Abstract
Germ-line mutations in BRCA1 breast cancer susceptibility gene account for a large proportion of hereditary breast cancer families and show considerable ethnic and geographical variations. The contribution of BRCA1 mutations to hereditary breast cancer has not yet been thoroughly investigated in Middle Eastern and North African populations. In this study, 16 Tunisian high-risk breast cancer families were screened for germline mutations in the entire BRCA1 coding region and exon–intron boundaries using direct sequencing. Six families were found to carry BRCA1 mutations with a prevalence of 37.5%. Four different deleterious mutations were detected. Three truncating mutations were previously described: c.798_799delTT (916 delTT), c.3331_3334delCAAG (3450 delCAAG), c.5266dupC (5382 insC) and one splice site mutation which seems to be specific to the Tunisian population: c.212 + 2insG (IVS5 + 2insG). We also identified 15 variants of unknown clinical significance. The c.798_799delTT mutation occurred at an 18% frequency and was shared by three apparently unrelated families. Analyzing five microsatellite markers in and flanking the BRCA1 locus showed a common haplotype associated with this mutation. This suggests that the c.798_799delTT mutation is a Tunisian founder mutation. Our findings indicate that the Tunisian population has a spectrum of prevalent BRCA1 mutations, some of which appear as recurrent and founding mutations.
Keywords: Hereditary breast cancer, BRCA1 mutations, Founder effect, Tunisians
Introduction
Breast cancer is the most common cause of cancer-related death among women worldwide with an estimated 1.15 million new cases and 411,000 breast cancer deaths around the world in 2002 [1, 2]. Breast cancer incidence has increased progressively in Middle Eastern and North African (MENA) populations over the last 10 years, probably due to more reliable data being collected from cancer registries and to easier access by patients to screening and diagnostic programs [3]. In MENA populations, breast cancer represents ~13–30% of newly diagnosed malignancies in women and occurs at a median age of 49–52 years as compared to 63 in industrialized nations [3–12]. Breast cancer in MENA populations is characterized by younger age at onset, advanced stage and poor prognosis [3–12]. Mastectomy is performed in more than 80% of MENA women with breast cancer [3].
Comparison of the clinical and tumor gene expression profiles of breast cancer between French and MENA patients showed minor but significant biological differences. A significant difference has been found in breast cancer diagnosis age (±10 years old) between French and MENA patients. This difference was more marked in Tunisian patients with a more than 13-year-old diagnosis difference. Moreover, a higher frequency of aggressive tumors was found among MENA patients as defined by the SBR histo-prognostic grade III, tumor size at diagnosis and lymph node metastasis [13].
Breast cancer is an extremely complex, heterogeneous and multi-factorial disease characterized by a progressive multistep process caused by interactions of both genetic and non-genetic factors. A family history of breast cancer is the most prominent risk factor for the development of the disease [14]. It is estimated that about 5–10% of all breast cancers may arise from hereditary predisposition [15, 16]. Investigation of multiple-case families in which breast cancer segregates with mendelian patterns of inheritance led to the identification of the tumor suppressor genes BRCA1 (MIM 113705) [17] and BRCA2 (MIM 600185) [18], which account for a substantial proportion of early-onset breast cancer but a much smaller proportion of late-onset disease [19, 20]. Germ-line BRCA1 mutations are involved in about 30% of site-specific female breast cancer families, 75% of breast/ovarian cancer families, but not in male breast cancer families [21–23]. Germ-line BRCA2 mutations are involved in about 30% specific female breast cancer families, 20–25% of breast/ovarian cancer families, and 50–80% of male/female breast cancer families [19, 20, 22–24]. These figures are deduced from studies on European and American populations and they are expected to be different for other populations. Cumulative risk of cancer in BRCA germ-line mutated carriers is highly dependant on the mutation type, the genetic background and acquired environmental factors. Life-time cumulative cancer risk in BRCA1 mutation carriers is about 80–85% for female breast cancer, 50% for ovarian cancer, 12–40% for bilateral breast cancer [19, 25–28]. Life-time cumulative cancer risk in BRCA2 mutation carriers is about 80–85% for female breast cancer (with a later breast cancer onset), 10–25% for ovarian cancer, 6–10% for male breast cancer [19, 23–28].
The frequency and spectrum of mutations within BRCA1/2 genes vary widely among populations. In some ethnic or geographically isolated groups, founder mutations can explain the majority of inherited breast and ovarian cancer cases [29–32], whereas in other populations, germ-line mutations are randomly scattered throughout the coding sequence [33].
The analysis of the BRCA1/2 genes as well as other genes conferring increased risk of hereditary breast cancer (e.g. TP53, PTEN, STK11, CDH1) was poorly studied in the MENA populations. Few reports, including from our group, reported few BRCA1/2 mutations associated with hereditary breast cancers in a small number of families from MENA countries [34–39].
The aim of this study was to investigate the mutational spectrum and frequency of germ-line BRCA1 mutations as well as to explore the existence of population-specific recurrent or founder mutations, if any, in Tunisian breast cancer families.
Patients and methods
Patients and families
The current study was conducted on breast cancer families in the middle coast of Tunisia with the cooperation of two main cancer treatment centers in Sousse, Tunisia: departments of Radiation Oncology and Medical Oncology of Farhat Hached teaching Hospital, Sousse, Tunisia.
Selection criteria of familial cases were based on age at diagnosis and the number of affected first- and second-degree relatives with breast or ovarian cancer. These families’ characteristics are presented in Table 1. The inclusion criteria were (i) Three or more breast and/or ovarian cancers in first or second degree relatives one of which developed at an early age <35 years or (ii) two cases with young age at diagnosis (<35 years) and/or bilateral breast cancers and/or multiple cancers including ovarian and breast cancers. Blood samples were taken from at least one affected woman from each family. Twenty-four patients belonging to 16 families were recruited. One family presented a case of ovarian cancer and no families with male breast cancer were found. All families were apparently unrelated. Written informed consent was obtained from all subjects. Ethics committee approval was obtained from Weill Cornell Medical College in Qatar and from Farhat Hached teaching hospital of Sousse in Tunisia.
Table 1.
