Abstract
Completion of genome sequences for many organisms allows a reasonably complete definition of the complement of extracellular matrix (ECM) proteins. In mammals this “core matrisome” comprises ∼300 proteins. In addition there are large numbers of ECM-modifying enzymes, ECM-binding growth factors, and other ECM-associated proteins. These different categories of ECM and ECM-associated proteins cooperate to assemble and remodel extracellular matrices and bind to cells through ECM receptors. Together with receptors for ECM-bound growth factors, they provide multiple inputs into cells to control survival, proliferation, differentiation, shape, polarity, and motility of cells. The evolution of ECM proteins was key in the transition to multicellularity, the arrangement of cells into tissue layers, and the elaboration of novel structures during vertebrate evolution. This key role of ECM is reflected in the diversity of ECM proteins and the modular domain structures of ECM proteins both allow their multiple interactions and, during evolution, development of novel protein architectures.
In mammals, there are ∼300 core extracellular matrix (ECM) proteins and many more ECM-associated proteins. Their evolution was key during the transition to multicellularity, the arrangement of cells into tissues, and the elaboration of vertebrate structures.
The term extracellular matrix (ECM) means somewhat different things to different people (Hay 1981, 1991; Mecham 2011). Light and electron microscopy show that extracellular matrices are widespread in metazoa, underlying and surrounding many cells, and comprising distinct morphological arrangements. The initial biochemical studies on extracellular matrix concentrated on large, structural extracellular matrices such as cartilage and bone. In the 1980s, the availability of model systems such as the Engelbreth-Holm-Swarm (EHS) sarcoma opened the way to biochemical analyses of basement membranes and led to the discovery of the different group of ECM proteins that make up basement membranes. Biochemistry of native ECM was, and still is, impeded by the fact that the ECM is, by its very nature, insoluble and is frequently cross-linked. Furthermore, ECM proteins tend to be large, and early work was frequently on proteolytic fragments. The application of molecular biology to studies of ECM proteins and their genes uncovered many previously unknown ECM molecules and defined their structures. The protein chemistry and molecular biology revealed that ECM proteins are typically made up of repeated domains, often encoded in the genome as separate exonic units. The completion of the sequences of many genomes now allows description of the entire list of proteins and, potentially, the definition of the complete repertoire of ECM proteins, based on homologies with known ECM proteins. Comparative analyses of the genomes of different organisms allow deductions about the evolution of this repertoire, which we term the matrisome. Newer methods such as mass spectrometry are also beginning to allow more detailed biochemical characterization of extracellular matrices. In this article, we will give an overview of the mammalian matrisome and briefly discuss certain aspects of the evolution of the matrisome and of the ECM.
DEFINITION OF THE MATRISOME
In analyzing the structure and functions of extracellular matrices, one would like to have a complete “parts list”—a list of all the proteins in any given matrix and a larger list of all the proteins that can contribute to matrices in different situations (the “matrisome”). As mentioned, the biochemistry of ECM is challenging because of the insolubility of most ECMs. However, the availability of complete genome sequences coupled with our accumulated knowledge about ECM proteins now makes it possible to come up with a reasonably complete list of ECM proteins. ECM proteins typically contain repeats of a characteristic set of domains (see figures and Table 1) (LamG, TSPN, FN3, VWA, Ig, EGF, collagen prodomains, etc.). Many of these domains are not unique to ECM proteins but their arrangements are highly characteristic. That is, the architecture of ECM proteins is diagnostic—they are built from assemblies of many ancient, and a few more recent, protein domains, each of which is typically encoded by one or a few exons in the genome. ECM proteins represent one of the earliest recognized and most elaborate examples of exon (domain) shuffling during evolution (Engel 1996; Patthy 1999; Hohenester and Engel 2002; Whittaker et al. 2006; Adams and Engel 2007). This characteristic of ECM proteins allows bioinformatic sweeps of the proteome encoded by any given genome, using a list of 50 or so domains to identify a list of candidate ECM proteins. Negative sweeps of that list using domains from other protein families (e.g, tyrosine kinases, which share FN3 and Ig domains with ECM proteins) and screens for transmembrane domains allow refinement of the list. A very few known ECM proteins do not have readily recognizable domains (e.g., elastin, dermatopontin, and some dentin matrix proteins) although, increasingly, even those are now being incorporated into protein analysis sites such as SMART and InterPro, allowing their routine capture in the sweeps. Using such methods plus manual annotation, we have been able to define a robust list of the proteins defining the mammalian matrisome by analysis of the human and mouse genomes (Naba et al. 2011). We call this list of “core” ECM proteins the core matrisome. It comprises 1%–1.5% of the mammalian proteome (without considering the contribution of alternatively spliced isoforms (prevalent in transcripts of matrisome genes). This list comprises almost 300 proteins, including 43 collagen subunits, three dozen or so proteoglycans, and around 200 glycoproteins.
Table 1.
Extracellular matrix proteoglycans
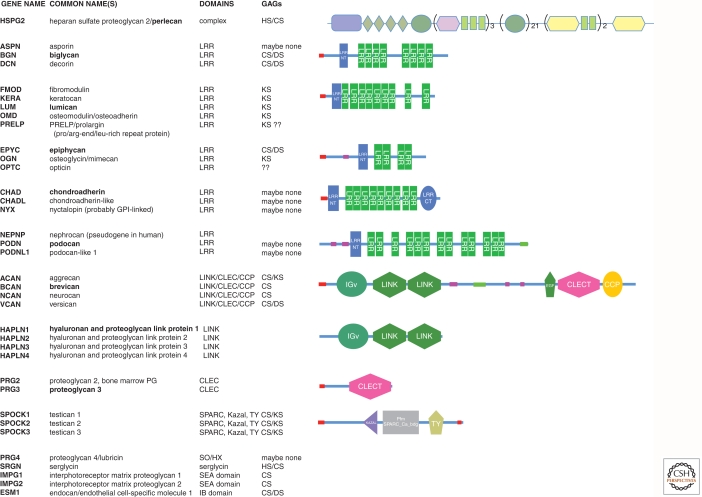 |
Table lists known ECM proteoglycans and shows representative domain structures.
There are five subfamilies of small leucine-rich repeat (LRR) proteoglycans based on sequence homologies (Merline et al. 2009; Schaefer and Schaefer 2010)—A representative structure is shown for one member (bold) from each subfamily. LRR repeat regions in other proteins are involved in protein–protein interactions.
The hyalectans (aggrecan, brevican, neurocan, and versican) all share a similar structure with Ig-LINK-LINK at their amino terminus and EGF(1 or 2)-CLEC-CCP at their carboxyl terminus with variable lengths of sequence between these two sets of domains. The central intervening sequence bears the GAG side chains and is almost 2000 amino acids long in aggrecan, and in versican it varies from a few to almost 2700 amino acids through alternative splicing. The LINK domains in both hyalectans and in the four link proteins bind to hyaluronan with nanomolar affinity and the EGF-CLEC-CCP cluster, which is similar to that in selectins, binds carbohydrates.
The domain structures were extracted from the SMART database (http://smart.embl-heidelberg.de). The domain structure of perlecan, the major proteoglycan of basement membranes, includes multiple domains in N—C order (SEA, LDLa, Ig, LamB, EGF, Ig, LamG, EGF). The glycosaminoglycan (GAG) content of each proteoglycan is based on the reviews cited above (HS, heparan sulfate; CS, chondroitin sulfate; DS, dermatan sulfate; KS, keratan sulfate).
