Abstract
The relative contribution of a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS)4 and ADAMTS5 to aggrecan degradation under oncostatin M (OSM) stimulation, the role of the ancillary domains of the aggrecanases on their ability to cleave within the chondroitin sulfate (CS)-2 region, the role of hyaluronidases (HYAL) in stimulating aggrecan release in the absence of proteolysis, and the identity of the hyaluronidase involved in OSM-mediated cartilage breakdown were investigated. Bovine articular cartilage explants were cultured in the presence of interleukin-1β (IL-1β), tumor necrosis factor α (TNFα) and/or OSM, or treated with trypsin and/or hyaluronidase. Aggrecan was digested with various domain-truncated isoforms of ADAMTS4 and ADAMTS5. Aggrecan and link protein degradation and release were analyzed by immunoblotting. Aggrecanase and HYAL gene expression were determined. ADAMTS4 was the most inducible aggrecanase upon cytokine stimulation, whereas ADAMTS5 was the most abundant aggrecanase. ADAMTS5 was the most active aggrecanase and was responsible for the generation of an OSM-specific degradation pattern in the CS-2 region. Its ability to cleave at the OSM-specific site adjacent to the aggrecan G3 region was enhanced by truncation of the C-terminal thrombospondin domain, but reduced by further truncation of both the spacer and cysteine-rich domains of the enzyme. OSM has the ability to mediate proteoglycan release through hyaluronan degradation, under conditions where HYAL-2 is the predominant hyaluronidase being expressed. Compared to other catabolic cytokines, OSM exhibits a unique potential at degrading the proteoglycan aggregate, by promoting early robust aggrecanolysis, primarily through the action of ADAMTS5, and hyaluronan degradation.
Keywords: Cartilage catabolism, proteoglycan aggregate, ADAMTS, hyaluronidase, oncostatin M
Introduction
The large aggregating chondroitin sulfate proteoglycan, aggrecan, is the major non-collagenous component of the cartilage extracellular matrix (ECM) (Dudhia, 2005). It consists of a large multi-domain core protein to which numerous polyanionic chondroitin sulfate (CS) and keratan sulfate (KS) glycosaminoglycans (GAG) chains are covalently attached. In the tissue, numerous aggrecan molecules associate with a single hyaluronan (HA) filament, with each interaction being further stabilized by a link protein (LP). The resulting proteoglycan aggregates are trapped within the collagen network in the cartilage ECM and provide the tissue with its ability to resist compressive loads.
The aggrecan core protein is composed of three globular domains: the N-terminal G1 and G2 domains and the C-terminal G3 domain. The G1 domain is responsible for the association of aggrecan with HA (Fosang and Hardingham, 1989; Watanabe et al., 1997). A short extended region, termed the interglobular domain (IGD), separates the G1 from the G2 domain. Although, the G2 domain shares structural similarities with the HA-binding region of the G1 domain, it lacks the ability to bind HA (Watanabe et al., 1997). The extended region between the G2 and G3 domains is dedicated to substitution by sulfated GAG side chains. A KS-attachment region is adjacent to the G2 domain, and is followed by the long CS-attachment region comprising the CS-1 and CS-2 domains. The G3 domain, which is structurally distinct from the G1 and G2 domains, is involved in the secretion, glycosylation and interaction of aggrecan with other ECM components (Aspberg et al., 1997; Chen et al., 2002).
In arthritic diseases, cartilage undergoes irreversible destruction in response to various catabolic stimuli. Under such conditions, aggrecan molecules are known to be rapidly degraded and released from the cartilage matrix, followed by the degradation of the matrix collagens. The primary cause of aggrecan degradation is attributed to proteolysis by members of the matrix metalloproteinase (MMP) and a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) families of enzymes. ADAMTS4 and ADAMTS5 are considered to be the major aggrecanases of articular cartilage, and are multidomain metalloproteinases synthesized as proenzymes that require proteolytic cleavage to become fully active. In addition to their N-terminal prodomain, they are both composed of a catalytic domain, a disintegrin (Dis) domain, a thrombospondin motif (TS1), a cysteine-rich (CysR) and a spacer (Sp) domain. In comparison to ADAMTS4, ADAMTS5 contains an additional C-terminal thrombospondin motif (TS2). These proteinases can cleave aggrecan at specific sites within a proteinase-sensitive region of the aggrecan core protein located within the IGD. Both enzymes cleave at the Glu373↓Ala374 bond (Sandy et al., 1991; Tortorella et al., 2000; Tortorella et al., 2001). Cleavage at this site results in the loss of the part of the aggrecan molecule bearing the KS- and CS-attachment domains, while the G1 domain remains attached to the HA filament and LP in the tissue. Moreover, five additional cleavages attributed to aggrecanases, have been described within the CS-2 region (Loulakis et al., 1992; Tortorella et al., 2000; Tortorella et al., 2001; Durigova et al., 2008b). Cleavage at these sites contributes to the C-terminal truncation of aggrecan, the loss of the GAG chains, and the loss of tissue function.
While the main focus in studies of cartilage breakdown has been the proteolytic degradation of aggrecan, several studies have suggested that non-proteolytic mechanisms, such as HA degradation, can also contribute to the loss of cartilage integrity and joint function (Fosang et al., 1991; Sztrolovics et al., 2002a). In vivo, HA degradation can occur through the action of free radicals (Yamazaki et al., 2003) or members of the hyaluronidase family (Flannery et al., 1998; Csoka et al., 2001). Such an event, whether it is accompanied by proteolysis of aggrecan or not, could allow the diffusion of aggrecan G1, LP and HA fragments from the cartilage matrix.
Cartilage matrix degradation is known to be enhanced by cytokines, such as interleukin-1β (IL-1β) or tumor necrosis factor α (TNFα), which stimulate aggrecan degradation by MMPs and aggrecanases (Mort and Billington, 2001). We have previously reported that oncostatin M (OSM) is also a potent pro-catabolic cytokine capable of mediating aggrecan degradation at all five originally established ADAMTS-derived cleavage sites within the aggrecan IGD and CS-2 region (Durigova et al., 2008a), and a sixth site corresponding to cleavage at the Glu2047-Ala2048 bond in the proximity of the G3 domain (Durigova et al., 2008b). In addition, the release of aggrecan G1, LP, and HA, and HA degradation was also observed in the presence of this cytokine (Durigova et al., 2008a).
In the present study, the mechanisms involved in the early stages of OSM-mediated breakdown of the proteoglycan aggregate were investigated. The relative roles of ADAMTS4 and/or 5 in aggrecan degradation and the role of the ancillary domains of the aggrecanases on their ability to cleave within the CS-2 region were elucidated. Furthermore, the ability of hyaluronidases to stimulate proteoglycan release in the absence of aggrecan proteolysis, as well as the identity of the hyaluronidase expressed in the presence of OSM, was determined.
Materials and Methods
Sources of tissue and cells
Bovine articular cartilage from metacarpophalangeal joints of skeletally mature (18–24 months old) animals was obtained at a local abattoir. Pieces of full-depth cartilage were collected, washed twice for 15 min in Dulbecco’s modified Eagle medium (DMEM) buffered with 44 mM NaHCO3 (Sigma-Aldrich, St. Louis, MO, USA), 20 mM HEPES, pH 7.4, containing the antibiotics: 100 U/ml penicillin G sodium, 100 µg/ml streptomycin sulfate and 150 ng/ml gentamycin (medium A); and supplemented with 0.25 µg/ml Fungizone (Gibco Invitrogen, Paisley, UK).
Chondrocyte isolation and culture
Cartilage pieces (5 mm3) maintained in medium A were initially treated with trypsin (Sigma, 5 mg/g of cartilage) for 30 min at 37°C. Trypsin was then inactivated by incubation in medium A containing 10% fetal calf serum (FCS) (PAA Laboratories, Etobicoke, Ont., Canada). Digested tissue was transferred into medium A+10% FCS, containing bacterial collagenase (Sigma, 7.2 mg/g cartilage), bovine testes hyaluronidase (Sigma, 4 mg/g cartilage) and DNase I (Sigma, 0.4 mg/g cartilage), and incubated overnight at 37°C with constant stirring. Undigested residual cartilage was removed by filtering the digestion mixture through a 70 µm nylon mesh strainer, and cells recovered by centrifugation. Cell pellets were washed twice with medium A supplemented with 0.25 µg/ml Fungizone, and the chondrocytes were then resuspended in medium A. The cell viability was between 90–95%. Chondrocytes were plated in 12-well culture plates (Corning Costar, Lowell, MA, USA) at a density of 2×106 cells/well in 2.5 ml medium A+10% FCS per well. A 24h pre-incubation period was followed by a 3-day treatment in DMEM in the presence of different cytokines, as described below.
Cartilage explant culture
Cartilage explants in 12-well culture plates (Corning Costar, 100 mg tissue (wet weight) per well per 2 ml of medium A) were allowed to equilibrate for 48 h at 37°C, then the medium was changed and the explants were cultured in the presence of various cytokines or enzymes for two or four days. Cytokines were prepared in medium A containing 0.1 mg/ml BSA: human recombinant IL-1β (5 ng/ml), TNFα (10 ng/ml) and OSM (10 ng/ml) (R&D, Minneapolis, MN, USA). Cartilage explants were collected at the end of the culture period, snap frozen in liquid nitrogen and stored at −80°C. Culture media were analyzed for sulfated GAG content by the dimethylmethylene blue (DMMB) colorimetric assay (Farndale et al., 1986), then stored at −20°C.
Enzyme treatments
Digestion of cartilage explants by trypsin and hyaluronidase were carried out at 37°C in 50 mM Tris-HCl, 100 mM NaCl, 1 mM CaCl2, pH 7.5 or 20 mM sodium acetate, 150 mM NaCl, pH 6, respectively. For trypsin digests, cartilage explants were incubated for 2 h in the presence of 0.5 µg trypsin (TPCK-treated, Sigma). Trypsin action was inhibited by incubation for an additional 15 min in the presence of 1 mM Pefabloc (Roche, Basel, Switzerland). Digestions by hyaluronidase (from Streptomyces hyalurolyticus, MP Biomedicals, Irvine, CA, USA) were carried out for 8 h in the presence of 20 U of enzyme. For the combined treatments, the 2-h incubation with trypsin was followed by 30-min incubation in the presence of 5 µg/ml soybean trypsin inhibitor (SBTI, Sigma), after which the buffer was replaced for subsequent 6-h hyaluronidase digestion. At the end of the enzymatic treatments, tissue and culture media were collected.