Families’ characteristics of our cohort
| Family ID | Sex of proband | Phenotype and age at diagnosis | Family history of breast and ovarian cancers and age at diagnosis | Family history of other cancers |
|---|---|---|---|---|
| F1 | Female | Br30 | M Br 35, PC Br 35 | Leukemia |
| F2 | Female | Br 34 | S Br 20 | Leukaemia |
| F3 | Female | Br 34 | MA Br 45, MA Br ? | None |
| F4 | Female | Br 35 | MA Br 50, GM Br? | Brain |
| F5 | Female | Br 46 | S Br 42, M Br 65 | None |
| F7 | Female | Br 36 | S Br 40, M Br ?, GM Br 50 | None |
| F8 | Female | BBr 38 | M Br 38, S Br 43 | None |
| F9 | Female | Br 42 | S Br 35 | Prostate |
| F10 | Female | Br 34 | M Br33 | None |
| F11 | Female | Br 53 | S Br 40, S Br 60 | None |
| F12 | Female | Br 29 | S Br 39 | None |
| F13 | Female | Br 61 | S Br 28 | None |
| F14 | Female | Br 47 | S Br 35, N Br 30 | Cervical, liver |
| F15 | Female | Br 42 | M Br 40 | None |
| F16 | Female | Br35 | M Br?, MA Br? | Colon |
| F17 | Female | Br 49 | MA Br 52, MA Br 68, PC Br 42, PC Br 35, PC Ov 62 | Colon, Nasopharynx |
Br breast cancer, BBr bilateral breast cancer, OV ovarian cancer, M mother, S sister, GM grand-mother, MA maternal aunt, PC paternal cousin, N Niece
Methods
Genomic DNA was extracted from peripheral blood leukocytes using the standard salting out procedure [40]. As for mutation screening, the BRCA1 gene coding region and exon–intron boundaries were analyzed using direct DNA sequencing. The exons were amplified in 20 μl with 100 ng DNA, 1× reaction buffer, 200 μM dNTPs, 1.5 mM MgCl2, 0.8 μM primers (designed by Centre Jean Perrin, sequences available on request) and 1 unit Taq polymerase (except primers, all other reagents from Promega, France). PCR was performed in an thermocycler (Biometra, Germany) with an initial denaturation at 94°C for 5 min, followed by 30 cycles of (94°C 20 s, 54°C 20 s, 72°C 20 s), except for exon 7 (15 cycles of 94°C 20 s, 60°C 10 s, 72°C 20 s then 25 cycles of 94°C 20 s, 56°C 15 s, 72°C 20 s) and 23 (5 cycles of 94°C 20 s, 57°C 20 s, 72°C 20 s then 30 cycles of 94°C 20 s, 53°C 20 s, 72°C 20 s). Exon 11 was analysed in nine overlapping PCR fragments. After amplification, the PCR products were subjected to electrophoresis in a 2% agarose gel. The product was cut from the gel and purified using QIAquick gel extraction kit (Qiagen, CA). Both DNA strands were sequenced using BigDyeDeoxy terminator cycle sequencing kit (BD V3.1, Applied Biosystems) according to the manufacturer’s instructions. The product was purified on a separation column (AutoSeqTM G-50, Amersham Biosciences), and the templates were sequenced on an automated ABI-PRISM 310 Genetic Analyzer (Applied Biosystems). Sequence analysis was performed using Seqman software (DNAstar, Madison, WI).
Results
In the current study, the whole BRCA1 gene screening was carried out on 24 patients belonging to 16 unrelated high-risk breast cancer families from the middle coast of Tunisia. These families’ characteristics are presented in Table 1. The median age of onset disease was 41 years, range 29–65.
Twenty-three cases were diagnosed with site-specific breast cancer and one with bilateral breast cancer. BRCA1 gene mutation analysis was performed in these cases. BRCA1 gene’s entire coding region and its exon–intron boundaries were examined by direct sequencing.
Our results revealed six breast cancer families carrying deleterious BRCA1 mutations with a frequency of 37.5% (6/16). In this study, a total of 19 alterations were identified in the BRCA1 gene. An overview of the mutational spectrum is listed in Table 2 with both the Human Genome Organization (HUGO) approved systematic nomenclature and the Breast Cancer Information Core (BIC) traditional nomenclature.
Table 2.
BRCA1 Germline mutations identified in a cohort of 16 breast cancer families from the middle coast of Tunisia
| Exon/intron | Systematic nomenclature | BIC traditional nomenclature | Amino acid change | Mutation type |
|---|---|---|---|---|
| Deleterious mutations | ||||
| 11 | c798_799delTT | 916delTT | Stop285 | Frameshift |
| 11 | c3331_3334delCAAG | 3450delCAAG | Stop1115 | Frameshift |
| 20 | c.5266dupC | 5382insC | Stop1829 | Frameshift |
| 5 | c.212 + 2insG | IVS5 + 2insG | – | Splicing |
| Sequence variants of unknown significance | ||||
| 8 | c.442-58delT | IVS8-58delT | – | Intronic variant |
| 11 | c.1067 A > G | 1186A > G | Q356R | Missense |
| 11 | c.1456 T > C | 1575T > C | F486L | Missense |
| 11 | c.1648A > C | 1767A > C | N550H | Missense |
| 11 | c.2077 G > A | 2196G > A | D693N | Missense |
| 11 | c.2082C > T | 2201C > T | S694S | Silent |
| 11 | c.2311 T > C | 2430T > C | L771L | Silent |
| 11 | c.2529C > T | 2640C > T | R841W | Missense |
| 11 | c.2612C > T | 2731C > T | P871L | Missense |
| 11 | c.3113A > G | 3232A > G | E1038G | Missense |
| 11 | c.3119A > G | 3238A > G | S1040N | Missense |
| 11 | c.3548C > T | 3667A > G | K1183R | Missense |
| 13 | c.4308T > C | 4427T > C | S1436S | Silent |
| 16 | c.4837A > G | 4956A > G | S1613G | Missense |
| 18 | c.5117 G > C | 5236G > C | G1706A | Missense |
Four distinct deleterious BRCA1 mutations were identified. Three are frame-shift mutations including two small deletions (c.798_799delTT, c.3331_3334delCAAG) and one small insertion (c.5266dupC), all resulting in frame-shifts that cause premature protein termination at codon 285, 1115 and 1829, respectively. The fourth mutation was an intron 5 splice site insertion (c.212 + 2insG). The 15 remaining identified variants were missense (11), silent (3) and intronic (1) mutations classified as polymorphisms or unknown variants in the BIC database (Table 2). Among these variants three were unique (observed only once): (F486L), c.1648A/C (N550H) and c.5117 G/C (G1706A).