This core matrisome list does not include mucins, secreted C-type lectins, galectins, semaphorins, and plexins and certain other groups of proteins that plausibly do associate with the ECM but are not commonly viewed as ECM proteins; lists of these “ECM-affiliated” proteins are given in Naba et al. (2011). The core matrisome list also does not include ECM-modifying enzymes, such as proteases, or enzymes involved in cross-linking, or growth factors and cytokines, although these are well known to bind to ECMs (see below).
Two useful databases provide information on the expression and distribution of various ECM proteins (http://www.matrixome.com/bm/Home/home/home.asp, The Matrixome Project, maintained by Kiyotoshi Sekiguchi and http://www.proteinatlas.org/;Human Protein Atlas) (Ponten et al. 2008; Uhlen et al. 2010). A third database (MatrixDB, http://matrixdb.ibcp.fr/) (Chautard et al. 2009, 2010) collates information about interactions among ECM proteins.
COLLAGENS
Collagens are found in all metazoa and provide structural strength to all forms of extracellular matrices, including the strong fibers of tendons, the organic matrices of bones and cartilages, the laminar sheets of basement membranes, the viscous matrix of the vitreous humor, and the interstitial ECMs of the dermis and of capsules around organs. Collagens are typified by the presence of repeats of the triplet Gly-X-Y, where X is frequently proline and Y is frequently 4-hydroxyproline. This repeating structure forms stable, rodlike, trimeric, coiled coils, which can be of varying lengths. A primordial collagen exon encoded six of these triplets (18 amino acids) encoded in 54 base pairs and, during evolution, this original motif has been duplicated, modified, and incorporated into many genes (Fig. 1A). Collagen subunits assemble as homotrimers or as restricted sets of heterotrimers and, in general, collagen subunits are very restricted in the partnerships they can form, although occasional promiscuity has been noted (for more details, see Ricard-Blum 2011; Yurchenco 2011).
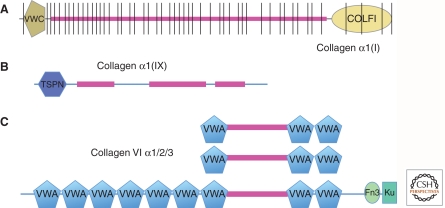
Figure 1.
Examples of collagen structures. (A) Collagen I is a fibrillar collagen with a continuous collagen domain of around 1000 amino acids (fuschia) comprising Gly-X-Y repeats that form a triple helix. It is encoded by multiple exons (note vertical lines) that are variants of a primordial exon encoding six such repeats. The collagen domain is flanked by amino- and carboxy-terminal noncollagenous domains that are removed by proteolysis to allow fibrillogenesis of the mature collagen. The VWC domain in this and other fibrillar collagens can be alternatively spliced and binds bone morphogenetic proteins (BMPs). (B) Collagen IX is a FACIT collagen (fibril-associated collagen with interrupted triple helix); the interruptions in the collagen domain allow bending. This and other FACIT collagens associate with fibrillar collagens and their amino-terminal domains extend out from the fibrils and presumably function as protein-binding domains. (C) Collagen VI is a heterotrimer of three related subunits, one of which is much longer and forms globular heads at each end. VWA domains are commonly protein-binding domains and probably allow interactions with other proteins during the formation of short fibrils by collagen VI.
Some of these genes are viewed as collagens, sensu stricto, whereas others that contain only short collagen segments are often referred to as “collagen-like” or “collagen-related.” The distinction is to some extent arbitrary because many proteins viewed as “true” collagens also contain significant portions made up of other domains. The original type I collagen of bones and tendons consists almost entirely of a long (∼1000 amino acids) and rigid uninterrupted collagen triple helix (plus terminal noncollagenous prodomains that are removed during biosynthetic processing of the protein; Fig. 1A). The rodlike trimers assemble into higher-order oligomers and fibrils and become cross-linked by various enzymatic and nonenzymatic reactions conferring considerable structural strength. Several other collagens with similar fibrillar structure are found in various tissues. Many other collagen types have interruptions in the Gly-X-Y repeating structure, introducing flexibility into the molecules. All collagen genes also encode additional noncollagenous domains, some of which are the characteristic collagen N and C prodomains, whereas others are domains shared with other ECM proteins and retained in the mature proteins (Fig. 1B,C). These additional protein domains confer specific binding affinities, allowing collagen molecules to interact with each other and with other proteins to assemble the various structures. The diversity of collagen structures, genes, and assemblies is discussed by Ricard-Blum (2011) and the assembly of type IV collagen into the laminar structure of basement membranes is reviewed by Yurchenco (2011). Other reviews of the collagen family cover additional aspects (Eyre and Wu 2005; Robins 2007; Gordon and Hahn 2009).
Among the collagen-like or collagen-related proteins (see table in Ricard-Blum 2011), a few are membrane proteins; others, such as complement component C1q and related proteins are secreted but their main functions do not involve ECM and they are not considered as part of the ECM or matrisome; yet others, such as the collagen-like domain of acetylcholinesterase, serve to anchor other proteins into the ECM, and some, such as EMIDs, are true ECM proteins. It is worth keeping in mind the possibility that the presence of collagen-like domains could act to bind some of these non-ECM proteins to the ECM, at least part of the time; in that sense they are ECM-associated.
PROTEOGLYCANS
Proteoglycans are interspersed among the collagen fibrils in different ECMs. Rather than providing structural strength, they confer additional properties. Proteoglycans are glycoproteins with attached glycosaminoglycans (GAGs; repeating polymers of disaccharides with carboxyl and sulfate groups appended). The addition of GAGs confers on proteoglycans a high negative charge, leading them to be extended in conformation and able to sequester both water and divalent cations such as calcium. These properties confer space-filling and lubrication functions. GAGs, especially heparan sulfates, also bind many secreted and growth factors into the ECM (see Sarrazin et al. 2011 for more details).
There are around three dozen extracellular matrix proteoglycans encoded in mammalian genomes; they fall into several families (Table 1) (see also Iozzo and Murdoch 1996). The two largest are those based on LRR repeats (Merline et al. 2009; Schaefer and Schaefer 2010) and those containing LINK and C-type lectin domains (hyalectans). Many of the LRR proteoglycans bind to various collagens and to growth factors and the hyalectan family members bind to various ECM glycoproteins such as tenascins, and through the LINK domain, to hyaluronic acid. These binding functions contribute to regulation of protein complexes in the ECM.
In addition, there are around a dozen proteoglycans that do not fall into these two families (e.g., lubricin/PRG4, endocan/ESM1, serglycin, and three testicans related to SPARC/osteonectin; see Table 1). Perhaps the most significant of all is perlecan (HSPG2), a multidomain protein that is a core proteoglycan of all basement membranes (see Table 1 and below). There are also many examples of proteins falling into other categories (e.g., some collagens, agrin, betaglycan, CD44, and other glycoproteins) that are sometimes or always modified by attachment of GAGs, which could lead one to consider them also as proteoglycans. The boundary between proteoglycans and glycoproteins is thus somewhat a matter of definition. The consensus view is to consider as proteoglycans those that have a significant fraction of their total mass made up by GAGs.
There are also two small families of integral membrane proteoglycans: glypicans (Filmus et al. 2008) and syndecans (Couchman 2010; Xian et al. 2010), both of which bear heparan sulfate side chains as does CD44, and there are a few additional transmembrane chondroitin sulfate proteoglycans. Further details of structure and functions of various heparan sulfate proteoglycans are discussed by Bishop et al. (2007) and Sarrazin et al. (2011).
