14:4084–4099 [DOI] [PubMed]
- 4.Hegedűs T, Serohijos AWR, Dokholyan NV, He L, Riordan JR. Computational studies reveal phosphorylation-dependent changes in the unstructured R domain of CFTR. J Mol Biol. 2008;378:1052–1063. doi: 10.1016/j.jmb.2008.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker JMR, Hudson RP, Kanelis V, Choy W-Y, Thibodeau PH, Thomas PJ, et al. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat Struct Mol Biol. 2007;14:738–745. doi: 10.1038/nsmb1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serohijos AWR, Hegedűs T, Aleksandrov AA, He L, Cui L, Dokholyan NV, et al. Phenylalanine-508 mediates a cytoplasmic–membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc Natl Acad Sci USA. 2008;105:3256–3261. doi: 10.1073/pnas.0800254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kongsuphol P, Cassidy D, Hieke B, Treharne KJ, Schreiber R, Mehta A et al (2009) Mechanistic insight into control of CFTR by AMPK. J Biol Chem 284:5645–5653 [DOI] [PMC free article] [PubMed]
- 8.Treharne KJ, Xu Z, Chen JH, Best OG, Cassidy DM, Gruenert DC et al (2009) Inhibition of protein kinase CK2 closes the CFTR Cl− channel, but has no effect on the cystic fibrosis mutant DeltaF508-CFTR. Cell Physiol Biochem 24:347–360 [DOI] [PMC free article] [PubMed]
- 9.King JDJ, Fitch AC, Lee JK, McCane JE, Mak DO, Foskett JK, et al. AMP-activated protein kinase phosphorylation of the R domain inhibits PKA stimulation of CFTR. Am J Physiol Cell Physiol. 2009;297:C94–C101. doi: 10.1152/ajpcell.00677.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hegedűs T, Aleksandrov AA, Mengos A, Cui L, Jensen TJ, Riordan JR. Role of individual R domain phosphorylation sites in CFTR regulation by protein kinase A. Biochim Biophys Acta. 2009;1788:1341–1349. doi: 10.1016/j.bbamem.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Lewis HA, Buchanan SG, Burley SK, Conners K, Dickey M, Dorwart M, et al. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kongsuphol P, Hieke B, Ousingsawat J, Almaca J, Viollet B, Schreiber R et al (2009) Regulation of Cl− secretion by AMPK in vivo. Pflugers Arch 457:1071–1078 [DOI] [PubMed]
- 13.Carling D (2004) The AMP-activated protein kinase cascade: a unifying system for energy control. Trends Biochem Sci 29:18–24 [DOI] [PubMed]
- 14.Hallows KR, McCane JE, Kemp BE, Witters LA, Foskett KJ. Regulation of channel gating by AMP-activated protein kinase modulates cystic fibrosis transmembrane conductance regulator activity in lung submucosal cells. J Biol Chem. 2003;278:998–1004. doi: 10.1074/jbc.M210621200. [DOI] [PubMed] [Google Scholar]
- 15.Mehta A. CFTR: more than just a chloride channel. Pediatr Pulmonol. 2005;39:292–298. doi: 10.1002/ppul.20147. [DOI] [PubMed] [Google Scholar]
- 16.Hallows KR, Raghuram V, Kemp BE, Witters LA, Foskett KJ. Inhibition of cystic fibrosis transmembrane conductance regulator by novel interaction with the metabolic sensor AMP-activated protein kinase. J Clin Invest. 2000;105:1711–1721. doi: 10.1172/JCI9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.UniProt Consortium (2008) The universal protein resource. Nucleic Acids Res 36:D190–D195 [DOI] [PMC free article] [PubMed]