Among the 19 BRCA1 alterations detected in our study, the splice site mutation (c.212 + 2insG) has not been reported previously in the BIC database and is therefore considered to be novel. The deleterious mutations were located only in three exons of BRCA1 gene with one mutation in exon 5 (c.212 + 2insG), two mutations in exon 11 (c.798_799delTT, c.3331_3334delCAAG) and one mutation in exon 20 (c.5266dupC).
The c.798_799delTT was shared by three apparently unrelated families and it accounted for 3 of the 16 (18%) BRCA1 mutated families. The remaining mutations, including c.3331_3334delCAAG, c.5266dupC and c.212 + 2insG, were unique (i.e., each one was detected only in one family).
From the six families with BRCA1 mutations, three (50%) had three breast cancer cases. The remaining three families (50%) had two cases of early-onset breast cancer, with a diagnosis age being ≤35 years. Five of these were families with site-specific breast cancer, and only one was a bilateral breast cancer family. The median age of onset disease was 42 years (ranged from 34–65) and 40 years (ranged from 29–61) among carriers and non-carriers, respectively.
Discussion
The prevalence and spectrum of BRCA1 mutations among Middle-Eastern and North African (MENA) breast and ovarian cancer families have not yet been thoroughly studied [39, 41]. In the current study, we have screened the BRCA1 gene in a cohort of 16 high-risk breast cancer families from the middle-coast of Tunisia. Interestingly, the prevalence is higher and the mutation spectrum is different as we observed only one of the four previously reported mutations [41].
BRCA1 mutations were identified in six families of our cohort with a notable 37.5% frequency. Four distinct deleterious BRCA1 mutations were identified in our study: c.798_799delTT c.3331_3334delCAAG, c.5266dupC and c.212 + 2insG.
The c.798_799delTT was the most commonly observed mutation. It was shared by five patients belonging to three apparently unrelated families (F2, F8 and F10). It accounted for 3 of the 16 (18%) BRCA1 mutated families. First, this mutation was detected in two unrelated patients belonging to families F2 and F10 with site-specific breast cancer. The same mutation was also observed in two Algerian families [39]. Analyzing five microsatellite markers in and flanking the BRCA1 locus showed a common haplotype associated with this mutation in these four carriers (data not shown) [39]. Then this mutation was targeted and found in family 8. The index case, diagnosed with bilateral breast cancer at the age of 38 years, was a carrier of the c.798_799delTT mutation. In this family the mutation co-segregates with the disease and the same mutation was identified in the proband’s mother and sister diagnosed with breast cancer at ages 38 and 43, respectively (Fig. 1). These findings suggest that the c.798_799delTT mutation is a Tunisian and North African founder mutation. It has been cited four times in the BIC database but only once referring to a Spanish origin (Galician). To our knowledge, it has been reported in two breast-ovarian cancer families from northeastern France [42] and also in two breast-ovarian cancer families from southern Italy (Sicily) [43]. Recently, it has been identified in three unrelated families from the middle and south Sardinia area with many Phoenician and Carthaginian archaeological sites [44]. This restricted geographical distribution of the c.798_799delTT mutation in these close Mediterranean countries may suggest a common founder ancestor.
Fig. 1.
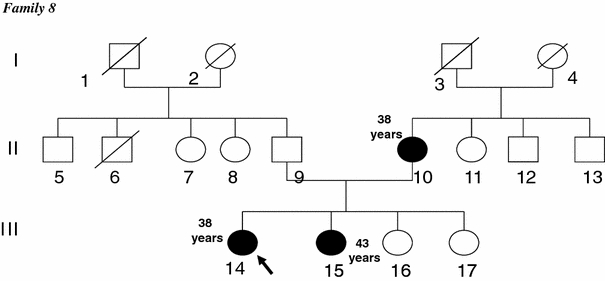
Pedigree of the family 8. Blackened circles indicate women affected with breast cancer. The proband was designed by an arrow. The index case, diagnosed with bilateral breast cancer at the age of 38 years, was a carrier of the c.798_799delTT mutation. In this family the mutation co-segregates with the disease and the same mutation was identified in the proband’s mother and sister diagnosed with breast cancer at ages 38 and 43, respectively
The c.5266dupC mutation in BRCA1 exon 20 is the second most frequently reported mutation in the BIC database. Though often referred to as a Jewish founder [45], this mutation is very prevalent in Central and Eastern Europe with a high frequency among different ethnic populations such as Poland (~34%) [46], Russia (14%) [31], Greece (31%) [47] and Brazil [48]. Studies using genetic markers suggest that the same haplotype is shared by 5382insC mutation carriers and have placed the origin of the mutation back to approximately 36 generations, in the Middle Ages, somewhere in the Baltic area [49–51]. More recent data support origin from a single common ancestor [52]. In our study, the c.5266dupC mutation has been found only once in a breast cancer family. The same mutation was additionally identified in a Tunisian family with a history of breast and ovarian cancer [41], which suggests this mutation contributed to both breast and ovarian phenotype in the Tunisian population. Having been detected in two unrelated families, it has a prevalence of 4% (2/48) in the mutation spectrum of BRCA1 gene. Altogether, our cumulative results suggest that c.5266dupC is also a potential recurrent mutation and may be one of the founder effects in the Tunisian population. Further studies will be necessary in order to estimate more accurately the population allele frequency and distribution of this mutation among Tunisians. Further haplotype studies of all the carriers are required to find out whether the c.5266dupC identified in our population has a common ancestor with the European mutation or whether high-frequency alleles have arisen independently more than once.
The identification of these founder and recurrent mutations in the population is an extremely important step towards the improvement of genetic counselling since molecular testing can be targeted to the founder and recurrent mutations allowing for a more rapid and less expensive test. Furthermore, families sharing identical mutations will make up a genetically homogenous subgroup allowing more significant studies of their clinical expression.