GLYCOPROTEINS
In addition to the collagens and proteoglycans that provide strength and space-filling functions (among others), there are around 200 complex glycoproteins in the mammalian matrisome (see Table 2 and Naba et al. 2011). These confer myriad functions including interactions allowing ECM assembly, domains and motifs promoting cell adhesion, and also signaling into cells and other domains that bind growth factors. The bound growth factors can serve as reservoirs that can be released (e.g., by proteolysis) or can be presented as solid-phase ligands by the ECM proteins (Hynes 2009).
Table 2.
Extracellular matrix glycoproteins
| Gene name | Protein names and synonyms |
|---|---|
| Basement membrane components | |
| Lama1-5 | 5 Laminin alpha subunits |
| Lamb1-4 | 3 Laminin beta subunits |
| Lamc1-3 | 3 Laminin gamma subunits |
| Nid1/2 | 2 Nidogens |
| Colq | Collagen-like tail subunit of asymmetric acetylcholinesterase |
| Major known ECM glycoproteins | |
| Eln | Elastin |
| Emilin1-3 | 3 Emilins, elastin microfibril interfacers |
| Emid1/2 | 2 EMI domain-containing proteins |
| Fbln1/2/5/7 | 4 Fibulins |
| Efemp 1/2 | Fibulins 3 and 4 |
| Fbn1/2 | 2 Fibrillins |
| Fn1 | Fibronectin |
| Fras1 | Fraser syndrome 1 homolog |
| Gldn | Gliomedin |
| Hmcn1/2 | Hemicentins 1 and 2 |
| Ibsp | Integrin-binding sialoprotein, BSP |
| Matn1-4 | 4 Matrilin proteins |
| Mfap1a/b - 5 | 6 Microfibrillar-associated proteins |
| Mmrn1 and 2 | 2 Multimerins |
| Npnt | Nephronectin |
| Papln | Papilin, proteoglycan-like sulfated glycoprotein |
| Postn | Periostin, osteoblast-specific factor |
| Sparc/Sparcl1 | Secreted acidic cysteine-rich glycoproteins SPARC and SPARC-like |
| Spp1/Srpx | 2 Secreted phosphoprotein 1, osteopontin |
| Tnc/n/r/x | 4–5 Tenascins |
| Thbs1-4 | 4 Thrombospondins—see also COMP/TSP5 |
| Comp/TSP5 | Cartilage oligomeric matrix protein (thrombospondin 5) |
| Nervous system-enriched ECM proteins | |
| Agrn | Agrin |
| Coch | Cochlin |
| Ntn1-5 | 4 Netrins |
| Ntng1/g2 | Netrins G1/G2 |
| Reln | Reelin |
| Slit1-3 | 3 Slit homologs |
| Sspo | SCO-spondin |
| Tecta/b | Tectorins α and β |
| Vascular ECM proteins | |
| Fga/b/g | Fibrinogen α/β/γ chains |
| Vtn | Vitronectin |
| Vwf | von Willebrand factor |
| ECM proteins of bones, cartilage, and teeth | |
| Ambn | Ameloblastin |
| Amelx | Amelogenin X chromosome |
| Bglap2 | Bone γ-carboxyglutamate protein 2 |
| Bglap-rs1 | Bone γ-carboxyglutamate protein-related sequence 1 |
| Cilp | Cartilage intermediate-layer protein, nucleotide pyrophosphohydrolase |
| Cilp2 | Cartilage intermediate-layer protein 2 |
| Dmp1 | Dentin matrix protein 1 |
| Dpt | Dermatopontin |
| Dspp | Dentin sialophosphoprotein |
| Mgp | Matrix Gla protein |
| CCN family proteins | |
| Cyr61 | Cysteine rich protein 61, CCN1 |
| Ctgf | Connective tissue growth factor, CCN2 |
| Nov | Nephroblastoma overexpressed gene, CCN3 |
| Wisp1-3 | 3 WNT1 inducible signaling pathway proteins, CCN4-6 |
| Growth-factor-binding proteins | |
| Bmper | BMP-binding endothelial regulator |
| Igfals | Insulin-like growth-factor-binding protein, acid labile subunit |
| Igfbp1-7 | 7 Insulin-like growth-factor-binding proteins |
| Igfbpl1 | Insulin-like growth-factor-binding protein-like 1 |
| Kcp | Kielin/chordin-like protein |
| Ltbp1-4 | 4 Latent transforming growth-factor β-binding proteins |
| Other possible ECM proteins | |
| Abi3bp | ABI gene family, member 3 (NESH)-binding protein |
| Adipoq | Adiponectin, C1Q, and collagen domain-containing protein |
| Aebp1 | AE-binding protein 1 |
| Bsph1 | Binder of sperm protein homolog 1 |
| Cdcp2 | CUB domain-containing protein 2 |
| Creld1/2 | Cysteine-rich with EGF-like domains 1 and 2 |
| Crim1 | Cysteine-rich transmembrane BMP regulator 1 (chordin like) |
| Crispld1/2 | Cysteine-rich secretory protein LCCL domain-containing 1 and 2 |
| Cthrc1 | Collagen triple helix repeat containing 1 |
| Ddx26b | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| Dmbt1 | Deleted in malignant brain tumors 1 |
| Ecm1 | Extracellular matrix protein 1 |
| Ecm2 | Extracellular matrix protein 2, female organ, and adipocyte specific |
| Edil3 | EGF-like repeats and discoidin I-like domains 3 |
| Egflam | EGF-like, fibronectin type III, and laminin G domains |
| Fgl1/2 | Fibrinogen-like proteins 1 and 2 |
| Fndc1/7/8 | 3 Fibronectin type III domain-containing proteins |
| Gas6 | Growth arrest specific 6 |
| Igsf10 | Immunoglobulin superfamily, member 10 |
| Lgi1-4 | 4 Leucine-rich repeat LGI family proteins |
| Lrg1 | Leucine-rich α-2-glycoprotein 1 |
| Mepe | Matrix extracellular phosphoglycoprotein with ASARM motif |
| Mfge8 | Milk fat globule-EGF factor 8 protein |
| Nell1/2 | NEL-like 1 and 2 |
| Oit3 | Oncoprotein-induced transcript 3 |
| Otog | Otogelin |
| Pcolce/Pcolce2 | 2 Procollagen C-endopeptidase enhancer proteins |
| Pxdn | Peroxidasin homolog |
| Rspo1-4 | 4 R-spondin homologs |
| Slamf6 | SLAM family member 6 |
| Smoc1 | SPARC-related modular calcium-binding proteins 1 and 2 |
| Sned1 | Sushi, nidogen, and EGF-like domains 1 |
| Spon1 | Spondin 1, (f-spondin) extracellular matrix protein |
| Spon2 | Spondin 2, extracellular matrix protein |
| Srpx2 | Sushi-repeat-containing protein, X-linked 2 |
| Svep1 | Sushi, von Willebrand factor type A, EGF, and pentraxin domain-containing 1 |
| Tgfbi | Transforming growth factor β-induced |
| Thsd4 | Thrombospondin type I domain-containing 4 |
| Tinag/Tinagl1 | Tubulointerstitial nephritis antigen/tubulointerstitial nephritis antigen-like 1 |
| Tnfaip6 | Tumor necrosis factor α-induced protein 6 |
| Tsku | Tsukushin |
| Vit | Vitrin |
| Vwa1-5 | 5 von Willebrand factor A domain-containing proteins |
| Vwc2/2l/e | 3 von Willebrand factor C domain-containing proteins |
| VwD/E | 2 von Willebrand factor D and EGF domain proteins |
| Zp1-4 | 4 Zona pellucida glycoproteins |
| Zp3r | Zona pellucida 3 receptor |
| Zpld1 | Zona pellucida-like domain-containing 1 |
| Novel predicted ECM proteins | |
| 2010321M09Rik | RIKEN cDNA 2010321M09 gene |
| 5430419D17Rik | RIKEN cDNA 5430419D17 gene |
| 6130401L20Rik | RIKEN cDNA 6130401L20 gene |
| 9230107M04Rik | RIKEN cDNA 9230107M04 gene |
| A930038C07Rik | RIKEN cDNA A930038C07 gene |
| AW551984 | Expressed sequence AW551984 |
| C330046G03Rik | RIKEN cDNA C330046G03 gene |
| D17H6S56E-3 | DNA segment, Chr 17, human D6S56E 3 |
| Gm106 | Predicted gene 106 |
| Gm414 | Predicted gene 414 |
| Gm6924 | Predicted gene 6924 |
Table lists known and predicted ECM glycoproteins other than collagens and proteoglycans, defined largely by their characteristic domain content. The table is split into rough categories—individual glycoproteins are listed in only a single category although many could be listed under more than one. The first page lists glycoproteins about which a significant amount is known; the second page lists glycoproteins about which relatively little to nothing has been reported concerning their role in the ECM.