SDS/PAGE and immunoblotting
Cartilage explants were extracted with 10 volumes (v/w) 4 M guanidinium chloride (GuCl), 100 mM sodium acetate, pH 6.0, containing proteinase inhibitors for 48 h at 4°C. Biotinylated SBTI (100 ng, used to monitor sample recovery) was added to 100 µl aliquots of tissue extracts and culture media, and the samples precipitated by addition of 9 volumes of ethanol. Following overnight precipitation at −20°C, the samples were recovered by centrifugation at 4°C, washed twice with 5 volumes of cold 75 % ethanol and then dried under vacuum. Samples were then resuspended in keratanase buffer (10 mM sodium acetate, pH 6.0) for subsequent keratanase and chondroitinase treatment (Durigova et al., 2008a). Loading of the media samples onto the gel was standardized in order to allow comparison of cartilage component loss from an equal amount of tissue. Cartilage matrix components were resolved by SDS/PAGE on Novex 4–12 % gradient NuPAGE Bis-Tris gels (Invitrogen) under reducing conditions, and transferred to nitrocellulose membranes for immunoblotting (Durigova et al., 2008a). Aggrecan and its degradation products were detected using rabbit polyclonal antibodies, recognizing G1, human G3 (Sztrolovics et al., 2002b) or bovine G3 (Roughley et al., 2003) domains of aggrecan, or the anti-neoepitope antibody (Durigova et al., 2008b) directed against the bovine ARLEIE-G3 aggrecan fragment and the bovine G1-DIPES matrix metalloproteinase product (Durigova et al., 2010). Link protein was detected using the mouse monoclonal antibody 8A4 (Developmental Studies Hybridoma Bank, University of Iowa, USA) (Caterson et al., 1985). An anti-rabbit Ig or anti-mouse Ig-biotinylated secondary antibody (Amersham, Amersham, UK) was used to detect the binding of the primary antibody, followed by incubation with a streptavidin-biotinylated horseradish peroxidase (HRP) complex (Amersham) and bands visualized by ECL.
Aggrecan preparation and aggrecanase digestion
Aggrecan was extracted with GuCl from fetal bovine articular cartilage or newborn human distal femoral cartilage, then purified by CsCl density centrifugation (Bayliss and Roughley, 1985; Roughley et al., 2003). Fetal aggrecan was used as a substrate because of its high content of intact aggrecan molecules relative to adult aggrecan. Recombinant human full-length aggrecanases, ADAMTS5-1 and ADAMTS4-1 and the domain deletion mutants: ADAMTS4-2, lacking the C-terminal Sp domain; ADAMTS4-3, lacking the Sp and CysR domains; ADAMTS4-4, lacking the Sp, CysR and TS domains; ADAMTS4-5, lacking the Sp, CysR, TS and Dis domains; ADAMTS5-2, lacking its terminal TS2 domain; ADAMTS5-3, lacking the TS2 and Sp domains; ADAMTS5-4, lacking the TS2, Sp and CysR domains; ADAMTS5-5, lacking the TS2, Sp, CysR and TS1 domains, and ADAMTS5-6 consisting solely of the catalytic domain were expressed and purified as described previously (Kashiwagi et al., 2004; Gendron et al., 2007). All aggrecan digestions were performed at 37°C in 100 µl 50 mM Tris–HCl buffer, pH 7.5, containing 250 mM NaCl and 5 mM CaCl2 (aggrecanase buffer) using 200 µg of purified aggrecan and 0.2, 1 or 5 nM aggrecanase. The reaction was stopped by heating at 75°C for 10 min, samples precipitated with ethanol, treated with keratanase II and chondroitinase ABC, and proteins analyzed by SDS/PAGE and immunoblotting, as described above.
RNA isolation, reverse transcription (RT) and PCR
Total RNA was extracted from frozen bovine explants by a modification of the method of Chomczynski and Sacchi (Chomczynski and Sacchi, 1987). Frozen cartilage explants (~100 mg) were homogenized using a stainless steel Tissumizer (Tekmar, Cincinnati, OH, USA) in 2 ml of 4 M guanidinium isothiocyanate, 25 mM sodium citrate, pH 7.0. Sodium sarcosyl (final concentration 0.5%) and 0.1 M 2-mercaptoethanol were added sequentially, and the mixture was left on ice for 1 hr with frequent vortexing, 12 µg of carrier tRNA (from E. coli) was added and the sample extracted with phenol-chloroform. Following centrifugation, RNA was precipitated from the upper aqueous phase overnight at −20°C with an equal volume of isopropanol. The RNA was recovered by centrifugation and washed with 75% ethanol.
Total RNA from cultured chondrocytes was isolated using an RNaqueous-4PCR kit (Ambion, Austin, TX, USA) and treated with DNase I (Ambion), to avoid any PCR amplification of genomic DNA, according to manufacturer’s protocol. RNA (1 µg) was used to synthesize cDNA using random hexamer primers (Invitrogen) and Omniscript reverse transcriptase (Qiagen, Hilden, Germany), according to the manufacturer’s recommendations. Primers for bovine hyaluronidases and oncostatin M (Table 1) were designed within a single exon using Primer3 software. These primers were initially tested on bovine genomic DNA and were observed to give bands of equal intensity upon analysis on an agarose gel. Non-quantitative PCR amplification for bovine hyaluronidases was carried out for 40 cycles (30 s at 95°C, 60 s at 55°C, 70 s at 72°C). Amplification products were analyzed by electrophoresis on a 1.6 % agarose gel. Real-time PCR amplification for ADAMTS4, ADAMTS5, HYAL-2, SPAM-1 and GAPDH mRNA (Table 1) was performed in 96-well plates using an ABI Prism 7500 instrument (Applied Biosystems, Foster City, CA, USA). All primers/probe sets were shown to give comparable amplification efficiencies. Each PCR reaction (25 µl) contained 50 ng cDNA, 12.5 µl TaqMan Universal PCR Master Mix (Applied Biosystems), 900 nM of each primer and 50 nM of TaqMan probe. After initial activation at 50°C for 2 min and 95°C for 10 min, the samples were subjected to 40 amplification cycles (denaturation at 95°C for 15 s followed by an anneal/extension step at 60°C for 1 min). Relative mRNA expression was calculated using the comparative ΔΔCt method. Fold change in gene expression compared to controls were then calculated as 2−ΔΔCt. In addition, −ΔCt values were used to assess the relative amount of each target mRNA compared to GAPDH mRNA levels.
Table 1.
PCR primer and probe sequences.
| Amplified target |
Primer / Probe sequence 5'-3' | Product size (bp) |
|---|---|---|
| GAPDH |
Forward: GGCTGCTTTTAATTCTGGCAAA Reverse:AATCATACTGGAACATGTAGACCATGTA 5'FAM/3'TAMRA:TGGACATCGTCGCCATCAATGACC 1 5'FAM/3'MGB-NFQ: ACATCGTCGCCAT CAA 2 |
93 |
| ADAMTS4 |
Forward: CCCCATGTGCAACGTCAAG Reverse: AGTCTCCACAAATCTGCTCAGTGA 5'FAM/3'TAMRA: AGCCCCCGAAGGGCTAAGCGC |
94 |
| ADAMTS5 |
Forward: GCCTCCATGCAGCCTTCA Reverse: CATGACGATTCCAAGTTCTGTGAAGA 5'FAM/3'MGB-NFQ: CGAAATTGGACATCTG |
82 |
| GAPDH |
Forward: CCATCTTCCAGGAGCGAGAT Reverse : CCATCCACAGTCTTCTGGT |
346 |
| HYAL-1 |
Forward: CTGGGACACCAAGGACATTT Reverse: AGTGCTGCAGGCAGGTAGAT |
336 |
| HYAL-2 |
Forward: GAAGGGACACGTGGAACACT Reverse: CTGGACACGAAAGCTGACAA |
481 |
|
Similar to SPAM-1 |
Forward: TACAGCACCCCCTCTCATTC Reverse : ACGTTGTCGATTGGCATGTA |
306 |
| SPAM-1 |
Forward: TTCTGCTTCCGTGTTGTTTG Reverse : CCACGCTGTCATTTGGTATG |
342 |
| OSM |
Forward: CCGGAGATCAGGAGACT Reverse: AGGTCCATACAGGGCCAACT |
298 |
The 5’FAM/3’TAMRA probe and GAPDH set of primers were used to normalize ADAMTS4 data.
The 5’FAM/3’MGB-NFQ probe and GAPDH set of primers were used to normalize ADAMTS5 data.
Gel filtration
A column (95 cm × 1 cm) was packed with Sephacryl S-1000 resin (Pharmacia, Uppsala, Sweden) and equilibrated in 50 mM Tris/HCl, 150 mM NaCl, pH 7.4. The void volume (Vo, 36 ml) and total volume (Vt, 75 ml) of the column were determined using proteoglycan aggregate and K3Fe(CN)6, respectively. For analysis of the HA size distribution, a sample of untreated tissue was digested overnight at 56°C with proteinase K (1 mg proteinase K/mg tissue) in 50 mM Tris/HCl, pH 7.6, containing 1 mM EDTA, 1 mM iodoacetamide and 10 µg/ml pepstatin A. The proteinase K was then inactivated by incubation at 100 °C for 5 min. A 0.5 ml aliquot of either tissue digest or IL-1/OSM-treated cartilage culture media sample was mixed with 0.5 ml 50 mM Tris/HCl, 150 mM NaCl, pH 7.4 + 0.5 % BSA, loaded onto the column, then eluted using 50 mM Tris/HCl, 150 mM NaCl, pH 7.4 at a flow rate of 6 ml/h. HA content was measured using a competitive HA binding assay (Durigova et al., 2008a).