In our set, the c.3331_3334delCAAG frame shift mutation is found in one family with a 6% frequency. According to data from the BIC database, this mutation has been cited 40 times mainly with Western European, Caucasian ethnicities but only in two families of Latin American/Caribbean ancestry and one family with Egyptian ethnicity (BIC). It has been reported to occur in Spain, Portugal [53, 54], Australia, Canada, South America (Chilean population) [55] and it is also a founder mutation in Hispanic families from Colombia [56] (BIC). These observations raise the question whether BRCA1: c.3331_3334delCAAG is an ancient mutation that has arisen once, or has occurred several times in human history. The large geographical distribution of this mutation could suggest independent origins as demonstrated for the 4184del4 BRCA1 mutation reported to have at least three independent origins [50]. A similar finding was observed for the 1135insA BRCA1 mutation that does not have a single origin and was found to occur in three distinct haplotype backgrounds [57]. This suggests that 4184del4 and 1135insA mutations may have appeared independently several times in the human genome and may therefore be relative “hot spots” within the BRCA1 gene [50, 57], this may also be the case for the c.3331_3334delCAAG mutation. Because of the complexity of the genomic structure of BRCA1 and the scattering of the mutations throughout the gene, a screening of the whole sequence is necessary and still remains a serious technical challenge especially in developing countries. For that reasons a “pre-screen” based on the most frequent BRCA1 mutations seen in the BIC could be considered as a first step, prior to complete gene sequence analysis [57].
The c.798_799delTT, 5266dupC and c.3331_3334delCAAG mutations have been reported in different populations. The identification of these mutations in Tunisian patients can be explained by the widespread allelic heterogeneity arising from the several different colonizations that Tunisia underwent through history especially because of its location at the northernmost bulge of Africa to mark the division between the eastern and western Mediterranean Sea. It is highly conceivable that these mutations could have migrated to Tunisia and their introduction in our population might have some specific historical origins. Further genealogical studies can trace the origins of these mutations and define exactly when and how they were introduced into the Tunisian population. Determining the origins of our identified mutations may be a helpful criterion in identifying high-risk women regarding their origins or ethnic background.
The c.212 + 2insG mutation has not been described in the BIC database and to our knowledge has not been reported in any other population studies so far. Therefore, it is considered to be novel. In general, splice-site alterations lead to skipping of the complete exon following the acceptor or preceding the donor site in which the mutation is located.
For genetic counseling purposes, it is extremely important to differentiate between deleterious and polymorphic splice-site alterations. Analysis at the mRNA level will allow us to identify whether this variant leads to an aberrant splicing and can contribute to a correct and definitive clinical interpretation.
Apart from the deleterious mutations, we have also identified 15 BRCA1 variants classified as polymorphisms or variants with unknown significance (Table 2). These variants are known to occur worldwide and in populations from various geographical areas. Among these 15 variants, only Q356R and F486L were not described in our previous study [41]. Three were unique: F486L, N550H and G1706A (observed only once). Interestingly, in the present study, the F486L and N550H variants were only identified in a healthy woman belonging to F17 family. The association of the Y179C, F486L and N550H variants together in cis with breast cancer has been suggested by Augello et al. with similar cases in Sicilian and Spanish patients [43, 58, 59]. In our set, the Q356R variant, not described in the previous study, occurred with a 16% frequency. Dunning et al. reported that the BRCA1Arg356 allele has a higher genotype distribution in healthy controls than in breast cancer patients and may thus play a protective role in breast cancer [60]. A recent study has also reported similar findings [61]. These findings need deeper investigations. Further studies are required to compare their frequencies in cases and controls and thus find out whether some variants or haplotypes are breast cancer risk modifiers.
The prevalence of BRCA1 mutations reported in our study (37.5%) is approximately 2.5 times higher than that reported initially (15.6%) [41]. Furthermore, the two spectra emerging from screening in families from the northern and central parts of Tunisia are not stringently similar. This suggests that significant differences in the frequency and spectrum of BRCA1 mutations between the different regions of the country cannot be excluded. However, the higher detection rate observed in the present study (37.5%) can be partly explained by the existence of the founder mutation. To better define the true BRCA1 mutations frequency and spectrum in the Tunisian population, we extended the study up to 48 families by combining our data with our previously published results involving 32 families which were recruited in the same manner and selected using the same criteria as here [41]. As a result, a cumulative mutation analysis from these 48 Tunisian breast/ovarian cancer families demonstrated deleterious BRCA1 mutations in 11 families with a notable frequency of 23% (11/48). Altogether, seven different deleterious mutations were detected in the BRCA1 gene. It is noteworthy that all found mutations are located only in three exons of BRCA1 (exons 5, 11 and 20). Moreover, more than half (57%) of the identified mutations are located in exon 11 which spans 60% of the entire BRCA1 coding sequence. Because of the scattering of the BRCA1 mutations throughout the gene, a screening of the whole sequence is necessary but still remains a serious technical challenge especially in developing countries. In Tunisia, in the future planning for the effective population screening, a pre-screening of these three exons could be considered as a first step, prior to complete the whole sequence analysis. Mutation screening can be performed by different techniques which are less laborious and expensive than direct sequencing. The most used techniques are DHPLC (Denaturing High Performance Liquid Chromatography), PTT (Protein Truncation Test) and SSCP (Single Strand Conformational Polymorphism) [44, 47, 56, 59].
Unlike the spectra observed in genetically homogenous populations such as Iceland, Norway and Poland, the heterogeneous BRCA1 mutation spectrum observed in our series of families is consistent with the genetic heterogeneity and ethnic diversity of the Tunisian population and also may reflect the existence of multiple distinct founders.
For the likelihood of having BRCA1 mutations, two criteria are of importance: age at diagnosis, and the number and types of cancer in the family. To establish the best criteria in selecting families with a high probability of carrying BRCA1 mutations in our population, we combined our results with those of a previously-published study about the contribution of BRCA mutations to breast cancer in Tunisia [34]. Three out the 36 families studied in this report were excluded from our cumulative analysis (one family had a BRCA2 mutation and two families presented male breast cancer). The stratification of families into three subgroups allowed a more detailed analysis. Hereditary site-specific female breast cancer families accounted for the large majority of this sample (69.5%; 34 out of 49 families). However, only six families were found to carry deleterious BRCA1 mutations, accounting for the lowest mutation frequency (17.6%, 6/34). Families presenting bilateral breast cancer accounted only for 8% (4/49). In this subgroup, we found that 25% (1/4) of families were attributed to deleterious BRCA1 mutations. In contrast, a higher rate of BRCA1 pathogenic mutations was detected in hereditary breast/ovarian cancer families (36%, 4/11). This subgroup accounted for 22.5% (11/49) of our sample. When taking the affected relative number into account, we noticed that among mutation carriers only 36% had three or more affected relatives. These observations are not in agreement with previous studies showing that the frequency of the mutation increased with the number of affected family members.