The best-studied ECM glycoproteins are the laminins (11 genes; 5α, 3β, 3γ) and fibronectins (1 gene encoding multiple splice isoforms). These are reviewed in detail by Aumailley et al. (2005) and Yurchenco (2011) and by Schwarzbauer and DeSimone (2011), respectively. Also well studied are the thrombospondins and tenascins, reviewed by Bentley and Adams (2010) and Adams and Lawler (2011) and by Chiquet-Ehrismann and Turner (2011), respectively. The structures of these glycoproteins are well known and exemplify the typical multiple repeating domain structure and extended multimeric forms of ECM proteins (Fig. 2). The same is true for fibulins (de Vega et al. 2009) and nidogens (Ho et al. 2008; Yurchenco 2011) and many others. Two subgroups of ECM glycoproteins have been studied particularly in the context of the nervous system (netrins, slits, reelin, agrin, SCO-spondin—see article by Barros et al. [2011] and Fig. 3) and the hemostatic system (von Willebrand factor, vitronectin, and fibrinogen—a facultative ECM protein) (Bergmeier et al. 2008; Bergmeier and Hynes 2011). These two biological systems also involve roles for more widely distributed ECM proteins such as thrombospondins, fibronectins, laminins, collagens, proteoglycans, etc. Similarly, the matrices of other tissues typically contain both ubiquitous and tissue-restricted ECM glycoproteins. Another group of ECM glycoproteins that has been studied in the context of disease and the regulation of transforming growth factor beta (TGF-β) functions includes the fibrillins and LTBPs (Ramirez and Dietz 2009; Ramirez and Rifkin 2009; see article by Munger and Sheppard 2011).
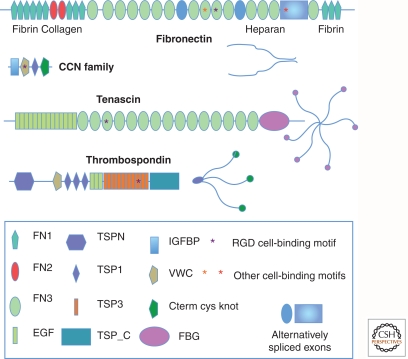
Figure 2.
Examples of characteristic ECM glycoprotein structures. Note the multidomain structure of these ECM glycoproteins. Each domain is typically encoded by a single exon or a small set of exons. This has allowed shuffling of domains into different combinations during evolution. Individual domains are specialized for binding different proteins, as indicated for fibronectin. Some domains are alternatively spliced, as noted for fibronectin, and is also true for tenascin (not shown). Cell-binding motifs such as RGD and LGV are indicated by asterisks. Fibronectin dimerizes through disulfide bonding at the carboxyl terminus, whereas thrombospondin and tenascin form trimers and hexamers, respectively, through coiled coil domains and disulfide bonds near the amino terminus. The appearances of the intact protein multimers (as would be seen by electron microscopy) are diagrammed. Note that growth factor-binding domains (IGFBP, VWC, and others) are included in many ECM proteins. CYR61 is shown as a representative member of the CCN family (see Table 2), small ECM proteins that contain integrin-binding motifs and growth-factor-binding domains (IGFBP and VWC) and are known to regulate growth factor functions (Chen and Lau 2009) as are the larger proteins shown.
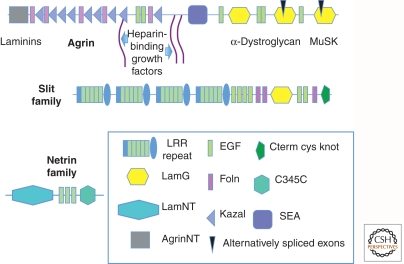
Figure 3.
Glycoproteins with special roles in the nervous system. These three proteins are involved in synapse formation (Agrin) and in axonal guidance (Slits and Netrins). Sites for binding other ECM proteins (laminins), growth factors, and cell-surface receptors (α-dystroglycan and MuSK) are indicated for agrin. Slit also contains known GF-binding domains (Foln). Agrin has two small alternatively spliced exons that markedly affect its functions, a characteristic of many ECM proteins. Unlike many ECM proteins, the major receptors for these three proteins are not integrins. Slit family proteins bind to Robo receptors, whereas Netrins bind to Unc5 and DCC receptors. The functions of these ECM-receptor pairs in the CNS are discussed in Barros et al. (2011) but they also function in other aspects of cell and tissue pattern regulation during development. They are evolutionarily ancient and are conserved in all bilaterial phyla.
However, as can be seen in Table 2, there are multiple other ECM glycoproteins about which much less (in some cases, almost nothing) is known. These include some enormous glycoproteins with impressive arrays of domains, such as SCO-spondin (59 domains of seven types) and hemicentin-1, also known as fibulin-6 (61 domains of six types), and many that are affected in disease (Aszódi et al. 2006; Nelson and Bissell 2006; Bateman et al. 2009). It will be of considerable interest to learn the distributions and functions of this diverse set of ECM glycoproteins and we can expect that the approaches that have been effective for the better-studied proteins will provide many insights into the roles of those less well known and novel.
ECM-BOUND GROWTH AND SECRETED FACTORS
As mentioned above and elsewhere (Hynes 2009; Ramirez and Rifkin 2009; Rozario and DeSimone 2010), many growth factors bind to ECM proteins and must be considered also as constituents of extracellular matrices. One popular idea is that growth and other secreted factors bind to GAGs, especially heparan sulfates. Although this is undoubtedly true, there are clear examples of growth factors binding to specific domains of ECM proteins. Fibronectin binds specifically to a variety of growth factors (VEGF, HGF, PDGF, etc.; Rahman et al. 2005; Wijelath et al. 2006; Lin et al. 2010) and the VWC/chordin and follistatin domains found in many ECM proteins (see Figs. 1–3) are known to bind BMPs (Wang et al. 2008; Banyai et al. 2010). TGF binds specifically to TB domains in LTBPs, which bind in turn to fibrillins and to fibronectin-rich matrices (Ramirez and Rifkin 2009; Munger and Sheppard 2011). These ECM-TGF interactions have significant consequences for genetic diseases; mutations in fibrillins affect the regulation of TGF-β function in Marfan’s syndrome and in other diseases (Ramirez and Dietz 2009).