Results
OSM-mediated aggrecan degradation and release
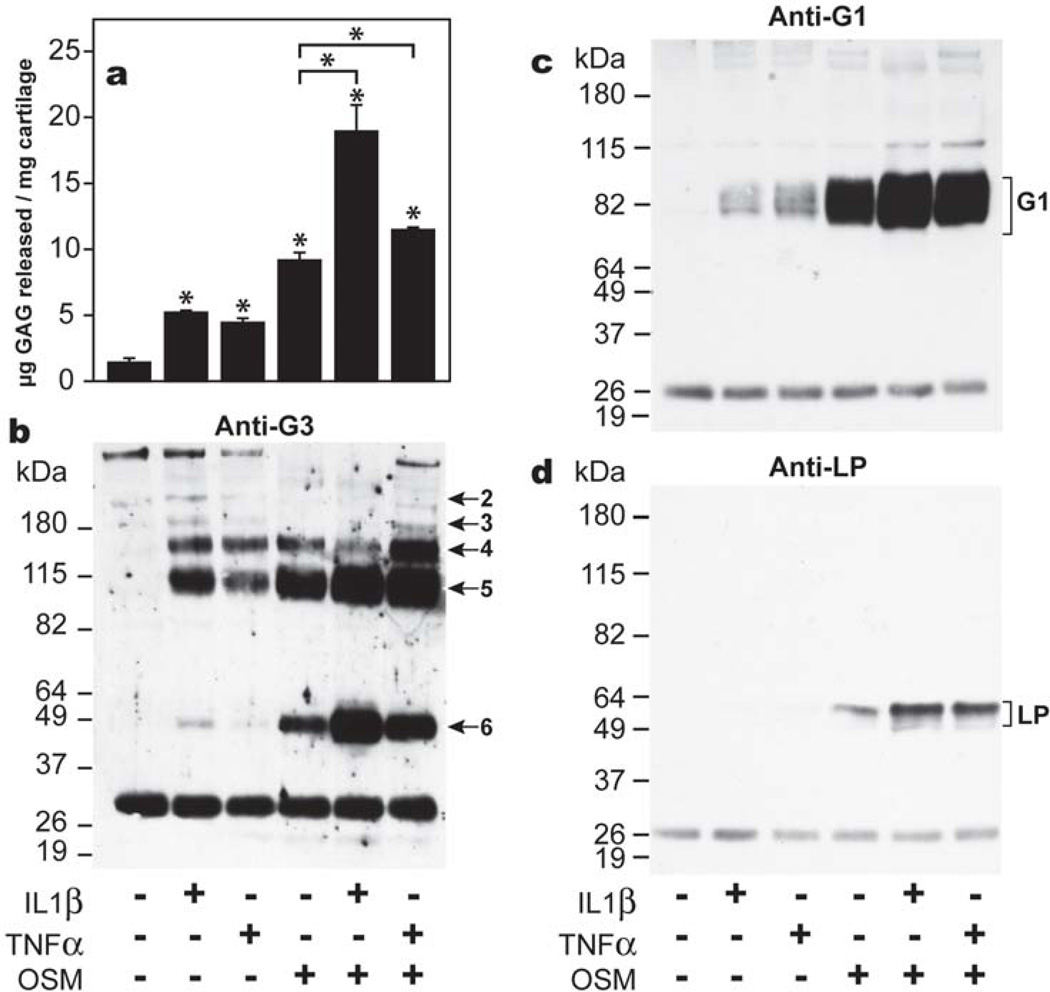
Initial experiments were carried out to assess and confirm whether the unique abilities of OSM to mediate aggrecan degradation and release from bovine articular cartilage were detectable after two days of culture. The extent of aggrecan release in response to cytokine treatment was monitored by GAG release (Fig. 1, panel a), and aggrecan proteolysis in the IGD and CS-2 domains were monitored by immunoblotting using an anti-G3 (Fig.1, panel b) or anti-G1 (Fig.1, panel c) antibody. Increased levels of sulfated GAG release were detected with all cytokine treatments compared to untreated samples. However, the most dramatic increase in GAG release was observed when cartilage was treated with OSM alone or IL-1β/OSM and TNFα/OSM mixtures (Fig.1, panel a). Immunoblotting analysis using an anti-aggrecan G3 antibody confirmed that aggrecan had undergone aggrecanase-mediated degradation. Four G3-containing aggrecan fragments (fragments 2–5), characteristic of aggrecanase action in the CS-2 region (Roughley et al., 2003), were observed in the culture medium for cartilage treated with IL-1β and TNFα. OSM alone or in combination with IL-1β or TNFα mediated aggrecan degradation at the same four sites, but also produced an additional aggrecanase-generated G3-containing fragment (fragment 6, Fig. 1, panel b), whose high abundance was previously shown to be characteristic of OSM (Durigova et al., 2008b). In the presence of IL-1β or TNFα low levels of free G1 domains were detectable in the culture medium, while in the presence of OSM alone or in combination with these two cytokines increased levels of free G1 domains were released from the tissue (Fig. 1, panel c). The molecular size of the G1 domains is consistent with aggrecan cleavage at the aggrecanase-mediated Glu373↓Ala374 site in the IGD (Sztrolovics et al., 2002b; Roughley et al., 2003). To verify that aggrecan G1 release was occurring simultaneously with LP release, immunoblotting of LP in the culture media was performed. An identical profile to that of aggrecan G1 in the presence of OSM was observed (Fig. 1, panel d), as expected for the release of G1-LP-HA complexes. Thus, the characteristic features of OSM previously reported after 8 days of culture (Durigova et al., 2008a) can be observed after 2 days, so validating this time point for the analysis of aggrecanase and hyaluronidase expression in response to OSM.
Fig. 1. Aggrecanolysis in response to cytokine stimulation.
Bovine articular cartilage explants were cultured in the presence of IL-1β, TNFα and OSM, alone or in combination for two days, when culture media were analyzed for GAG content by the DMMB assay (a), generation of aggrecan G3 (b) and G1 (c) -containing fragments, and LP release (d) by immunoblotting. The different cytokine treatments are indicated below the figures. The migration positions of known aggrecanase-generated products by cleavage in the IGD (G1) (Panel c) and CS-2 (Panel b) region (fragments 2–6) of aggrecan, and the position of LP (Panel d) are indicated on the right. Migration positions of molecular weight markers (kDa) are indicated on the left. The ~26 kDa band represents biotinylated SBTI used as a loading control. * indicates p<0.02 for GAG release as determined by unpaired t-test compared to control or as indicated by the brackets.
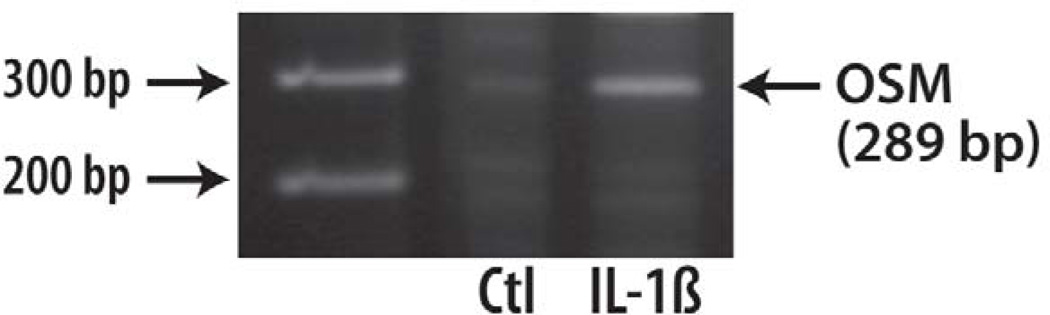
While fragment 6 was thought to be a unique product of OSM stimulation (Durigova et al., 2008b), a small amount of this aggrecan G3 component was produced under IL-1β treatment alone (Fig. 1 panel b). However it was shown that IL-1β drives the expression of OSM by chondrocytes (Fig. 2), suggesting the participation of this cytokine in the generation of the final G3 product.
Fig. 2. OSM expression by chondrocytes.
Bovine articular chondrocytes were cultured in the absence (Ctl) or presence of IL-1β for 24 h. Total RNA was extracted and subjected to RT-PCR to determine OSM mRNA expression. The position of the expected PCR product is indicated by an arrow. A DNA molecular ladder was loaded on the gel. The 200 and 300 bp marker positions are indicated on the left.
ADAMTS4 and ADAMTS5 gene expression in response to cytokine stimulation
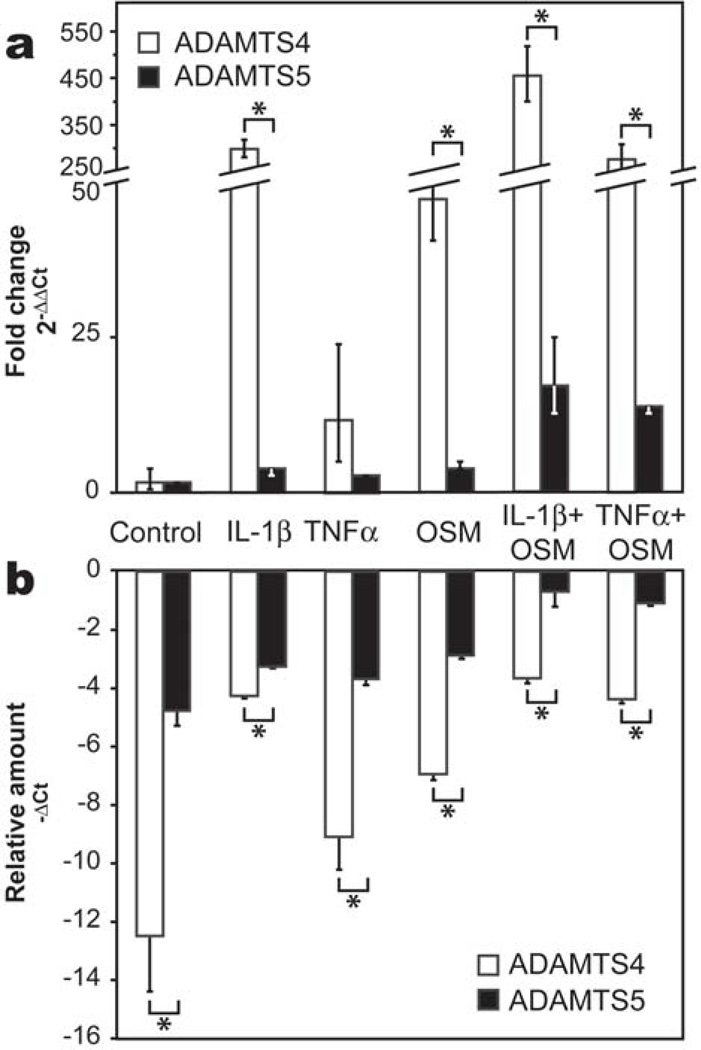
Quantitative analysis of expression levels of ADAMTS4 and 5 mRNA in response to cytokine treatment revealed that ADAMTS4 was by far the most strongly induced of the two by all cytokines (Fig. 3, panel a). Expression levels of both ADAMTS4 and ADAMTS5 were synergistically up-regulated in the presence of combinations of OSM with IL-1β or TNFα, while relatively little modulation in ADAMTS5 expression was observed in the presence of IL-1β, TNFα or OSM alone. IL-1β+OSM and TNFα+OSM treatments induced a 454-fold and 276-fold increase in ADAMTS4 expression, while these combinations induced only a 17- and 13-fold increase in ADAMTS5 gene expression, respectively. The relative amounts of ADAMTS4 and ADAMTS5 mRNA in the tissue were assessed by comparing the −ΔCt values of each target gene (Fig. 3, panel b), where the least negative −ΔCt value reflects the highest amount of the target gene. For the cytokines alone or OSM/cytokine combinations, the −ΔCt value for ADAMTS5 mRNA was always greater than that of ADAMTS4. In addition, ADAMTS5 reached similar levels of expression to GAPDH after stimulation with IL-1β+OSM or TNFα+OSM. Thus, while ADAMTS4 is the most inducible aggrecanase in cytokine-stimulated bovine cartilage explants, the level of its mRNA expression in the tissue is always lower than that of ADAMTS5.
Fig. 3. ADAMTS4 and ADAMTS5 mRNA expression in response to cytokine treatment.