Most studies on different populations indicate that the frequency of BRCA1 mutations is higher in early onset breast cancer and that the mutation carriers are relatively younger than those non-mutation carriers at the age of cancer diagnosis [62, 63]. In this cumulative analysis, we notified that there was no significant difference between BRCA1 mutation carriers and non-carriers at the age of cancer diagnosis (46 vs. 43.8 years).
Interestingly, among carriers, the mean age of cancer diagnosis was more than a decade lower in the breast cancer families than in the breast-ovarian cancer families (43 vs. 53.5 years). These results suggest that, in the Tunisian population, the presence of ovarian cancer, is more important than the family scale in the prediction of BRCA1 mutations and that diagnosis age <45 years may be a useful tool to distinguish breast (but not ovarian) cancer families having a greater likelihood of carrying BRCA1 mutations.
Our cumulative mutation analysis showed that in 77% of the affected families, the disease could not be explained by BRCA1 mutations and that the lowest rates occurred in breast cancer families (17% in site-specific breast cancer and 25% in bilateral breast cancer). This indicates that more efforts are necessary to better understand which kind of families should be more appropriately considered for BRCA1 screening, particularly among those characterized by more than three female breast cancer cases. This also indicates that BRCA2 screening in these families should take priority, and also suggests that low penetrance genes are likely to contribute to susceptibility to hereditary breast/ovarian cancer in Tunisia [64].
In summary, our data extend the previous ones about hereditary breast and ovarian cancer in Tunisia and highlighted the importance of the first major breast cancer susceptibility gene in the aetiology of breast cancer in our population. Our study revised the frequency of deleterious BRCA1 mutations and consistently enlarged its spectrum in Tunisia. These studies seem to indicate that the BRCA1 mutational spectrum is rather broad and heterogeneous in the Tunisian population. Some of these mutations appear specific and recurrent or founding ones and also reflect the existence of multiple distinct founders. In the current study, we described for the first time an example of a founder mutation in the Tunisian population (c.798_799delTT). Our cumulative mutation analysis showed that genetic testing of Tunisian high-risk breast cancer patients should be first targeted to BRCA1 gene’s exons 5, 11 and 20 where all identified mutations are located and where it is about 23% more likely to detect a mutation. In the Tunisian population, the likelihood of finding a BRCA1 mutation is higher in breast-ovarian cancer families (36%). Early-onset and bilateral breast cancer, regardless of the family scale, may also be useful tools to distinguish breast cancer families having a greater likelihood of carrying BRCA1 mutations with an 18.4% detection rate.
To conclude, the identification of specific and recurrent/founder mutations suggests that any given population should develop a mutation database for its programme of breast cancer screening. It is very important to collect more national data concerning BRCA1 mutations in order to get the best estimation of the mutation distribution (especially for recurrent/founder mutations) but it is also important to evaluate if there are any regional differences even in a small country like Tunisia. It is also hoped that similar mutation surveys in other North African countries are completed so that the information can be compared and the most common mutations, playing a role in the pathogenesis of breast carcinoma, identified. This will facilitate genetic testing throughout the region.
Acknowledgments
This publication was made possible by a grant from the Qatar National Research Fund (NPRP08-083-3-031). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Qatar National Research Fund. This work was also supported by the Ministry of Higher Education and Scientific Research and the Ministry of Public Health of the Republic of Tunisia and by IncoMed grant (ICA-CT-2002-10005). We wish to thank Mr. Rdissi Adel for proof-reading and revising this manuscript.
Conflict of interest
The authors declare no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Parkin D, Bray F, Ferlay J, Pasani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.GLOBOCAN . International agency for research on cancer (IARC) France: Lyon; 2002. [Google Scholar]
- 3.El Saghir NS, Khalil MK, Eid T, El Kinge AR, Charafeddine M, Geara F, Seoud M, Shamseddine AI. Trends in epidemiology and management of breast cancer in developing Arab countries: a literature and registry analysis. Int J Surg. 2007;5:225–233. doi: 10.1016/j.ijsu.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Gharbi O, Landolsi A, Nouira M, Ben FL, Bibi M, Korbi S, Bakir D, Kallel L, Khairi H, Ben Ahmed S. Breast cancer in elderly women in Tunisia: retrospective study of 106 patients older than 65 years. Tunis Med. 2003;81:696–701. [PubMed] [Google Scholar]
- 5.Ben Ahmed S, Aloulou S, Bibi M, Landolsi A, Nouira M, Ben Fatma L, Kallel L, Gharbi O, Korbi S, Khaïri H, Kraïem C. Breast cancer prognosis in Tunisian women: analysis of a hospital series of 729 patients. Sante Publique. 2002;14:231–241. doi: 10.3917/spub.023.0231. [DOI] [PubMed] [Google Scholar]
- 6.Maalej M, Frikha H, Ben Salem S, Daoud J, Bouaouina N, Ben Abdallah M, Ben Romdhane K. Breast cancer in Tunisia: clinical and epidemiological study. Bull Cancer. 1999;86:302–306. [PubMed] [Google Scholar]