It seems virtually certain that the known examples of growth factor binding to ECM, including directly to ECM proteins, presage many more such cases, and this aspect of ECM function is in great need of further investigation. The ECM can act as a reservoir or sink of such factors and there are many examples of this for chemokines and for many of the most important developmental signals (e.g., VEGFs, Wnts, Hhs, BMPs, and FGFs). Such factors form gradients that control pattern formation during developmental processes and it is clear that some of those gradients are markedly affected by ECM binding (Yan and Lin 2009). Indeed, it seems probable that many more gradients incorporate ECM binding as part of their regulation. Investigation of this concept will be greatly aided by our current fairly complete inventory of ECM proteins and their constituent domains.
MODIFIERS OF ECM STRUCTURE AND FUNCTION
Another aspect of ECM function is that ECM proteins and the fibrils into which they assemble are subsequently often significantly modified. Collagens have long been known to become cross-linked by disulfide bonding, transglutaminase cross-linking, and through the action of lysyl oxidases and hydroxylases (Eyre and Wu 2005; Robins 2007; Ricard-Blum 2011). Laminins and other basement membrane proteins also become cross-linked by disulfide bonding (see Yurchenco 2011 for further details) and the same is true of fibronectin, which also undergoes further processing to a state characterized by insolubility in deoxycholate (DOC) (Choi and Hynes 1979; Schwarzbauer and DeSimone 2011). The exact basis for this insolubility is not known, but fibronectin and other ECM proteins are also substrates for transglutaminase 2, which undoubtedly contributes to the insolubility of ECM (Lorand and Graham 2003; Iismaa et al. 2009).
Proteolytic enzymes also modify the ECM—indeed, procollagen propeptidases are necessary to process collagens so that they can polymerize. Collagens and other ECM proteins are also substrates for matrix metalloproteases (MMPs) (Page-McCaw et al. 2007; Cawston and Young 2010), ADAMs (Murphy 2008) and ADAMTS proteases (Porter et al. 2005; Apte 2009), and many other proteolytic enzymes (elastases, cathepsins, various serine esterase proteases, etc.) can also act on many ECM proteins (see article by Lu et al. 2011). These various proteolytic processes play roles in ECM turnover and are thought to release ECM-bound growth factors and also to expose cryptic activities in the ECM (Mott and Werb 2004; Ricard-Blum 2011), including the release of antiangiogenic inhibitors (Bix and Iozzo 2005; Nyberg et al. 2005; Hynes 2007). Similarly, enzymes that degrade GAGs, such as heparanases and sulfatases, can also alter the properties of ECM proteoglycans (see articles by Lu et al. 2011; Sarrazin et al. 2011). The remodeling of ECM by these various processes has major effects on development and pathology (Daley et al. 2008; Kessenbrock et al. 2010; Lu et al. 2011). Lists of these ECM-modifying enzymes can be found in the reviews cited and in Naba et al. (2011).
CELLULAR RECEPTORS FOR EXTRACELLULAR MATRIX
For the ECM to affect cellular functions, it is obvious that there must be receptors for ECM proteins. The major receptors are the integrin family, comprising 24 αβ heterodimers (Fig. 4). These have been extensively reviewed elsewhere and specific aspects are covered in other articles in this collection (Schwartz 2010; Campbell and Humphries 2011; Geiger and Yamada 2011; Huttenlocher and Horwitz 2011; Watt and Fujimura 2011; Wickström et al. 2011). Another receptor for ECM proteins is dystroglycan, which binds to laminin, agrin, and perlecan in basement membranes as well as to the transmembrane neurexins (Barresi and Campbell 2006). Each of these dystroglycan ligands contains LamG domains, which bind to dystroglycan in a glycosylation-dependent manner (see Fig. 3), probably by binding carbohydrate side chains on dystroglycan. Mutations in dystroglycan or its associated proteins in the membrane or the cytoskeleton (or in laminin) can all produce various forms of muscular dystrophy, because of the loss of the transmembrane connection to the basement membrane surrounding the muscle cells. Other cellular receptors for ECM include GPVI on platelets and the DDR (discoidin domain receptor) tyrosine-kinases, all of which are receptors for collagens (Leitinger and Hohenester 2007), the GPIb/V/IX complex, which forms a receptor for von Willebrand factor on platelets (Bergmeier et al. 2008; Bergmeier and Hynes 2011), and CD44, which binds to hyaluronan and is expressed on many cells. As noted in Figure 3, Slits bind to Robo receptors of the Ig superfamily and netrins bind to Unc5-related tyrosine-kinase receptors or to DCC, an Ig superfamily receptor, whereas agrin binds to the MuSK tyrosine-kinase receptor. Thus, although integrins comprise the dominant class of ECM receptors and are present on most cells, numerous other receptors for ECM proteins are expressed on specific cell types.
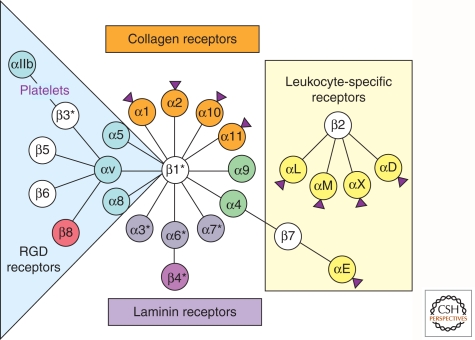
Figure 4.
Integrin receptors for ECM proteins. The diversity of integrin subunits and their interactions. Shown are the mammalian integrins, separated by color coding into subsets of closely related subunits. The RGD- binding (blue) and laminin-binding (purple) subclasses are evolutionarily very ancient and found in all metazoan phyla, but they have diverged into clades in the vertebrate lineage. The α4/α9 clade (green) is vertebrate-specific. Two subclasses of chordate α subunits have inserted I domains (purple arrowheads); they include collagen-specific integrins (orange) and a set of α subunits confined to leukocytes (yellow). Some subunits show alternatively spliced isoforms (*). The leukocyte integrins bind predominantly to cell-surface counter-receptors, whereas integrins containing either the β1 or αv subunits bind predominantly to extracellular matrix (ECM) proteins, although within each class there are exceptions to these generalizations and it is worth noting that most integrins are capable of binding multiple ligands, and there are many others beyond those shown here (Humphries et al. 2006). Many, if not all, αv integrins are also capable of activating TGF-β. Most β subunits are highly related (white) and bind to talin and related proteins (Campbell and Humphries 2011; Wickström et al. 2011), whereas the β4 subunit instead binds to intermediate filaments through specific linker proteins and the β8 subunit binds to band 4.1 proteins instead of to talin. (figure modified from Hynes 2002).
In addition to binding extracellular ligands, these ECM receptors provide transmembrane links to the cytoskeleton and to signal transduction pathways. The cytoplasmic domains of ECM receptors assemble large and dynamic complexes of proteins, which regulate cytoskeletal assembly and activate many signaling cascades within cells (Geiger and Yamada 2011). In the case of integrins, these submembranous complexes also regulate the extracellular affinity of the receptors (so-called “inside-out” signaling) and the same may be true of other classes of ECM receptors. It has become clear that the signaling functions of ECM adhesion receptors are at least as complicated as those of canonical growth factor receptors and that engagement of ECM receptors provides signals regulating cellular survival, proliferation, and differentiation as well as adhesive and physical connections involved in cell shape, organization, polarity, and motility.