Bovine articular cartilage explants were cultured in the presence of IL-1β, TNFα and OSM, alone or in combination for two days, when message levels of ADAMTS4 and ADAMTS5 were analyzed by real-time PCR. (a) Induction of ADAMTS4 and ADAMTS5 was determined by the ΔΔCt method and expressed as fold change (2−ΔΔCt) in target gene expression relative to unstimulated control samples. (b) Relative amounts of ADAMTS4 and ADAMTS5 mRNA were assessed from ΔCt values for the target genes and GAPDH. Results are expressed as −ΔCt which increases with message abundance. * indicates p<0.01 as determined by unpaired t-test.
Aggrecan degradation by truncated forms of ADAMTS4 and ADAMTS5
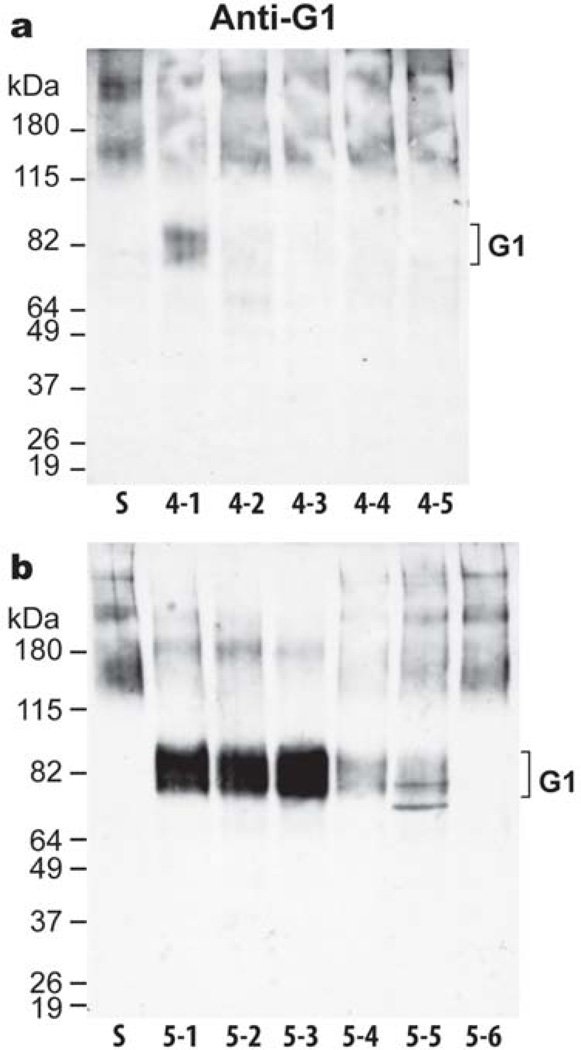
It is known that the activities of both aggrecanases are modulated by truncation within the various C-terminal domains of the enzymes (Flannery et al., 2002; Gao et al., 2002; Patwari et al., 2005; Zeng et al., 2006), and that such truncation occurs in vivo. Therefore, to investigate whether the degradation pattern mediated by OSM and its unique ability to promote increased proteolysis at an additional site (site 6) in the CS-2 domain is mediated through modulation of aggrecanase truncation, bovine aggrecan was digested with various domain deletion forms of ADAMTS4 and ADAMTS5. Analysis of aggrecan cleavage in the IGD, revealed that for ADAMTS4, only the full-length protease (ADAMTS4-1) was able to cleave the Glu373-Ala374 bond (Fig. 4, panel a). Full-length ADAMTS5 (ADAMTS5-1) was also able to cleave at the Glu373-Ala374 bond, and truncation of the C-terminal TS domain (ADAMTS5-2) or Sp domain (ADAMTS5-3) did not affect the ability of the enzyme to cleave at this site (Fig. 4, panel b). In contrast, truncation of the CysR (ADAMTS5-4) and the second TS domain (ADAMTS5-5) greatly reduced the aggrecanolytic activity at this site. Finally, the removal of the Dis domain (ADAMTS5-6) completely abolished aggrecan cleavage in the IGD. A small amount of a slightly faster migrating G1 component was observed following ADAMTS5-5 digestion. The possibility that this could be a G1-DIPES product equivalent to that generated by MMP cleavage was ruled out by its lack of reaction with a specific anti-DIPES antibody. Overall, these results showed that there is a vast difference in the ability of both aggrecanases to cleave within the aggrecan IGD, with ADAMTS5 being much more potent at this cleavage and less affected by C-terminal truncation.
Fig. 4. Cleavage within the aggrecan IGD by truncated forms of ADAMTS4 and ADAMTS5.
Fetal bovine aggrecan (lane S) was digested for 16h with 5 nM (a) recombinant ADAMTS4-1, full-length (lane 4-1); ADAMTS4-2, lacking the Sp domain (lane 4-2); ADAMTS4-3, lacking the Sp and CysR domains (lane 4-3); ADAMTS4-4, lacking the Sp, CysR and TS domains (lane 4-4); ADAMTS4-5, lacking the Sp, CysR, TS and Dis domains (lane 4-5) or (b) ADAMTS5-1, full-length (lane 5-1); ADAMTS5-2, lacking the TS2 domain (lane 5-2); ADAMTS5-3, lacking the TS2 and Sp domains (lane 5-3); ADAMTS5-4, lacking the TS2, Sp and CysR domains (lane 5-4); ADAMTS5-5, lacking the TS2, Sp, CysR and TS1 domains (lane 5-5); and ADAMTS5-6, lacking the TS2, Sp, CysR, TS1 and Dis domains (lane 5-6). Digested samples were then analyzed by immunoblotting using an aggrecan anti-G1 antibody. The migration position of aggrecanase-generated G1 is indicated on the right. Migration positions of molecular weight markers (kDa) are indicated on the left.
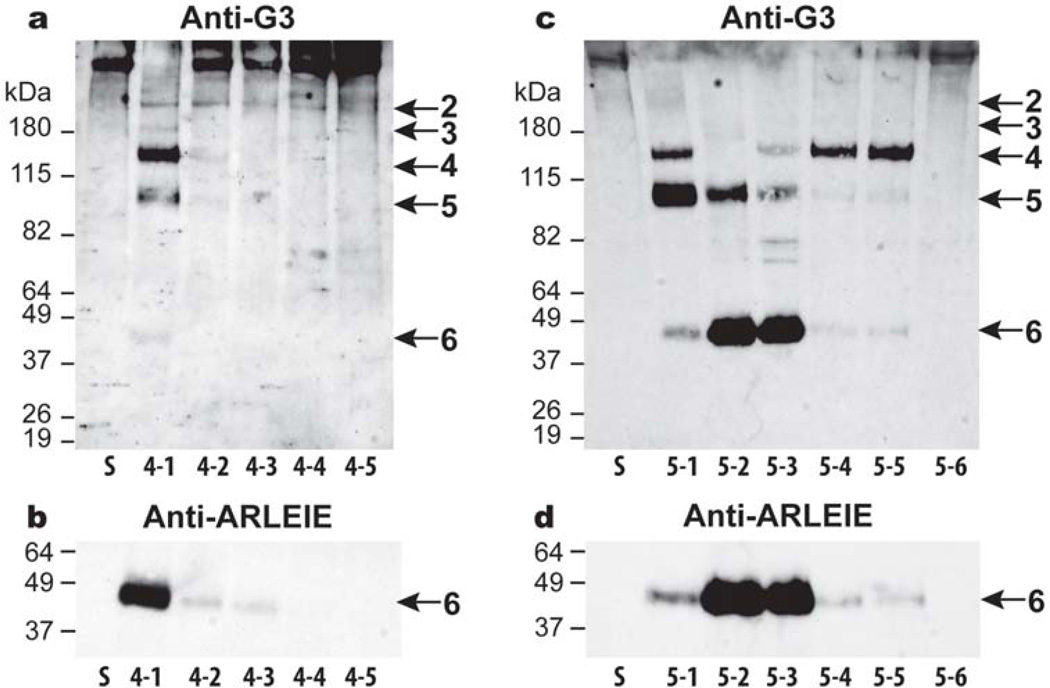
ADAMTS5 was also found to be more active than ADAMTS4 in the CS-2 domain and less affected by the truncation of its C-terminal domains. The full-length ADAMTS4-1 cleaved at all five aggrecanase-derived sites in the CS-2 domain including site 6, with cleavage at sites 4 and 5 being predominant (Fig. 5, panel a). ADAMTS4-2, lacking the C-terminal Sp domain, was much less potent at cleaving at these sites, and further truncation of ADAMTS4 prevented aggrecan cleavage in the CS-2 domain. Immunoblotting with an anti-neoepitope antibody (anti-ARLEIE) directed against the new N-terminus of fragment 6 confirmed that ADAMTS4-1 was the most potent form of this enzyme at generating this fragment (Fig. 5, panel b). Digestion by ADAMTS5 isoforms (Fig. 5, panel c) showed that fragment 6 is more readily generated by ADAMTS5 rather than ADAMTS4. Aggrecan digestion by domain deletion isoforms of ADAMTS5 demonstrated that sites 4, 5 and 6 are preferential cleavage sites for ADAMTS5. Generation of fragments 5 and 6 is dependent on the presence of the CysR domain, as in the absence of this domain, fragment 4 becomes the final major degradation product. In addition, removal of the C-terminal TS domain, appears to promote aggrecanase activity at site 6. Immunoblotting using the anti-ARLEIE antibody confirmed that fragment 6 is preferentially generated by isoforms ADAMTS5-2 and 5-3, lacking the C-terminal TS and Sp domains, respectively, but containing the CysR domain (Fig. 5, panel d). Thus, not only is ADAMTS5 the most potent aggrecanase at degrading within the CS-2 domain of aggrecan, but the unique ability of this enzyme to readily cleave at the additional site 6 is promoted by C-terminal truncation.
Fig. 5. Cleavage within the aggrecan CS-2 domain by truncated forms of ADAMTS4 and ADAMTS5.
Fetal bovine aggrecan (lane S) was digested for 16 h with 5 nM (a and b) recombinant ADAMTS4-1, full-length (lane 4-1); ADAMTS4-2, lacking the Sp domain (lane 4-2); ADAMTS4-3, lacking the Sp and CysR domains (lane 4-3); ADAMTS4-4, lacking the Sp, CysR and TS domains (lane 4-4); ADAMTS4-5, lacking the Sp, CysR, TS and Dis domains (lane 4-5) or (c and d) ADAMTS5-1, full-length (lane 5-1); ADAMTS5-2, lacking the TS2 domain (lane 5-2); ADAMTS5-3, lacking the TS2 and Sp domains (lane 5-3); ADAMTS5-4, lacking the TS2, Sp and CysR domains (lane 5-4); ADAMTS5-5, lacking the TS2, Sp, CysR and TS1 domains (lane 5-5); and ADAMTS5-6, lacking the TS2, Sp, CysR, TS1 and Dis domains (lane 5-6). Digested samples were then analyzed by immunoblotting using an aggrecan anti-G3 (a and c) or anti-ARLEIE antibody (b and d). Blots incubated with the anti-ARLEIE antibody were allowed to develop for different exposure times to maximize the detection of the 4-1 product while allowing comparison of the relative activities of the ADAMTS5 forms. The migration positions of aggrecanase-generated products are indicated on the right (2–6). Migration positions of molecular weight markers (kDa) are indicated on the left.