- 7.Bener A, Ayub H, Kakil R, Ibrahim W. Patterns of cancer incidence among the population of Qatar: a worldwide comparative study. Asian Pac J Cancer Prev. 2008;9:19–24. [PubMed] [Google Scholar]
- 8.Maalej M, Hentati D, Messai T, Kochbati L, El May A, Mrad K, Romdhane KB, Ben Abdallah M, Zouari B. Breast cancer in Tunisia in 2004: a comparative clinical and epidemiological study. Bull Cancer. 2008;95:E5–E9. doi: 10.1684/bdc.2008.0584. [DOI] [PubMed] [Google Scholar]
- 9.Omar S, Khaled H, Gaafar R, Zekry AR, Eissa S, el-Khatib O. Breast cancer in Egypt: a review of disease presentation and detection strategies. East Mediterr Health J. 2003;9:448–463. [PubMed] [Google Scholar]
- 10.Atoum MF, Al-Hourani HM. Lifestyle related risk factors for breast cancer in Jordanian females. Saudi Med J. 2004;25:1245–1248. [PubMed] [Google Scholar]
- 11.Alam AA. Knowledge of breast cancer and its risk and protective factors among women in Riyadh. Ann Saudi Med. 2006;26:272–277. doi: 10.5144/0256-4947.2006.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Kuraya K, Schraml P, Sheikh S, Amr S, Torhorst J, Tapia C, Novotny H, Spichtin H, Maurer R, Mirlacher M, Simon R, Sauter G. Predominance of high-grade pathway in breast cancer development of Middle East women. Mod Pathol. 2005;18:891–897. doi: 10.1038/modpathol.3800408. [DOI] [PubMed] [Google Scholar]
- 13.Chalabi N, Bernard-Gallon DJ, Bignon YJ, Consortium BreastMed, Kwiatkowski F, Agier M, Vidal V, Laplace-Chabaud V, Sylvain-Vidal V, Bertholet V, De Longueville F, Lacroix M, Leclercq G, Remacle J, Sibille C, Zammateo N, Ben Jaafar N, Sefiani A, Ouldim K, Mégarbané K, Jalkh N, Mahfoudh W, Troudi W, Ben Ammar-El Gaïed A, Chouchane L. Comparative clinical and transcriptomal profiles of breast cancer between French and South Mediterranean patients show minor but significative biological differences. Cancer Genomics Proteomics. 2008;5:253–261. [PubMed] [Google Scholar]
- 14.Pharoah PD, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BA. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31:33–36. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- 15.Szabo CI, King MC. Inherited breast and ovarian cancer. Hum Mol Genet. 1995;4:1811–1817. doi: 10.1093/hmg/4.suppl_1.1811. [DOI] [PubMed] [Google Scholar]
- 16.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 17.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 18.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 19.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjäkoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Risch HA, McLaughlin JR, Cole DE, Rosen B, Bradley L, Fan I, Tang J, Li S, Zhang S, Shaw PA, Narod SA. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J Natl Cancer Inst. 2006;98:1694–1706. doi: 10.1093/jnci/djj465. [DOI] [PubMed] [Google Scholar]
- 21.Brose MS, Rebbeck TR, Calzone KA, Stopfer JE, Nathanson KL, Weber BL. Cancer risk estimates for BRCA1 mutation carriers identified in a risk evaluation program. J Nat Cancer Inst. 2002;94:1365–1372. doi: 10.1093/jnci/94.18.1365. [DOI] [PubMed] [Google Scholar]
- 22.King MC, Marks JH, Mandell JB, Group NewYork breast cancer study. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 23.Liede A, Karlan BY, Narod SA. Cancer risks for male carriers of germline mutations in BRCA1 or BRCA2: a review of the literature. J Clin Oncol. 2004;22:735–742. doi: 10.1200/JCO.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 24.Giordano SH. A review of the diagnosis and management of male breast cancer. Oncologist. 2005;10:471–479. doi: 10.1634/theoncologist.10-7-471. [DOI] [PubMed] [Google Scholar]
- 25.Satagopan JM, Offit K, Foulkes W, Robson ME, Wacholder S, Eng CM, Karp SE, Begg CB. The lifetime risks of breast cancer in Ashkenazi Jewish carriers of BRCA1 and BRCA2 mutations. Cancer Epidemiol Biomarkers Prev. 2001;10:467–473. [PubMed] [Google Scholar]
- 26.Ford D, Easton DF, Stratton M, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The breast cancer linkage consortium. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simchoni S, Friedman E, Kaufman B, Gershoni-Baruch R, Orr-Urtreger A, Kedar-Barnes I, Shiri-Sverdlov R, Dagan E, Tsabari S, Shohat M, Catane R, King MC, Lahad A, Levy-Lahad E. Familial clustering of site-specific cancer risks associated with BRCA1 and BRCA2 mutations in the Ashkenazi Jewish population. Proc Natl Acad Sci USA. 2006;103:3770–3774. doi: 10.1073/pnas.0511301103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Begg CB, Haile RW, Borg A, Malone KE, Concannon P, Thomas DC, Langholz B, Bernstein L, Olsen JH, Lynch CF, Anton-Culver H, Capanu M, Liang X, Hummer AJ, Sima C, Bernstein JL. Variation of breast cancer risk among BRCA1/2 carriers. JAMA. 2008;299:194–201. doi: 10.1001/jama.2007.55-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClain MR, Nathanson KL, Palomaki GE, Haddow JE. An evaluation of BRCA1 and BRCA2 founder mutations penetrance estimates for breast cancer among Ashkenazi Jewish women. Genet Med. 2005;7:34–39. doi: 10.1097/01.GIM.0000151156.14983.08. [DOI] [PubMed] [Google Scholar]