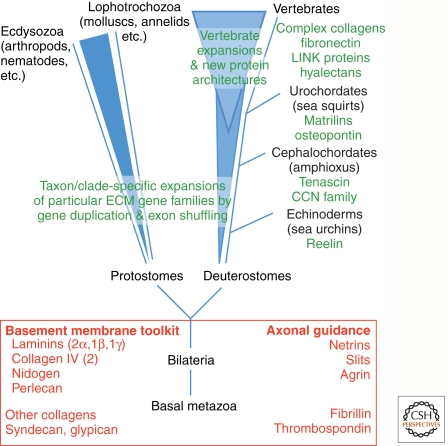
EVOLUTION OF THE MATRISOME AND THE EXTRACELLULAR MATRIX
The ∼300 proteins that make up the core matrisome in mammals are a mixture of very ancient proteins and some much newer ones (Fig. 5). Comparative analyses of the genomes of different taxa have revealed that some ECM proteins are shared by almost all metazoa, even simple organisms such as sponges, coelenterates, and cnidaria (Huxley-Jones et al. 2007; Ozbek et al. 2010). Most notable are the proteins that make up the core of basement membranes—type IV collagens (2 subunits), laminin (4 genes, 2 α, 1β, and 1γ), nidogen, and perlecan (1 gene each)—see Yurchenco 2011. We call this set of genes the basement membrane toolkit and it is found in all protostome and deuterostome genomes and must therefore have been present in the common ancestor of all bilateria (Hynes and Zhao 2000; Whittaker et al. 2006). Many, but not all, of these genes are also found in more primitive metazoan organisms such as cnidaria and sponges (Putnam et al. 2007; Chapman et al. 2010; Srivasatava et al. 2010). It is plausible to argue that the evolution of multilayered organisms with their different cell layers separated by basement membranes was dependent on this basement membrane toolkit that has been maintained ever since. Fibrillar collagens are also found in early metazoa, including Hydra and sponges. Interestingly, another collagen, the paralog of collagens XV and XVIII is also ancient, being found in both protostomes and deuterostomes, although the key functions of this class of collagens are not fully understood. Most other collagens are later evolutionary developments, for example the cuticular collagens of Caenorhabditis elegans (Hutter et al. 2000) and the complex collagens with VWA and FN3 domains (see Fig. 1C and Ricard-Blum 2011) found in vertebrates. Also found in all bilateria are the neuronal guidance ECM proteins, netrins, slits, and agrin (Fig. 3).
Figure 5.
Evolution of ECM proteins. The figure outlines the main phylogenetic lineages (although the branch lengths are not drawn to scale), and illustrates the evolution of complexity of the matrisome and ECM during evolution. The inferred basal bilaterian had a core of ECM proteins including the basement membrane toolkit and some other ECM proteins (not all of which are shown) that have been retained in later-developing taxa, including the two main branches of metazoa (protostomes and deuterostomes). More primitive taxa had some, but not all, of these ECM proteins. During evolution of protostomes, there was modest expansion of the number of ECM genes/proteins mostly comprising taxon-specific expansions of ECM protein families by gene duplication and divergence, with some exon shuffling. A similar modest expansion occurred during evolution of the deuterostome lineage—first known acquisitions of novel ECM proteins of interest are noted in green. During evolution of the vertebrate subphylum, there was a major increase in ECM protein diversity, probably related to two whole genome duplications that occurred in that lineage. This expansion included expansion and diversification of preexisting ECM protein families, and also the development of novel protein architectures by shuffling of domains and the inclusion of novel domains (e.g., FN1, FN2, LINK). Some examples of such novel ECM proteins are indicated. As discussed in the text, this large expansion and diversification of the matrisome in vertebrates is presumably linked to novel structures such as neural crest and endothelial-lined vasculature as well as connective tissues such as cartilage, bones, and teeth, and also the development of more complex nervous and immune systems.
One characteristic feature of the evolution of ECM proteins, as for other genes, is an increase in numbers of homologous genes as one ascends the tree of life (Fig. 5). Thus, mammals have six type IV collagen genes (see Ricard-Blum 2011), two nidogen genes, and 11 laminin genes (see Yurchenco 2011) that have arisen by gene duplications and subsequent divergence without altering the basic structures of the proteins. This diversification accompanies the diversification of basement membranes in vertebrates. Similar evolution by duplication and diversification from a primordial gene shared by all bilateria is seen in the case of thrombospondins (see Adams and Lawler 2011), although in this case the diversification has involved more extensive evolution of the domain architecture than is the case for the basement membrane toolkit. This suggests that thrombospondins have evolved to fulfill a more diverse set of functions, whereas basement membranes have retained many of their basic structure-function requirements during the more than half a billion years of their evolution.
Other ECM proteins, in contrast, are more recent developments. Two clear examples are tenascins and fibronectins (Tucker and Chiquet-Ehrismann 2009; Chiquet-Ehrismann and Tucker 2011). Both are restricted to chordates, as are many of the more complex collagen genes. A tenascin gene is found in all the chordate genomes that have been sequenced and vertebrates have expanded the tenascin family. Tenascins represent a novel architectural assembly of preexisting domains (EGF and FN3; see Fig. 2). In contrast, fibronectin contains domains that do not appear until quite late in evolution; whereas FN3 domains are ancient, being found in cell-surface receptors in all metazoa, FN1 and FN2 domains are restricted to chordates. The earliest fibronectin-like gene so far reported (although lacking the precise, characteristic domain organization of vertebrate fibronectin) appears in urochordates (ascidians, sea squirts) whereas vertebrates all have the canonical structure found in mammals (see Fig. 2) (Hynes 1990; Schwarzbauer and DeSimone 2011). Once assembled, this gene appears to have been strongly selected (it is essential for life) and has remained unchanged. Reelin, a protein that controls aspects of brain development in mammals also appears to be a deuterostome-specific gene (Whittaker et al. 2006), using one old domain (EGF) and two new ones (Reeler and BNR). Analyses of proteoglycans reveal a similar story. Whereas perlecan is ancient (as are the transmembrane proteoglycans, syndecan and glypican), proteoglycans containing the LINK domain are confined to deuterostomes, indeed largely to vertebrates (there are two genes containing that domain in sea urchins) (Whittaker et al. 2006).
In general, it seems clear that the fraction of the proteome that is ECM proteins has expanded disproportionately during the evolution of the deuterostome lineage, both by duplication and divergence of existing genes and by the appearance of novel gene architectures and even some new domains. It is interesting to speculate on the reasons for this. One obvious explanation is the development of cartilage, bones, and teeth in vertebrates and that undoubtedly accounts for some of the elaboration of novel collagens, proteoglycans, and ECM glycoproteins. However, proteins such as tenascins, fibronectin, and reelin (as well as other neural ECM proteins) have no obvious strong connections to the development of structural ECMs and it is tempting to hypothesize that their emergence was more closely tied to the emergence of novel structures such as the neural crest, endothelial-lined vasculature, and more complex nervous systems. Consonant with this model of key roles for ECM proteins in evolution, the matrisome is one of the most plastic and rapidly evolving compartments of the proteome.