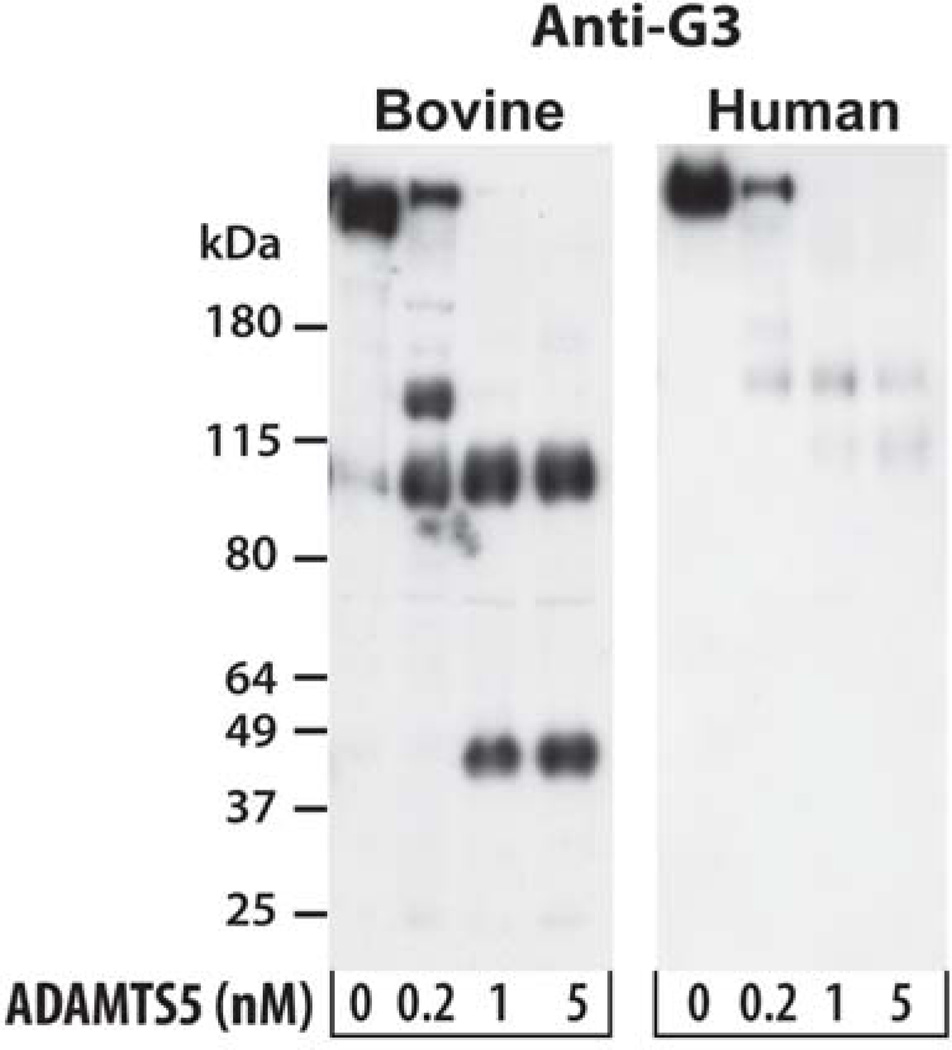
Since the sequence in the region of the sixth aggrecanase cleavage site is only partially conserved between bovine and human aggrecan (Durigova et al., 2008b), the generation of G3 fragments in ADAMTS5-2 digests of human aggrecan was investigated. An antibody raised against five peptides representing regions throughout the human G3 domain was used. In both species, the G3 domain is initially liberated following cleavage at sites in the CS2 region (Fig. 6). In the case of bovine aggrecan the site 6 cleavage product accumulates and remains resistant to further degradation. In marked contrast, human aggrecan is cleaved at positions 2–4 but then appears to undergo further, much more extensive, degradation to products which are too small to be detected on this gel system.
Fig. 6. G3 domain analysis of ADAMTS5 cleavage products of human and bovine aggrecan.
Fetal bovine aggrecan (left panel) and newborn human aggrecan (right panel) were digested for 6 hours with the indicated concentration of ADAMTS5-2. Digested samples were then analyzed by immunoblotting using an anti-bovine or anti-human aggrecan G3 antiserum. Migration positions of molecular weight markers (kDa) are indicated on the left.
Hyaluronan degradation in the presence of OSM
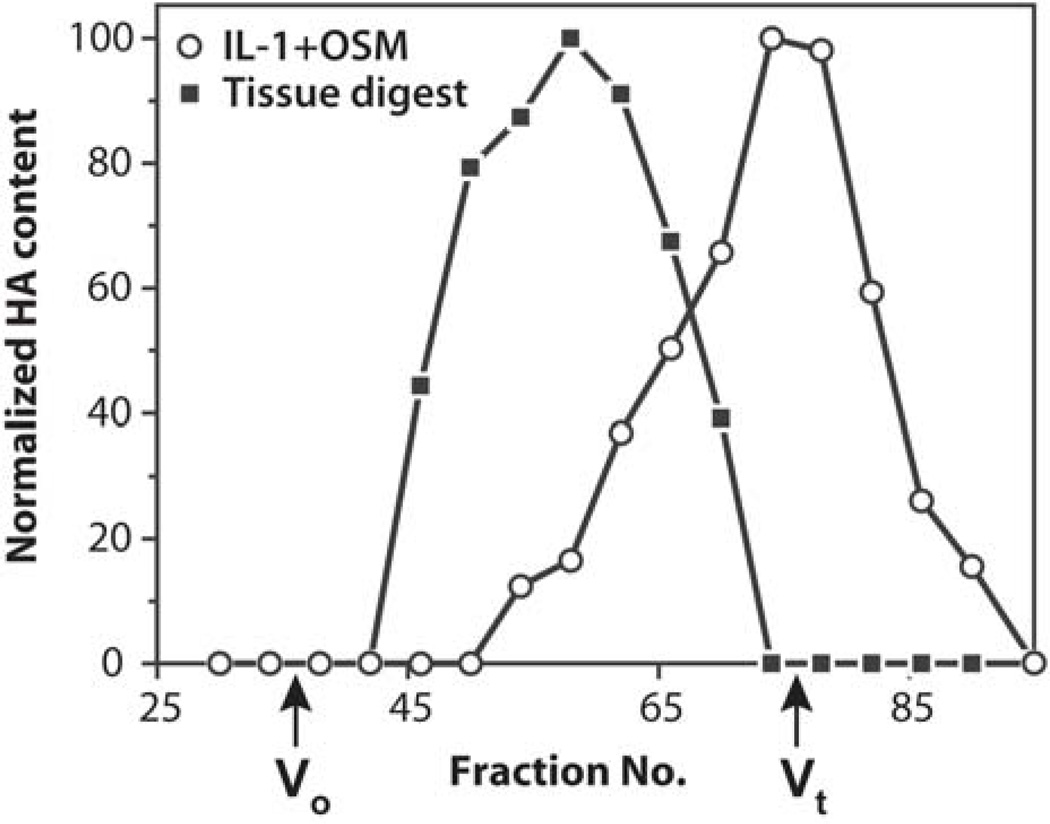
The concomitant increased release of free aggrecan G1 domains and intact LP from the cartilage upon IL-1β+OSM stimulation suggested that they were released as intact G1-LP complexes. As these components interact in the tissue with HA, it is likely that their release is due to HA cleavage. Therefore the size of HA, both in untreated tissue and culture media collected from IL-1β+OSM-treated tissue, was analyzed by gel filtration (Fig. 7). HA in cartilage digested with proteinase K exhibited a broad range of sizes with a Kav of 0.56, which corresponds to very large chains. In contrast, HA in the IL-1β+OSM-treated culture medium eluted at the Vt of the column. These results confirmed that HA has undergone extensive cleavage in the presence of the cytokines prior to its release from the tissue.
Fig. 7. Analysis of HA size.
Samples of proteinase K cartilage digest (solid squares) and culture media from IL-1β+OSM-treated cartilage (open circles) were analyzed by gel filtration on a Sephacryl S-1000 column. HA content in collected fractions was measured and the highest value for each analysis was set as 100 %. The void volume (Vo) and total volume (Vt) of the column are indicated.
Proteoglycan aggregate release in response to protease and hyaluronidase action
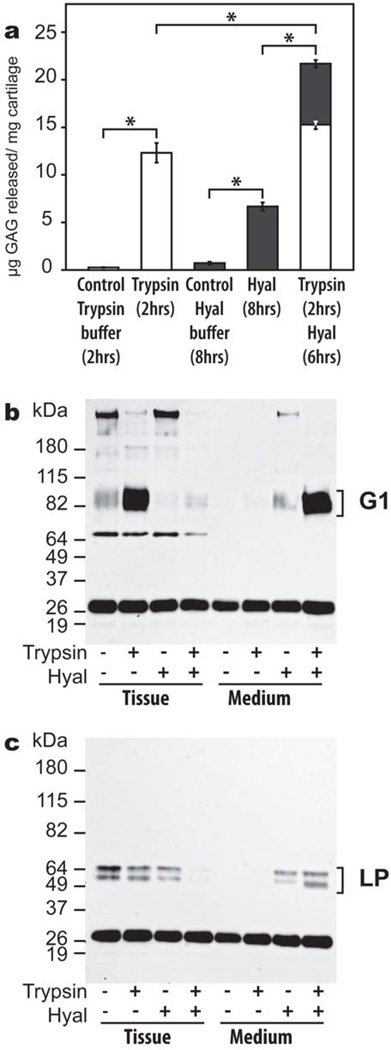
Hyaluronan cleavage in the tissue is mostly ascribed to non-proteolytic mechanisms, such as the action of hyaluronidases, which may occur simultaneously with aggrecan proteolysis. To demonstrate that hyaluronidase action can result in aggrecan-LP-HA release in the presence or absence of proteolysis, cartilage explants were treated with trypsin to induce limited proteolysis of aggrecan, and/or hyaluronidase to degrade HA. Trypsin treatment caused aggrecan degradation, resulting in a 60-fold increase in GAG release after 2 hours (Fig. 8, panel a). In comparison, an 8-hour digestion by a specific hyaluronidase caused a 12-fold increase in GAG release. Sequential treatment of cartilage by trypsin and hyaluronidase showed that GAG release could be enhanced by a combination of proteolysis and HA degradation. Thus, HA cleavage can mediate GAG loss from the tissue whether or not prior proteolysis has occurred. Immunoblot analysis demonstrated that intact aggrecan and very low amounts of free G1 were detected in the control tissue, while these components were absent from the corresponding culture media (Fig. 8, panel b). Trypsin treatment resulted in the accumulation of free G1 domains in the tissue, but caused no visible release of the G1 domain into the culture media. In contrast, hyaluronidase treatment had no effect on G1 generation in the tissue, but the endogenous G1 domains and some intact aggrecan were released. The sequential treatment with trypsin and hyaluronidase showed that all aggrecan G1 domain previously generated by trypsin could be released into the culture media by hyaluronidase action. Furthermore, immunoblot analysis showed that hyaluronidase action alone can induce some LP release from the tissue, whereas combination of proteolysis and HA fragmentation causes the release of all of the tissue LP (Fig. 8 panel c). These results mimic the effects of OSM-mediated aggrecan G1 and LP release from cartilage, suggesting that this cytokine acts through two concurrent mechanisms: aggrecan proteolysis by aggrecanases and HA cleavage by hyaluronidases.