- 30.Kadouri L, Hubert A, Rotenberg Y, Hamburger T, Sagi M, Nechushtan C, Abeliovich D, Peretz T. Cancer risks in carriers of the BRCA1/2 Ashkenazi founder mutations. J Med Genet. 2007;44:467–471. doi: 10.1136/jmg.2006.048173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sokolenko AP, Mitiushkina NV, Buslov KG, Bit-Sava EM, Iyevleva AG, Chekmariova EV, ESh Kuligina, Ulibina YM, Rozanov ME, Suspitsin EN, Matsko DE, Chagunava OL, Trofimov DY, Devilee P, Cornelisse C, Togo AV, Semiglazov VF, Imyanitov EN. High frequency of BRCA1 5382insC mutation in Russian breast cancer patients. Eur J Cancer. 2006;42:1380–1384. doi: 10.1016/j.ejca.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 32.Antoniou AC, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjäkoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF. Breast and ovarian cancer risks to carriers of the BRCA1 5382insC and 185delAG and BRCA2 6174delT mutations: a combined analysis of 22 population based studies. J Med Genet. 2005;42:602–603. doi: 10.1136/jmg.2004.024133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuhausen SL, Hilmi Ozcelik H, Southey MS, et al. BRCA1 and BRCA2 mutation carriers in the breast cancer family registry: an open resource for collaborative research. Breast Cancer Res Treat. 2009;116:379–386. doi: 10.1007/s10549-008-0153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Troudi W, Uhrhammer N, Sibille C, Dahan C, Mahfoudh W, Bouchlaka Souissi C, Jalabert T, Chouchane L, Bignon YJ, Ben Ayed F, Ben Ammar Elgaaied A. Contribution of the BRCA1 and BRCA2 mutations to breast cancer in Tunisia. J Hum Genet. 2007;52:915–920. doi: 10.1007/s10038-007-0195-5. [DOI] [PubMed] [Google Scholar]
- 35.Monastiri K, Ben Ahmed S, Presneau N, Bignon JY, Chouchane L. Rapid detection of BRCA-1 germline mutations by the protein truncation test in Tunisian families. Tunis Med. 2002;80:515–518. [PubMed] [Google Scholar]
- 36.Mestiri S, Monastiri K, Ben Ahmed S, Bouaouina N, Presneau N, Bignon YJ, Khairi H, Chouchane L. Mutational analysis of breast/ovarian cancer hereditary predisposition gene BRCA1 in Tunisian women. Arch Inst Pasteur Tunis. 2000;77:11–15. [PubMed] [Google Scholar]
- 37.El-Harith el-HA, Abdel-Hadi MS, Steinmann D, Dork T. BRCA1 and BRCA2 mutations in breast cancer patients from Saudi Arabia. Saudi Med J. 2002;23:700–704. [PubMed] [Google Scholar]
- 38.Atoum MF, Al-Kayed SA. Mutation analysis of the breast cancer gene BRCA1 among breast cancer Jordanian females. Saudi Med J. 2004;25:60–63. [PubMed] [Google Scholar]
- 39.Uhrhammer N, Abdelouaheb A, Lafarge L, Feillel V, Ben Dib A, Bignon YJ. BRCA1 mutations in Algerian breast cancer patients: high frequency in young, sporadic cases. Int J Med Sci. 2008;5:197–202. doi: 10.7150/ijms.5.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Troudi W, Uhrhammer N, Romdhane KB, Sibille C, Amor MB, Khodjet El Khil H, Jalabert T, Mahfoudh W, Chouchane L, Ayed FB, Bignon YJ, Elgaaied AB. Complete mutation screening and haplotype characterization of BRCA1 gene in Tunisian patients with familial breast cancer. Cancer Biomark. 2008;4:11–18. doi: 10.3233/cbm-2008-4102. [DOI] [PubMed] [Google Scholar]
- 42.Muller D, Bonaiti-Pellié C, Abecassis J, Stoppa-Lyonnet D, Fricker JP. BRCA1 testing in breast and/or ovarian cancer families from northeastern France identifies two common mutations with a founder effect. Fam Cancer. 2004;3:15–20. doi: 10.1023/B:FAME.0000026819.44213.df. [DOI] [PubMed] [Google Scholar]
- 43.Russo A, Calò V, Agnese V, Bruno L, Corsale S, Augello C, Gargano G, Barbera F, Cascio S, Intrivici C, Rinaldi G, Gulotta G, Macaluso M, Surmacz E, Giordano A, Gebbia N, Bazan V. BRCA1 genetic testing in 106 breast and ovarian cancer families from Southern Italy (Sicily): a mutation analyses. Breast Cancer Res Treat. 2007;105:267–276. doi: 10.1007/s10549-006-9456-9. [DOI] [PubMed] [Google Scholar]
- 44.Palomba G, Loi A, Uras A, Fancello P, Piras G, Gabbas A, Cossu A, Budroni M, Contu A, Tanda F, Farris A, Orrù S, Floris C, Pisano M, Lovicu M, Santona MC, Landriscina G, Crisponi L, Palmieri G, Monne M. A role of BRCA1 and BRCA2 germline mutations in breast cancer susceptibility within Sardinian population. BMC Cancer. 2009;9:245. doi: 10.1186/1471-2407-9-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levy-Lahad E, Catane R, Eisenberg S, Kaufman B, Hornreich G, Lishinsky E, Shohat M, Weber BL, Beller U, Lahad A, Halle D. Founder BRCA1 and BRCA2 mutations in Ashkenazi Jews in Israel: frequency and differential penetrance in ovarian cancer and in breast-ovarian cancer families. Am J Hum Genet. 1997;60:1059–1067. [PMC free article] [PubMed] [Google Scholar]
- 46.Górski B, Jakubowska A, Huzarski T, Byrski T, Gronwald J, Grzybowska E, Mackiewicz A, Stawicka M, Bebenek M, Sorokin D, Fiszer-Maliszewska Ł, Haus O, Janiszewska H, Niepsuj S, Góźdź S, Zaremba L, Posmyk M, Płuzańska M, Kilar E, Czudowska D, Waśko B, Miturski R, Kowalczyk JR, Urbański K, Szwiec M, Koc J, Debniak B, Rozmiarek A, Debniak T, Cybulski C, Kowalska E, Tołoczko-Grabarek A, Zajaczek S, Menkiszak J, Medrek K, Masojć B, Mierzejewski M, Narod SA, Lubiński J. A high proportion of founder BRCA1 mutations in Polish breast cancer families. Int J Cancer. 2004;110:683–686. doi: 10.1002/ijc.20162. [DOI] [PubMed] [Google Scholar]
- 47.Konstantopoulou I, Rampias T, Ladopoulou A, Koutsodontis G, Armaou S, Anagnostopoulos T, Nikolopoulos G, Kamakari S, Nounesis G, Stylianakis A, Karanikiotis C, Razis E, Gogas H, Keramopoulos A, Gaki V, Markopoulos C, Skarlos D, Pandis N, Bei T, Arzimanoglou I, Fountzilas G, Yannoukakos D. Greek BRCA1 and BRCA2 mutation spectrum: two BRCA1 mutations account for half the carriers found among high-risk breast/ovarian cancer patients. Breast Cancer Res Treat. 2008;107:431–441. doi: 10.1007/s10549-007-9571-2. [DOI] [PubMed] [Google Scholar]