CONCLUDING REMARKS
We now have a reasonably complete inventory of ECM proteins and their associated modifiers. Some ECM proteins have been well studied and we have a good picture of their basic functions—other ECM proteins are virtually unstudied. Even in the case of the well studied proteins, many of the constituent domains, all of which are well conserved and must, therefore, have important functions, still lack assigned functions. Presumably, many of them, like those that we do understand, serve to bind other proteins in ways that contribute to ECM assembly, binding, and presentation of growth factors and interactions with cells to influence their behavior. There is now a pressing need to describe the changes in ECM composition in development and pathology, to better understand the interactions of individual domains, and to probe the cooperation of these multiprotein assemblies in modulating the functions of cells and tissues. The techniques for such analyses (biophysical, imaging, etc.) continue to advance and there is every prospect that studies of ECM structure and function will yield important insights into the crucial roles played by this vital component of metazoan organization, and genetic analyses and studies of human disease are revealing the biological relevance of individual ECM proteins and of specific interactions.
ACKNOWLEDGMENTS
We thank Charlie Whittaker and Sebastian Hoersch for their assistance and collaboration in the bioinformatic mining of genomes during our development of the ECM inventory discussed here. The work in our laboratory was supported by the National Cancer Institute and the Howard Hughes Medical Institute.
Footnotes
Editors: Richard O. Hynes and Kenneth M. Yamada
Additional Perspectives on Extracellular Matrix Biology available at www.cshperspectives.org
REFERENCES
- Adams J, Engel J 2007. Bioinformatic analysis of adhesion proteins. In Methods in molecular biology, pp. 147–172 Humana Press, New York: [DOI] [PubMed] [Google Scholar]
- Adams JC, Lawler J 2011. The thrombospondins. Cold Spring Harb Perspect Biol 10.1101/cshperspect. a009712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apte SS 2009. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: Functions and mechanisms. J Biol Chem 284: 31493–31497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aszódi A, Legate KR, Nakchbandi I, Fässler R 2006. What mouse mutants teach us about extracellular matrix function. Annu Rev Cell Dev Biol 22: 591–621 [DOI] [PubMed] [Google Scholar]
- Aumailley M, Bruckner-Tuderman L, Carter WG, Deutzmann R, Edgar D, Ekblom P, Engel J, Engvall E, Hohenester E, Jones JCR, et al. 2005. A simplified laminin nomenclature. Matrix Biol 24: 326–332 [DOI] [PubMed] [Google Scholar]
- Bányai L, Sonderegger P, Patthy L 2010. Agrin binds BMP2, BMP4 and TGFβ1. PLoS ONE 5: e10758 10.1371/journal.pone.0010758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barresi R, Campbell K 2006. Dystroglycan: From biosynthesis to pathogenesis of human disease. J Cell Sci 119: 199–207 [DOI] [PubMed] [Google Scholar]
- Barros CS, Franco SJ, Muller U 2011. Extracellular matrix: Functions in the nervous system. Cold Spring Harb Perspect Biol 3: a005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman JF, Boot-Handford RP, Lamandé SR 2009. Genetic diseases of connective tissues: Cellular and extracellular effects of ECM mutations. Nat Rev Genet 10: 173–183 [DOI] [PubMed] [Google Scholar]
- Bentley AA, Adams JC 2010. The evolution of thrombospondins and their ligand-binding activities. Mol Biol Evol 27: 2187–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmeier W, Hynes RO 2011. ECM proteins in hemostasis and thrombosis. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmeier W, Chauhan AK, Wagner DD 2008. Glycoprotein Ibalpha and von Willebrand factor in primary platelet adhesion and thrombus formation: Lessons from mutant mice. Thromb Haemost 99: 264–270 [DOI] [PubMed] [Google Scholar]
- Bishop JR, Schuksz M, Esko JD 2007. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 446: 1030–1037 [DOI] [PubMed] [Google Scholar]
- Bix G, Iozzo RV 2005. Matrix revolutions: “Tails” of basement-membrane components with angiostatic functions. Trends Cell Biol 15: 52–60 [DOI] [PubMed] [Google Scholar]
- Campbell ID, Humphries MJ 2011. Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol 3: a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawston TE, Young DA 2010. Proteinases involved in matrix turnover during cartilage and bone breakdown. Cell Tissue Res 339: 221–235 [DOI] [PubMed] [Google Scholar]
- Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, et al. 2010. The dynamic genome of Hydra. Nature 464: 592–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chautard E, Ballut L, Thierry-Mieg N, Ricard-Blum S 2009. MatrixDB, a database focused on extracellular protein–protein and protein–carbohydrate interactions. Bioinformatics 25: 690–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chautard E, Fatoux-Ardore M, Ballut L, Thierry-Mieg N, Ricard-Blum S 2010. MatrixDB, the extracellular matrix interaction database. Nucleic Acids Res 39: D235–D240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-C, Lau LF 2009. Functions and mechanisms of action of CCN matricellular proteins. Int J Biochem Cell Biol 41: 771–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiquet-Ehrismann R, Tucker RP 2011. Tenascins and the importance of adhesion modulation. Cold Spring Harb Perspect Biol 3: a004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi MG, Hynes RO 1979. Biosynthesis and processing of fibronectin in NIL.8 hamster cells. J Biol Chem 254: 12050–12055 [PubMed] [Google Scholar]
- Couchman JR 2010. Transmembrane signaling proteoglycans. Annu Rev Cell Dev Biol 26: 89–114 [DOI] [PubMed] [Google Scholar]
- Daley WP, Peters SB, Larsen M 2008. Extracellular matrix dynamics in development and regenerative medicine. J Cell Sci 121: 255–264 [DOI] [PubMed] [Google Scholar]
- de Vega S, Iwamoto T, Yamada Y 2009. Fibulins: Multiple roles in matrix structures and tissue functions. Cell Mol Life Sci 66: 1890–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J 1996. Domain organizations of modular extracellular matrix proteins and their evolution. Matrix Biol 15: 295–299 [DOI] [PubMed] [Google Scholar]
- Eyre DR, Wu J.-J 2005. Collagen cross-links. In Topics in current chemistry, Vol. 247, pp. 207–229 Springer-Verlag, Berlin [Google Scholar]
- Filmus J, Capurro M, Rast J 2008. Glypicans. Genome Biol 9: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B, Yamada KM 2011. Molecular architecture and function of matrix adhesions. Cold Spring Harb Perspect Biol 3: a005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MK, Hahn RA 2009. Collagens. Cell Tissue Res 339: 247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay ED, ed. 1981. Cell biology of extracellular matrix. Plenum Press, New York [Google Scholar]
- Hay ED, ed. 1991. Cell biology of extracellular matrix, 2nd ed. Plenum Press, New York [Google Scholar]
- Ho MSP, Böse K, Mokkapati S, Nischt R, Smyth N 2008. Nidogens—Extracellular matrix linker molecules. Microsc Res Tech 71: 387–395 [DOI] [PubMed] [Google Scholar]
- Hohenester E, Engel J 2002. Domain structure and organisation in extracellular matrix proteins. Matrix Biol 21: 115–128 [DOI] [PubMed] [Google Scholar]
- Humphries JD, Byron A, Humphries MJ 2006. Integrin ligands at a glance. J Cell Sci 119: 3901–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher A, Horwitz A 2011. Integrins in cell migration. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter H, Vogel BE, Plenefisch JD, Norris CR, Proenca RB, Spieth J, Guo C, Mastwal S, Zhu X, Scheel J, et al. 2000. Conservation and novelty in the evolution of cell adhesion and extracellular matrix genes. Science 287: 989–994 [DOI] [PubMed] [Google Scholar]