Fig. 8. Analysis of proteoglycan aggregate degradation and release from cartilage in response to trypsin and/or hyaluronidase treatment.
(a) Bovine articular cartilage explants were cultured in the presence of trypsin (open bars), hyaluronidase (Hyal, solid bars), or a combination of both enzymes, and GAG release into the culture media was analyzed. At the end of the culture period, cartilage tissue extracts and culture media were analyzed by immunoblotting using an anti-aggrecan G1 (b) or anti-LP (c) antibody. The migration position of aggrecanase-generated G1 and LP are indicated on the right. Migration positions of molecular weight markers (kDa) are indicated on the left. The ~26 kDa band represents biotinylated SBTI used as a loading control. The band at ~65 kDa in panel A represents non-specific staining of a component in the tissue extracts. * represents p<0.01 as determined by unpaired t-test.
Hyaluronidase expression in cytokine-stimulated chondrocytes
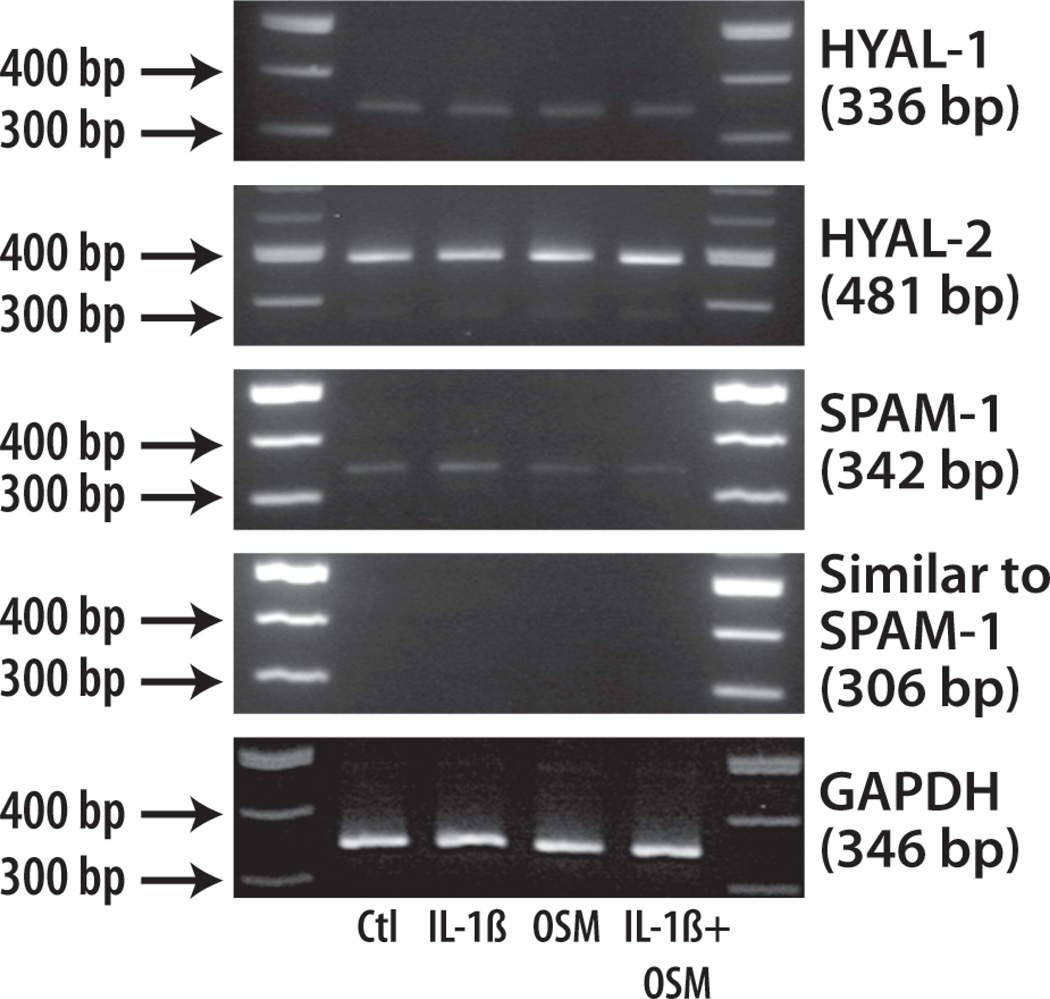
Previous studies have shown that hyaluronidases are important in HA degradation in cartilage (Flannery et al., 1998). There are five bovine hyaluronidase sequences available as a consequence of the bovine genome project in GenBank: HYAL-1 [NM_001017941], HYAL-2 [NM_174347], HYAL-3 [XM_868610], SPAM-1 (sperm adhesion molecule-1 or PH-20) [NM_001008413], similar to SPAM-1 [XM_586790]. Among these enzymes HYAL1, HYAL-2, SPAM-1 and similar to SPAM-1 are reported as the major hyaluronidases of articular cartilage. Therefore, their mRNA expression in chondrocytes in response to IL-1β, OSM or IL-1β+OSM treatment was investigated by RT-PCR (Fig. 9). No mRNA for similar to SPAM-1 could be detected, and very low amounts of HYAL-1 and SPAM-1 mRNA were present in the tissue. Their levels of expression did not vary upon any of the cytokine treatments. In contrast, more abundant expression of HYAL-2 mRNA was detected. This hyaluronidase was expressed by the chondrocytes for each of the cytokine treatments, but did not show any change in expression level when OSM was present. As HYAL-2 and SPAM-1 are known to exhibit extracellular activity at a neutral pH, they are the most likely candidates for HA degradation and release such as observed in the presence of OSM. However, real-time PCR analysis of HYAL-2 and SPAM-1 gene expression (data not shown) showed no major variation with any of the cytokine treatments.
Fig. 9. Hyaluronidase mRNA expression in adult bovine chondrocytes.
Articular chondrocytes were treated with selected cytokines for 3 days. Total RNA was extracted and subjected to RT-PCR to determine GAPDH, HYAL-1, HYAL-2, SPAM-1 and Similar to SPAM-1 gene expressions. The migration positions of molecular size markers are depicted on the left.
Discussion
The present study indicates that while the endogenous level of gene expression of ADAMTS4 in articular cartilage is low, it is very responsive to cytokine stimulation, with the highest increase occurring in the presence of OSM. In contrast, ADAMTS5 is constitutively expressed at higher levels than ADAMTS4 and its mRNA production is modulated to a lesser degree by the cytokine treatments. However, under all conditions, ADAMTS5 is the predominant aggrecanase mRNA expressed in this tissue. These results suggest that in adult bovine articular cartilage ADAMTS5 has the potential to be the major aggrecanase responsible for aggrecan turnover and maintenance of cartilage homeostasis, while both ADAMTS4 and ADAMTS5 participate in cytokine-induced aggrecan degradation. This conclusion is based on one representative time point (2 days), and it is possible that at earlier points the time course of mRNA expression for the two ADAMTS enzymes may differ (Ariyoshi et al., 2010). However these results should indicate the steady-state levels. Also it is important to note that many modulating influences are present between message expression and net proteolytic activity (Fosang and Rogerson, 2010), so that mRNA levels only represent the potential for overall proteolytic function.
Previous studies have reported conflicting data and results that differ from the present study with respect to both constitutive and cytokine-mediated expression of ADAMTS4 and ADAMTS5 in cartilage. Higher constitutive levels of ADAMTS4 mRNA compared to ADAMTS5 were detected in calf chondrocytes (Arai et al., 2004). Moreover, one study using bovine cartilage explants has suggested that ADAMTS5 is constitutively expressed, and only ADAMTS4 is being responsive to IL-1 or TNFα stimulation (Tortorella et al., 2001), while another has reported increase of ADAMTS5 gene expression and no impact on ADAMTS4 mRNA expression upon cytokine treatment (Little et al., 2002). Also, combinations of IL-1 or TNFα with OSM were potent inducers of both ADAMTS4 and ADAMTS5 gene expression in human cartilage in some studies (Song et al., 2007; Young et al., 2005), while in another study the TNFα/OSM combination had a strong positive effect on ADAMTS4 gene expression, but failed to increase ADAMTS5 mRNA levels (Hui et al., 2005). Such differences may reflect species, age or site variations in aggrecanase expression and care must be taken when extrapolating from one system to another. It is also apparent that when studying aggrecanase messages, stimulation index is not necessarily an accurate reflection of absolute message level.
In addition to their ability to modulate aggrecanase gene expression, proinflammatory cytokines are known to play a role in the regulation of aggrecanase activity. IL-1 has been shown to stimulate an activation process involving proteolytic removal of the C-terminal domain of the enzyme thereby differentially modulating ADAMTS4-mediated cleavage within the aggrecan IGD or CS-2 domain (Patwari et al., 2005). Similarly, C-terminal processing of ADAMTS5 appears to correlate with increased aggrecan degradation in OA (Yamanishi et al., 2002). Therefore, it has been proposed that proteolytic C-terminal processing is a potential mechanism of activity regulation for aggrecanases in vivo (Zeng et al., 2006; Flannery et al., 2002; Gao et al., 2002). While increased levels of ADAMTS5 relative to ADAMTS4 could be responsible for the robust aggrecanolysis observed in the presence of OSM, modulation of aggrecanase processing could also explain the characteristic aggrecan degradation pattern mediated by this cytokine. The Sp and CysR domains of aggrecanases contain GAG-binding motifs that modulate the affinity of the proteinases for their substrates (Kashiwagi et al., 2004; Gendron et al., 2007; Flannery et al., 2002; Zeng et al., 2006). Our study demonstrates that the presence of both domains is required for cleavage of the Glu373-Ala374 bond in the IGD by ADAMTS4, while the CysR domain was essential for ADAMTS5 activity. Differential truncation also modulated the ability of these enzymes to degrade the CS-2 domain. Overly truncated forms of ADAMTS5 (lacking the TS2, Sp, CysR and/or TS1 domains) exhibited a decreased ability to cleave this region, as very low levels of fragments 5 and 6 were generated. However, ADAMTS5 lacking the TS2/or Sp domains generated fragment 6 as their major degradation product. Thus, OSM may be involved in facilitating partial truncation of ADAMTS5, but not overtruncation, so promoting the efficiency of the enzyme to cleave the CS-2 region of aggrecan at cleavage sites 5 and 6.