- 48.da Costa EC, Vargas FR, Moreira AS, Lourenço JJ, Caleffi M, Ashton-Prolla P, Martins Moreira MA. Founder effect of the BRCA1 5382insC mutation in Brazilian patients with hereditary breast ovary cancer syndrome. Cancer Genet Cytogenet. 2008;184:62–66. doi: 10.1016/j.cancergencyto.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 49.Szabo CI, King MC. Population genetics of BRCA1 and BRCA2. Am J Hum Genet. 1997;60:1013–1020. [PMC free article] [PubMed] [Google Scholar]
- 50.Neuhausen SL, Mazoyer S, Friedman L, Stratton M, Offit K, Caligo A, Tomlinson G, Cannon-Albright L, Bishop T, Kelsell D, Solomon E, Weber B, Couch F, Struewing J, Tonin P, Durocher F, Narod S, Skolnick MH, Lenoir G, Serova O, Ponder B, Stoppa-Lyonnet D, Easton D, King MC, Goldgar DE. Haplotype and phenotype analysis of six recurrent BRCA1 mutations in 61 families: results of an international study. Am J Hum Genet. 1996;58:271–280. [PMC free article] [PubMed] [Google Scholar]
- 51.Backe J, Hofferbert S, Skawran B, Dörk T, Stuhrmann M, Karstens JH, Untch M, Meindl A, Burgemeister R, Chang-Claude J, Weber BH. Frequency of BRCA1 mutation 5382insC in German breast cancer patients. Gynecol Oncol. 1999;72:402–406. doi: 10.1006/gyno.1998.5270. [DOI] [PubMed] [Google Scholar]
- 52.Hamel N, Foretova L, Narod SA, Tihomirova L, Zajac V, Ciernikova S, Armaou S, Yannoukakos D, Greenwood C, Foulkes WD (2006) Investigating the origins of the BRCA1 mutation c.5385dupC. In: Abstracts of the 56th annual meeting of the American society of human genetics, New Orleans, Louisiana, USA, October 10–13, p 203
- 53.Blesa JR, García JA, Ochoa E. Frequency of germ-line BRCA1 mutations among Spanish families from a Mediterranean area. Hum Mutat. 2000;15:381–382. doi: 10.1002/(SICI)1098-1004(200004)15:4<381::AID-HUMU14>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 54.Peixoto A, Salgueiro N, Santos C, Varzim G, Rocha P, Soares MJ, Pereira D, Rodrigues H, Bento MJ, Fráguas A, Moura G, Regateiro F, Castedo S, Teixeira MR. BRCA1 and BRCA2 germline mutational spectrum and evidence for genetic anticipation in Portuguese breast/ovarian cancer families. Fam Cancer. 2006;5:379–387. doi: 10.1007/s10689-006-0009-5. [DOI] [PubMed] [Google Scholar]
- 55.Jara L, Ampuero S, Santibáñez E, Seccia L, Rodríguez J, Bustamante M, Martínez V, Catenaccio A, Lay-Son G, Blanco R, Reyes JM. BRCA1 and BRCA2 mutations in a South American population. Cancer Genet Cytogenet. 2006;166:36–45. doi: 10.1016/j.cancergencyto.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 56.Torres D, Rashid MU, Gil F, Umana A, Ramelli G, Robledo JF, Tawil M, Torregrosa L, Briceno I, Hamann U. High proportion of BRCA1/2 founder mutations in Hispanic breast/ovarian cancer families from Colombia. Breast Cancer Res Treat. 2007;103:225–232. doi: 10.1007/s10549-006-9370-1. [DOI] [PubMed] [Google Scholar]
- 57.Rudkin TM, Hamel N, Galvez M, Hogervorst F, Gille JJ, Møller P, Apold J, Foulkes WD. The frequent BRCA1 mutation 1135insA has multiple origins: a haplotype study in different populations. BMC Med Genet. 2006;7:15. doi: 10.1186/1471-2350-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Augello C, Bruno L, Bazan V, Calò V, Agnese V, Corsale S, Cascio S, Gargano G, Terrasi M, Barbera F, Fricano S, Adamo B, Valerio MR, Colucci G, Sumarcz E, Russo A, Gruppo Oncologico dell’Italia Meridionale (2006) Y179C, F486L and N550H are BRCA1 variants that may be associated with breast cancer in a Sicilian family: results of a 5-year GOIM (Gruppo Oncologico dell’Italia Meridionale) prospective study. Ann Oncol 17:vii30–vii33 [DOI] [PubMed]
- 59.Miramar MD, Calvo MT, Rodriguez A, Antón A, Lorente F, Barrio E, Herrero A, Burriel J, García de Jalón A. Genetic analysis of BRCA1 and BRCA2 in breast/ovarian cancer families from Aragon (Spain): two novel truncating mutations and a large genomic deletion in BRCA1. Breast Cancer Res Treat. 2008;112:353–358. doi: 10.1007/s10549-007-9868-1. [DOI] [PubMed] [Google Scholar]
- 60.Dunning AM, Chiano M, Smith NR, Dearden J, Gore M, Oakes S, Wilson C, Stratton M, Peto J, Easton D, Clayton D, Ponder BA. Common BRCA1 variants and susceptibility to breast and ovarian cancer in the general population. Hum Mol Genet. 1997;6:285–289. doi: 10.1093/hmg/6.2.285. [DOI] [PubMed] [Google Scholar]
- 61.Soucek P, Borovanova T, Pohlreich P, Kleibl Z, Novotny J. Role of single nucleotide polymorphisms and haplotypes in BRCA1 in breast cancer: Czech case-control study. Breast Cancer Res Treat. 2007;103:219–224. doi: 10.1007/s10549-006-9367-9. [DOI] [PubMed] [Google Scholar]
- 62.Tommasi S, Crapolicchio A, Lacalamita R, Bruno M, Monaco A, Petroni S, Schittulli F, Longo S, Digennaro M, Calistri D, Mangia A, Paradiso A. BRCA1 mutations and polymorphisms in a hospital-based consecutive series of breast cancer patients from Apulia, Italy. Mutat Res. 2005;578:395–405. doi: 10.1016/j.mrfmmm.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 63.Wen-Feng Li, Zhen Hu, Rao Nan-Yan, Song Chuang-Gui, Zhang Bin, Cao Ming-Zhi, Feng-Xi Su, Wang Yong-Sheng, Ping-Qing He, Gen-Hong Di, Shen Kun-Wei, Jiong Wu, Jin-Song Lu, Luo Jian-Min, Liu Xiao-Yi, Zhou Jie, Wang Lei, Zhao Lin, Liu Yan-Bing, Yuan Wen-Tao, Yang Lin, Shen Zhen-Zhou, Huang Wei, Sha Zhi-Ming. The prevalence of BRCA1 and BRCA2 germline mutations in high-risk breast cancer patients of Chinese Han nationality: two recurrent mutations were identified. Breast Cancer Res Treat. 2008;110:99–109. doi: 10.1007/s10549-007-9708-3. [DOI] [PubMed] [Google Scholar]
- 64.Akisik E, Yazici H, Dalay N. ARLTS1, MDM2 and RAD51 gene variations are associated with familial breast cancer. Mol Biol Rep. 2011;38:343–348. doi: 10.1007/s11033-010-0113-3. [DOI] [PubMed] [Google Scholar]