- Huxley-Jones J, Robertson DL, Boot-Handford RP 2007. On the origins of the extracellular matrix in vertebrates. Matrix Biol 26: 2–11 [DOI] [PubMed] [Google Scholar]
- Hynes RO 1990. Fibronectins. Springer-Verlag, New York [Google Scholar]
- Hynes RO 2002. Integrins: Bidirectional, allosteric signaling machines. Cell 110: 673–687 [DOI] [PubMed] [Google Scholar]
- Hynes RO 2007. Cell–matrix adhesion in vascular development. J Thromb Haemost 5(Suppl 1): 32–40 [DOI] [PubMed] [Google Scholar]
- Hynes RO 2009. The extracellular matrix: Not just pretty fibrils. Science 326: 1216–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO, Zhao Q 2000. The evolution of cell adhesion. J Cell Biol 150: F89–F96 [DOI] [PubMed] [Google Scholar]
- Iismaa SE, Mearns BM, Lorand L, Graham RM 2009. Transglutaminases and disease: Lessons from genetically engineered mouse models and inherited disorders. Physiol Rev 89: 991–1023 [DOI] [PubMed] [Google Scholar]
- Iozzo RV, Murdoch AD 1996. Proteoglycans of the extracellular environment: Clues from the gene and protein side offer novel perspectives in molecular diversity and function. FASEB J 10: 598–614 [PubMed] [Google Scholar]
- Kessenbrock K, Plaks V, Werb Z 2010. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 141: 52–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitinger B, Hohenester E 2007. Mammalian collagen receptors. Matrix Biol 26: 146–155 [DOI] [PubMed] [Google Scholar]
- Lin F, Ren X-D, Pan Z, Macri L, Zong W-X, Tonnesen MG, Rafailovich M, Bar-Sagi D, Clark RAF 2010. Fibronectin growth factor-binding domains are required for fibroblast survival. J Invest Dermatol 131: 84–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorand L, Graham R 2003. Transglutaminases: Crosslinking enzymes with pleiotropic functions. Nat Rev Mol Cell Biol 4: 140–156 [DOI] [PubMed] [Google Scholar]
- Lu P, Takai P, Weaver VM, Werb Z 2011. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecham R 2011. The extracellular matrix: An overview. Springer, Berlin [Google Scholar]
- Merline R, Schaefer R, Schaefer L 2009. The matricellular functions of small leucine- rich proteoglycans (SLRPs). J. Cell Commun Signal 3: 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott JD, Werb Z 2004. Regulation of matrix biology by matrix metalloproteinases. Curr Opin Cell Biol 16: 558–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger JS, Sheppard D 2011. Crosstalk among TGFβ signaling pathways, integrins, and the extracellular matrix. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G 2008. The ADAMs: Signalling scissors in the tumour microenvironment. Nat Rev Cancer 8: 929–941 [DOI] [PubMed] [Google Scholar]
- Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO 2011. The matrisome: In silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices (in revision). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Bissell MJ 2006. Of extracellular matrix, scaffolds, and signaling: Tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol 22: 287–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg P, Xie L, Kalluri R 2005. Endogenous inhibitors of angiogenesis. Cancer Res 65: 3967–3979 [DOI] [PubMed] [Google Scholar]
- Ozbek S, Balasubramanian PG, Chiquet-Ehrismann R, Tucker RP, Adams JC 2010. The evolution of extracellular matrix. Mol Biol Cell 21: 4300–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page-McCaw A, Ewald AJ, Werb Z 2007. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol 8: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patthy L 1999. Genome evolution and the evolution of exon-shuffling—A review. Gene 238: 103–114 [DOI] [PubMed] [Google Scholar]
- Pontén F, Jirström K, Uhlen M 2008. The human protein atlas—A tool for pathology. J Pathol 216: 387–393 [DOI] [PubMed] [Google Scholar]
- Porter S, Clark IM, Kevorkian L, Edwards DR 2005. The ADAMTS metalloproteinases. Biochem J 386: 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam N, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317: 86–94 [DOI] [PubMed] [Google Scholar]
- Rahman S, Patel Y, Murray J, Patel KV, Sumathipala R, Sobel M, Wijelath ES 2005. Novel hepatocyte growth factor (HGF) binding domains on fibronectin and vitronectin coordinate a distinct and amplified Met-integrin induced signalling pathway in endothelial cells. BMC Cell Biol 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Dietz HC 2009. Extracellular microfibrils in vertebrate development and disease processes. J Biol Chem 284: 14677–14681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Rifkin DB 2009. Extracellular microfibrils: Contextual platforms for TGFβ and BMP signaling. Curr Opin Cell Biol 21: 616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard-Blum S 2011. The collagen family. Cold Spring Harb Perspect Biol 3: a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins SP 2007. Biochemistry and functional significance of collagen cross-linking. Biochem Soc Trans 35: 849–852 [DOI] [PubMed] [Google Scholar]
- Rozario T, DeSimone D 2010. The extracellular matrix in development and morphogenesis: A dynamic view. Dev Biol 341: 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin S, Lamanna WC, Esko JD 2011. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol 3: a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer L, Schaefer RM 2010. Proteoglycans: From structural compounds to signaling molecules. Cell Tissue Res 339: 237–246 [DOI] [PubMed] [Google Scholar]
- Schwartz MA 2010. Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb Perspect Biol 2: a005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer JE, DeSimone DW 2011. Fibronectins, their fibrillogenesis and in vivo functions. Cold Spring Harb Perspect Biol 3: a005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466: 720–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RP, Chiquet-Ehrismann R 2009. Evidence for the evolution of tenascin and fibronectin early in the chordate lineage. Int J Biochem Cell Biol 41: 424–434 [DOI] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. 2010. Towards a knowledge-based human protein atlas. Nat Biotechnol 28: 1248–1250 [DOI] [PubMed] [Google Scholar]
- Wang X, Harris RE, Bayston LJ, Ashe HL 2008. Type IV collagens regulate BMP signalling in Drosophila. Nature 455: 72–77 [DOI] [PubMed] [Google Scholar]
- Watt FM, Fujiwara H 2011. Cell-extracellular matrix interactions in normal and diseased skin. Cold Spring Harb Perspect Biol 3: a005124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker CA, Bergeron K-F, Whittle J, Brandhorst BP, Burke RD, Hynes RO 2006. The echinoderm adhesome. Dev Biol 300: 252–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickström SA, Radovanac K, Fässler R 2011. Genetic analyses of integrin signaling. Cold Spring Harb Perspect Biol 3: a005116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijelath ES, Rahman S, Namekata M, Murray J, Nishimura T, Mostafavi-Pour Z, Patel Y, Suda Y, Humphries MJ, Sobel M 2006. Heparin-II Domain of fibronectin is a vascular endothelial growth factor-binding domain: Enhancement of VEGF biological activity by a singular growth factor/matrix protein synergism. Circ Res 99: 853–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian X, Gopal S, Couchman JR 2010. Syndecans as receptors and organizers of the extracellular matrix. Cell Tissue Res 339: 31–46 [DOI] [PubMed] [Google Scholar]
- Yan D, Lin X 2009. Shaping morphogen gradients by proteoglycans. Cold Spring Harb Perspect Biol 1: a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenco PD 2011. Basement membranes: Cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol 3: a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]