While ADAMTS5 digestion of bovine aggrecan results in the generation of a 50 kDa G3 fragment which is stable to the further actions of this protease, following cleavage in the CS2 region, the human aggrecan G3 domain is degraded into fragments that were undetectable using an antibody covering various regions of the domain. Thus it is possible that cleavage at the position equivalent to site 6 in bovine aggrecan may be occurring in the human, but the resultant product may be sensitive to further proteolysis by ADAMTS5. Importantly these findings suggest that ADAMTS5 generated fragments of the human G3 domain are being released from articular cartilage and once better characterized may play a role as biomarkers of tissue destruction.
In addition to early aggrecanolysis, OSM can also mediate the diffusion from the tissue of aggrecan G1, together with LP and HA. The loss of aggrecan G1 domain, LP and HA from cartilage cultures has also been previously detected in a variety of other cartilage explants systems (Fosang et al., 1991; Sztrolovics et al., 2002b; Yasumoto et al., 2003). These studies reported the release of these components in the absence of OSM, and while it is possible that the G1-LP-HA release from cartilage in response to a catabolic stimulus varies with species, site and/or age, it is also possible that this process could be modulated by endogenous OSM. Indeed, OSM is not only a monocyte/macrophage-specific cytokine, but can also be expressed by articular chondrocytes and its mRNA levels increase upon stimulation by IL-1.
Aggrecan G1 and LP release observed in the presence of OSM is accompanied by the degradation of HA, which could be attributed to the action of either free radicals or hyaluronidases. However, fragmentation of the LP within its N-terminal region, typical of free radical action was not detected in the presence of the cytokines (Roberts et al., 1987). Moreover, Sugimoto et al have shown that HA-degrading activity detected in bovine chondrocytes was not affected after addition of free radical scavengers to the cultures (Sugimoto et al., 2004). Thus, chondrocyte-derived hyaluronidases are likely responsible for the OSM-mediated HA degradation. Hyaluronidase-mediated degradation of HA in cartilage is known to occur intracellularly, through the action of lysosomal hyaluronidases, or extracellularly either at the cell surface or within the ECM (Hua et al., 1993; Embry and Knudson, 2003; Harada and Takahashi, 2007). Release of HA from cartilage in response to cytokine treatments has been demonstrated previously, and a role for extracellular hyaluronidase activity in this process has been proposed (Sztrolovics et al., 2002a; Sugimoto et al., 2004). In an intriguing study (Chockalingam et al., 2004) it was reported that when dead (freeze/thawed) bovine cartilage was incubated with recombinant ADAMTS4 or -5 aggrecan degradation occurred as expected but there was also release of the G1 domain, LP and low molecular weight HA from the tissue. Thus it is possible that aggrecanases can lead to release of hyaluronidases from the chondrocytes.
Among the hyaluronidases reported to be expressed in cartilage, HYAL-2 and SPAM-1 (PH-20) are the primary candidates for the extracellular degradation of HA, as both enzymes are known to be active at neutral pH and are expressed at the cell surface of chondrocytes (Cherr et al., 1996; Rai et al., 2001; El Hajjaji et al., 2005). However, high release and extensive HA degradation observed in the OSM cultures, suggest the presence of a hyaluronidase activity within the ECM (Durigova et al., 2008a). Studies have shown that membrane-anchored hyaluronidases could be released from the cell membrane after cleavage by phospholipases (Rai et al., 2001; Monzon et al., 2010). Thus, it is possible that membrane-bound HYAL-2 and/or SPAM-1 are released into the ECM through phospholipase action in the presence of OSM. In addition, it has been shown that proinflammatory cytokines such as IL-1β or TNFα can induce the gene expression and activity of both hyaluronidases in articular chondrocytes (Flannery et al., 1998; El Hajjaji et al., 2005).
In the present study chondrocytes were used as we were unable to detect sufficient hyaluronidase mRNA in cartilage extracts. While this limits the evaluation of in situ hyaluronidase expression in cartilage itself, the data do imply that the main hyaluronidase mRNA present in cytokine-treated chondrocytes is HYAL-2, suggesting it to be the primary hyaluronidase involved in OSM-mediated fragmentation of HA. However, proof of this proposal will require further experimental justification, for example by knock down techniques. While no modulation of its mRNA upon cytokine stimulation has been detected, increase of HYAL-2 activity, such as observed in the presence of OSM, may occur at the post-transcriptional level. Furthermore, it is apparent that hyaluronidase action alone is capable of causing proteoglycan loss from articular cartilage and thus participating in its degeneration.
Conclusions
In the present study, we have demonstrated that short-term exposure of adult bovine articular cartilage to IL-1β, TNFα and/or OSM mediates aggrecanolysis exclusively through aggrecanase action. While all cytokines stimulated ADAMTS4 mRNA expression to a greater degree than ADAMTS5 mRNA, ADAMTS5 mRNA was always the more abundant aggrecanase message expressed in the cartilage. ADAMTS5 was also far more active than ADAMTS4 at cleaving the aggrecan core protein, in both the IGD and CS-2 domains and partially truncated ADAMTS5 was more effective at generating the aggrecan degradation pattern attributable to OSM. Furthermore, in addition to extensive aggrecanolysis, OSM has been shown to mediate proteoglycan aggregate degradation through an additional mechanism involving HA degradation, which could be attributable to HYAL-2 action. Such hyaluronidase action can contribute to proteoglycan release even in the absence of proteolysis. These studies indicate that ADAMTS5 and HYAL-2 should form the focus of future work aimed at understanding the role of OSM in human cartilage catabolism in arthritic disorders.
Acknowledgements
This work was supported by the Shriners North America, the Canadian Institutes of Health Research (PJR and JSM grant MOP 49458), the Wellcome Trust (HN grant 075473) and the National Institutes of Health (HN grant AR40994). We would like to thank Patrick Soucy and Yeqing Geng for technical assistance and Guylaine Bédard for preparing the figures.
List of abbreviations
- ADAMTS
a disintegrin and metalloproteinase with thrombospondin motifs
- anti-ARLEIE
an antibody directed against the ARLEIE sequence within aggrecan
- CS-2
chondroitin sulfate domain-2
- CysR
cysteine-rich domain
- Dis
disintegrin domain
- ECM
extracellular matrix
- G1
globular domain 1
- G3
globular domain 3
- GAG
glycosaminoglycan
- HA
hyaluronan
- HYAL
hyaluronidase
- IGD
interglobular domain
- IL-1β
interleukin 1 beta
- MMP
matrix metalloproteinase
- LP
link protein
- OSM
oncostatin M
- OA
osteoarthritis
- SPAM-1
sperm adhesion molecule-1
- Sp
spacer domain
- TNFα
tumor necrosis factor alpha
- TS
thrombospondin domain
References
- Arai M, Anderson D, Kurdi Y, Annis-Freeman B, Shields K, Collins-Racie LA, Corcoran C, DiBlasio-Smith E, Pittman DD, Dorner AJ, Morris E, LaVallie ER. Effect of adenovirus-mediated overexpression of bovine ADAMTS-4 and human ADAMTS-5 in primary bovine articular chondrocyte pellet culture system. Osteoarthritis Cart. 2004;12:599–613. doi: 10.1016/j.joca.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Ariyoshi W, Knudson CB, Luo N, Fosang AJ, Knudson W. Internalization of aggrecan G1 domain neoepitope ITEGE in chondrocytes requires CD44. J Biol Chem. 2010;285:36216–36224. doi: 10.1074/jbc.M110.129270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspberg A, Miura R, Bourdoulous S, Shimonaka M, Heinegard D, Schachner M, Ruoslahti E, Yamaguchi Y. The C-type lectin domains of lecticans, a family of aggregating chondroitin sulfate proteoglycans, bind tenascin-R by protein-protein interactions independent of carbohydrate moiety. Proc Natl Acad Sci U S A. 1997;94:10116–10121. doi: 10.1073/pnas.94.19.10116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss MT, Roughley PJ. The properties of proteoglycan prepared from human articular cartilage by using associative caesium chloride gradients of high and low starting densities. Biochem J. 1985;232:111–117. doi: 10.1042/bj2320111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterson B, Baker JR, Christner JE, Lee Y, Lentz M. Monoclonal antibodies as probes for determining the microheterogeneity of the link proteins of cartilage proteoglycan. J Biol Chem. 1985;260:11348–11356. [PubMed] [Google Scholar]
- Chen L, Wu Y, Lee V, Kiani C, Adams ME, Yao Y, Yang BB. The folded modules of aggrecan G3 domain exert two separable functions in glycosaminoglycan modification and product secretion. J Biol Chem. 2002;277:2657–2665. doi: 10.1074/jbc.M101153200. [DOI] [PubMed] [Google Scholar]
- Cherr GN, Meyers SA, Yudin AI, VandeVoort CA, Myles DG, Primakoff P, Overstreet JW. The PH-20 protein in cynomolgus macaque spermatozoa: identification of two different forms exhibiting hyaluronidase activity. Dev Biol. 1996;175:142–153. doi: 10.1006/dbio.1996.0102. [DOI] [PubMed] [Google Scholar]
- Chockalingam PS, Zeng W, Morris EA, Flannery CR. Release of hyaluronan and hyaladherins (aggrecan G1 domain and link proteins) from articular cartilage exposed to ADAMTS-4 (aggrecanase 1) or ADAMTS-5 (aggrecanase 2) Arthritis Rheum. 2004;50:2839–2848. doi: 10.1002/art.20496. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Csoka AB, Frost GI, Stern R. The six hyaluronidase-like genes in the human and mouse genomes. Matrix Biol. 2001;20:499–508. doi: 10.1016/s0945-053x(01)00172-x. [DOI] [PubMed] [Google Scholar]