- 18.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 22.Trafny EA, Xuong NH, Adams JA, Eyck LFT, Taylor SS, Sowadski JM, Madhusudan EA. cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer. Protein Sci. 1994;3:176–187. doi: 10.1002/pro.5560030203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Townley R, Shapiro L. Crystal structures of the adenylate sensor from fission yeast AMP-activated protein kinase. Science. 2007;315:1726–1729. doi: 10.1126/science.1137503. [DOI] [PubMed] [Google Scholar]
- 24.Amodeo GA, Rudolph MJ, Tong L (2007) Crystal structure of the heterotrimer core of Saccharomyces cerevisiae AMPK homologue SNF1. Nature 449:492–495 [DOI] [PubMed]
- 25.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 26.Ye Y, Godzik A (2003) Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics 19(Suppl 2):246–255 [DOI] [PubMed]
- 27.Lin X, Murray JM, Rico MX, Wang DT, Chu Y, Zhou M, et al. Discovery of 2-pyrimidyl-5-amidothiophenes as potent inhibitors for AKT: synthesis and SAR studies. Bioorg Med Chem Lett. 2006;15:4163–4168. doi: 10.1016/j.bmcl.2006.05.092. [DOI] [PubMed] [Google Scholar]
- 28.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, Ross WS, Simmerling CL, et al. AMBER6. J Am Chem Soc. 1999;117:5179–5197. [Google Scholar]
- 29.Schrödinger LLC (2010) PyMOL. http://www.pymol.org
- 30.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22:195–201 [DOI] [PubMed]
- 32.Sanner M, Olson AJ, Spehner JC (1996) Reduced surface: an efficient way to compute molecular surfaces. Biopolymers 38:305–320 [DOI] [PubMed]
- 33.Tina KG, Bhadra R, Srinivasan N. PIC: Protein Interactions Calculator. Nucleic Acids Res. 2007;35:W473–W476. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin X, Townley R, Shapiro L (2007) Structural insight into AMPK regulation: ADP comes into play. Structure 15:1285–1295 [DOI] [PubMed]
- 35.Oliver AW, Paul A, Boxall KJ, Barrie ES, Aherne WG, Garret MD, et al. Trans-activation of the DNA-damage signalling protein kinase Chk2 by T-loop exchange. EMBO J. 2006;25:3179–3190. doi: 10.1038/sj.emboj.7601209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang X-B, Tabcharanie JA, Hou Y-X, Jensen TJ, Kartner N, Alon N et al (1993) Protein kinase A (PKA) still activates CFTR chloride channel after mutagenesis of all 10 PKA consensus phosphorylation sites. J Biol Chem 268:11304–11311 [PubMed]
- 37.Mio K, Ogura T, Mio M, Shimizu H, Hwang T-C, Sato C, et al. Three-dimensional reconstruction of human cystic fibrosis transmembrane conductance regulator chloride channel revealed an ellipsoidal structure with orifices beneath the putative transmembrane domain. J Biol Chem. 2008;283:30300–30310. doi: 10.1074/jbc.M803185200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkinson DJ, Strong TV, Mansoura MK, Wood DL, Smith SS, Collins FS, et al. CFTR activation: additive effects of stimulatory and inhibitory phosphorylation sites in the R domain. Am J Physiol. 1997;273:L127–L133. doi: 10.1152/ajplung.1997.273.1.L127. [DOI] [PubMed] [Google Scholar]
- 39.Vais H, Zhang R, Reenstra WW. Dibasic phosphorylation sites in the R domain of CFTR have stimulatory and inhibitory effects on channel activation. Am J Physiol. 2004;287:C737–C745. doi: 10.1152/ajpcell.00504.2003. [DOI] [PubMed] [Google Scholar]
- 40.Gadsby DC, Nairn AC (1999) Control of CFTR channel gating by phosphorylation and nucleotide hydrolysis. Physiol Rev 79:S77–S107 [DOI] [PubMed]
- 41.Lewarchik CM, Peters KW, Qi J, Frizzel RA (2008) Regulation of CFTR trafficking by its R domain. J Biol Chem 283:28401–28412 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structural similarities of AMPK subunit homologs (DOC 56 kb)