- Dudhia J. Aggrecan, aging and assembly in articular cartilage. Cell Mol Life Sci. 2005;62:2241–2256. doi: 10.1007/s00018-005-5217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durigova M, Nagase H, Mort JS, Roughley PJ. MMPs are less efficient than ADAMTS5 in cleaving aggrecan core protein. Matrix Biol. 2010 Nov 3; doi: 10.1016/j.matbio.2010.10.007. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durigova M, Roughley PJ, Mort JS. Mechanism of proteoglycan aggregate degradation in cartilage stimulated with oncostatin M. Osteoarthritis Cart. 2008a;16:98–104. doi: 10.1016/j.joca.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Durigova M, Soucy P, Fushimi K, Nagase H, Mort JS, Roughley PJ. Characterization of an ADAMTS-5-mediated cleavage site in aggrecan in OSM-stimulated bovine cartilage. Osteoarthritis Cart. 2008b;16:1245–1252. doi: 10.1016/j.joca.2008.02.013. [DOI] [PubMed] [Google Scholar]
- El Hajjaji H, Cole AA, Manicourt DH. Chondrocytes, synoviocytes and dermal fibroblasts all express PH-20, a hyaluronidase active at neutral pH. Arthritis Res Ther. 2005;7:R756–R768. doi: 10.1186/ar1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embry JJ, Knudson W. G1 domain of aggrecan cointernalizes with hyaluronan via a CD44-mediated mechanism in bovine articular chondrocytes. Arthritis Rheum. 2003;48:3431–3441. doi: 10.1002/art.11323. [DOI] [PubMed] [Google Scholar]
- Farndale RW, Buttle DJ, Barrett AJ. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta. 1986;883:173–177. doi: 10.1016/0304-4165(86)90306-5. [DOI] [PubMed] [Google Scholar]
- Flannery CR, Little CB, Hughes CE, Caterson B. Expression and activity of articular cartilage hyaluronidases. Biochem Biophys Res Commun. 1998;251:824–829. doi: 10.1006/bbrc.1998.9561. [DOI] [PubMed] [Google Scholar]
- Flannery CR, Zeng W, Corcoran C, Collins-Racie LA, Chockalingam PS, Hebert T, Mackie SA, McDonagh T, Crawford TK, Tomkinson KN, LaVallie ER, Morris EA. Autocatalytic cleavage of ADAMTS-4 (Aggrecanase-1) reveals multiple glycosaminoglycan-binding sites. J Biol Chem. 2002;277:42775–42780. doi: 10.1074/jbc.M205309200. [DOI] [PubMed] [Google Scholar]
- Fosang AJ, Hardingham TE. Isolation of the N-terminal globular protein domains from cartilage proteoglycans. Identification of G2 domain and its lack of interaction with hyaluronate and link protein. Biochem J. 1989;261:801–809. doi: 10.1042/bj2610801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosang AJ, Rogerson FM. Identifying the human aggrecanase. Osteoarthritis Cartilage. 2010;18:1109–1116. doi: 10.1016/j.joca.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Fosang AJ, Tyler JA, Hardingham TE. Effect of interleukin-1 and insulin like growth factor-1 on the release of proteoglycan components and hyaluronan from pig articular cartilage in explant culture. Matrix. 1991;11:17–24. doi: 10.1016/s0934-8832(11)80223-4. [DOI] [PubMed] [Google Scholar]
- Gao G, Westling J, Thompson VP, Howell TD, Gottschall PE, Sandy JD. Activation of the proteolytic activity of ADAMTS4 (aggrecanase-1) by C-terminal truncation. J Biol Chem. 2002;277:11034–11041. doi: 10.1074/jbc.M107443200. [DOI] [PubMed] [Google Scholar]
- Gendron C, Kashiwagi M, Lim NH, Enghild JJ, Thogersen IB, Hughes C, Caterson B, Nagase H. Proteolytic activities of human ADAMTS-5: comparative studies with ADAMTS-4. J Biol Chem. 2007;282:18294–18306. doi: 10.1074/jbc.M701523200. [DOI] [PubMed] [Google Scholar]
- Harada H, Takahashi M. CD44-dependent intracellular and extracellular catabolism of hyaluronic acid by hyaluronidase-1 and-2. J Biol Chem. 2007;282:5597–5607. doi: 10.1074/jbc.M608358200. [DOI] [PubMed] [Google Scholar]
- Hua Q, Knudson CB, Knudson W. Internalization of hyaluronan by chondrocytes occurs via receptor-mediated endocytosis. J Cell Sci. 1993;106:365–375. doi: 10.1242/jcs.106.1.365. [DOI] [PubMed] [Google Scholar]
- Hui W, Barksby HE, Young DA, Cawston TE, McKie N, Rowan AD. Oncostatin M in combination with tumour necrosis factor α induces a chondrocyte membrane associated aggrecanase that is distinct from ADAMTS aggrecanase-1 or-2. Ann Rheum Dis. 2005;64:1624–1632. doi: 10.1136/ard.2004.028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi M, Enghild JJ, Gendron C, Hughes C, Caterson B, Itoh Y, Nagase H. Altered proteolytic activities of ADAMTS-4 expressed by C-terminal processing. J Biol Chem. 2004;279:10109–10119. doi: 10.1074/jbc.M312123200. [DOI] [PubMed] [Google Scholar]
- Little CB, Hughes CE, Curtis CL, Jones SA, Caterson B, Flannery CR. Cyclosporin A inhibition of aggrecanase-mediated proteoglycan catabolism in articular cartilage. Arthritis Rheum. 2002;46:124–129. doi: 10.1002/1529-0131(200201)46:1<124::aid-art10121>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Loulakis P, Shrikhande A, Davis G, Maniglia CA. N-terminal sequence of proteoglycan fragments isolated from medium of interleukin-1-treated articular-cartilage cultures. Putative site(s) of enzymic cleavage. Biochem J. 1992;284:589–593. doi: 10.1042/bj2840589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzon ME, Fregien N, Schmid N, Falcon NS, Campos M, Casalino-Matsuda SM, Forteza RM. Reactive oxygen species and hyaluronidase 2 regulate airway epithelial hyaluronan fragmentation. J Biol Chem. 2010;285:26126–26134. doi: 10.1074/jbc.M110.135194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort JS, Billington CJ. Articular cartilage and changes in arthritis: matrix degradation. Arthritis Res. 2001;3:337–341. doi: 10.1186/ar325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwari P, Gao G, Lee JH, Grodzinsky AJ, Sandy JD. Analysis of ADAMTS4 and MT4-MMP indicates that both are involved in aggrecanolysis in interleukin-1-treated bovine cartilage. Osteoarthritis Cart. 2005;13:269–277. doi: 10.1016/j.joca.2004.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai SK, Duh FM, Vigdorovich V, Danilkovitch-Miagkova A, Lerman MI, Miller AD. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc Natl Acad Sci U S A. 2001;98:4443–4448. doi: 10.1073/pnas.071572898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CR, Mort JS, Roughley PJ. Treatment of cartilage proteoglycan aggregate with hydrogen peroxide. Relationship between observed degradation products and those that occur naturally during aging. Biochem J. 1987;247:349–357. doi: 10.1042/bj2470349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughley PJ, Barnett J, Zuo F, Mort JS. Variations in aggrecan structure modulate its susceptibility to aggrecanases. Biochem J. 2003;375:183–189. doi: 10.1042/BJ20030609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandy JD, Neame PJ, Boynton RE, Flannery CR. Catabolism of aggrecan in cartilage explants. Identification of a major cleavage site within the interglobular domain. J Biol Chem. 1991;266:8683–8685. [PubMed] [Google Scholar]
- Song RH, Tortorella MD, Malfait AM, Alston JT, Yang Z, Arner EC, Griggs DW. Aggrecan degradation in human articular cartilage explants is mediated by both ADAMTS-4 and ADAMTS-5. Arthritis Rheum. 2007;56:575–585. doi: 10.1002/art.22334. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Iizawa T, Harada H, Yamada K, Katsumata M, Takahashi M. Cartilage degradation independent of MMP/aggrecanases. Osteoarthritis Cart. 2004;12:1006–1014. doi: 10.1016/j.joca.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Sztrolovics R, Recklies AD, Roughley PJ, Mort JS. Hyaluronate degradation as an alternative mechanism for proteoglycan release from cartilage during interleukin-1beta-stimulated catabolism. Biochem J. 2002a;362:473–479. doi: 10.1042/0264-6021:3620473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztrolovics R, White RJ, Roughley PJ, Mort JS. The mechanism of aggrecan release from cartilage differs with tissue origin and the agent used to stimulate catabolism. Biochem J. 2002b;362:465–472. doi: 10.1042/0264-6021:3620465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorella MD, Pratta M, Liu RQ, Austin J, Ross OH, Abbaszade I, Burn T, Arner E. Sites of aggrecan cleavage by recombinant human aggrecanase-1 (ADAMTS-4) J Biol Chem. 2000;275:18566–18573. doi: 10.1074/jbc.M909383199. [DOI] [PubMed] [Google Scholar]
- Tortorella MD, Malfait A-M, Deccico C, Arner E. The role of ADAM-TS4 (aggrecanase-1) and ADAM-TS5 (aggrecanase-2) in a model of cartilage degradation. Osteoarthritis Cart. 2001;9:539–552. doi: 10.1053/joca.2001.0427. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Cheung SC, Itano N, Kimata K, Yamada Y. Identification of hyaluronan-binding domains of aggrecan. J Biol Chem. 1997;272:28057–28065. doi: 10.1074/jbc.272.44.28057. [DOI] [PubMed] [Google Scholar]
- Yamanishi Y, Boyle DL, Clark M, Maki RA, Tortorella MD, Arner EC, Firestein GS. Expression and regulation of aggrecanase in arthritis: the role of TGF-β. J Immunol. 2002;168:1405–1412. doi: 10.4049/jimmunol.168.3.1405. [DOI] [PubMed] [Google Scholar]
- Yamazaki K, Fukuda K, Matsukawa M, Hara F, Yoshida K, Akagi M, Munakata H, Hamanishi C. Reactive oxygen species depolymerize hyaluronan: involvement of the hydroxyl radical. Pathophysiology. 2003;9:215–220. doi: 10.1016/s0928-4680(03)00024-5. [DOI] [PubMed] [Google Scholar]
- Yasumoto T, Bird JL, Sugimoto K, Mason RM, Bayliss MT. The G1 domain of aggrecan released from porcine articular cartilage forms stable complexes with hyaluronan/link protein. Rheumatology (Oxford) 2003;42:336–342. doi: 10.1093/rheumatology/keg109. [DOI] [PubMed] [Google Scholar]
- Young DA, Lakey RL, Pennington CJ, Jones D, Kevorkian L, Edwards DR, Cawston TE, Clark IM. Histone deacetylase inhibitors modulate metalloproteinase gene expression in chondrocytes and block cartilage resorption. Arthritis Res Ther. 2005;7:R503–R512. doi: 10.1186/ar1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Corcoran C, Collins-Racie LA, LaVallie ER, Morris EA, Flannery CR. Glycosaminoglycan-binding properties and aggrecanase activities of truncated ADAMTSs: comparative analyses with ADAMTS-5, -9, -16 and -18. Biochim Biophys Acta. 2006;1760:517–524. doi: 10.1016/j.bbagen.2006.01.013. [DOI] [PubMed] [Google Scholar]