Abstract
The role of respiratory viruses in transmission of Streptococcus pneumoniae is poorly understood. Key questions such as which serotypes are most fit for transmission and disease, and whether influenza virus alters these parameters in a serotype specific manner have not been adequately studied. In a novel model of ferret transmission, we demonstrated that prior infection with influenza virus of donors enhanced pneumococcal transmission and disease. Nasal wash bacterial titers, the incidence of mucosal and invasive disease, and the percentage of contacts infected were all increased. Viral infection of contact ferrets increased their susceptibility to acquisition both in terms of percentage infected and distance over which they could acquire infection. These influenza mediated effects on colonization, transmission and disease were pneumococcal strain dependent. Overall, these data argue that human studies of the relationship between respiratory viral infections, acquisition of pneumococci, and development of disease need further study to be better understood.
Introduction
Streptococcus pneumoniae, the pneumococcus, is the most common etiologic agent causing otitis media, community acquired pneumonia, and invasive diseases such as sepsis, and meningitis [1]. Because pneumococci can express one of more than 90 different capsules, and the local and regional distribution of pneumococcal serotypes based on these capsules can differ considerably by geography, time period studied or age, prevention of pneumococcal disease by vaccination is complex. The regional nature of serotype distribution and concerns over serotype replacement are driving the development of new, higher valency conjugate vaccines to address shortcomings in presently utilized vaccines [2].
The interplay between serotype distributions, incidence of disease, and vaccine implementation is driving an intense interest in understanding patterns of pneumococcal epidemiology. An important, unanswered question in this context is which serotypes are most likely to cause disease? Typical study designs addressing this question may be confounded by differences in duration of carriage between strains and our lack of knowledge of the timing of invasion relative to onset of carriage. Although it is thought that prior influenza virus infection enhances the incidence and severity of bacterial disease (reviewed in [3] and [4]), the role of respiratory viruses such as influenza has not been assessed in recent studies of pneumococcal epidemiology. We have hypothesized that prior or concomitant infection with influenza viruses may favor particular serotypes or clonal types, or may alter the invasive disease potential in a strain specific manner [3].
A second major unanswered question in pneumococcal epidemiology is whether there are pneumococcal serotype/strain specific differences in transmission and whether these are modified by viral infections. Is prevalence of particular serotypes determined more by the ability to persist in the nasopharynx, or by an advantage in transmission? Does influenza alter this dynamic, enhancing transmission and favoring certain serotypes/strains? Limited data from small longitudinal studies conducted in the early 20th century suggest that respiratory viruses enhance transmission of S. pneumoniae [5,6]. Answering these questions definitively in humans would definitely require large longitudinal studies with frequent sampling of the cohorts. Because such studies would be expensive and logistically difficult, we sought to develop an animal model which was reflective of both human disease and transmission to address relevant hypotheses. In this report, we developed a novel transmission model of S. pneumoniae in ferrets. We then evaluated the ability of different serotypes of S. pneumoniae to colonize, cause secondary bacterial infections in mice and ferrets, and to transmit between ferrets.
Methods
Infectious agents
The pneumococcal isolates were chosen to belong to different serotypes/clonal types and have similar invasive disease potential in mice as described in [9], with the exception of TIGR4, which is a highly virulent type 4 clinical strain that is commonly used in murine models [20]. BHN78 (type 14, ST124), BHN54 (7F, ST191), and BHN60 (9V, ST838) are invasive isolates, and BHN97 (19F, ST425) is a carrier isolate. TIGR4 (type 4) is a human clinical strain that is commonly used in mouse models of pneumococcal disease [20]. BHN54 and BHN97 were engineered to express luciferase as described [21]. The St. Jude strain of mouse adapted influenza virus A/Puerto Rico/8/34 (H1N1; PR8), generated by reverse genetics [22], and influenza virus A/Sydney/5/97 (H3N2) were grown in Madin-Darby canine kidney (MDCK) cells.
Animal models
Eight week old BALB/cbJ mice (Jackson Laboratories, Bar Harbor Maine) were used in a dual infection model as described [23]. Young adult (7–8 week old) outbred ferrets serologically negative for influenza were bred in the Animal Resources Center at St. Jude Children’s Research Hospital and used in a dual infection model as described [9]. Bacterial and viral titers were determined from lung homogenates or nasal washes as described [9,24]. All experiments were conducted in biosafety level 2 facilities in a manner in accordance with the guidelines of the Committee on Care and Use of Laboratory Animals.
Bioluminescent imaging
Anesthetized mice or ferrets were imaged for 1 minute (mice) or 2 minutes (ferrets) by an IVIS CCD camera (Xenogen Corp.).Total photon emission from selected and defined areas within the images of each animal was quantified by the LivingImage software package version 2.20 (Xenogen Corp.), as described elsewhere [9,21,24].
Transmission model
Ferrets were infected with influenza virus (1×105 TCID50) or S. pneumoniae (1×107 CFU) intranasally in a volume of 400 μl sterile PBS while under anesthesia with isoflurane. Pairs of donor ferrets were housed alone in open caging in closed cubicles for 3 days (pneumo only and contact flu groups) and 6 days if donor ferrets sequentially received influenza virus then pneumococcus (donors flu and both flu) prior to introduction of contact ferrets. Pairs of contact ferrets which were either naïve to any infectious agents (pneumo only and donors flu) or had been infected 3 days prior with influenza virus (contacts flu and both flu) were then placed in the same cage as the donor ferrets, in a separate cage in the same cubicle 3 feet apart, or in a separate cage in a facing cubicle 10 feet apart (with intervening doors left open) overnight (for 14–16 hours). At this point the donor ferrets were removed to a separate cubicle and all doors were closed and no further contact allowed between groups of ferrets. There is no pressure differential between cubicles to create airflow from one group of ferrets to another, but air is pulled across the face of both cubicles at a high rate (8–10 exchanges per hour). The environment is controlled at 24°C and 30% humidity with a 12h:12h light:dark cycle. Care was taken to insure that inadvertent transmission by animal care attendants or through common source exposures was not possible.
Microarray analysis
Comparative genomic hybridizations were carried out using a reference design as described elsewhere [10,25], comparing the presence or absence of specific genes linked to invasion as well as 41 accessory regions (ARs) defined in [10] between the isolates with 2 reference strains (TIGR4 and R6). Analyses were performed with GenePix Pro (v6.0, MDS Analytical Technologies) and R Project for Statistical Computing (v2.4.0) using a Bayesian linear model as described [10] Genes were considered absent if they had a p-value of < 0.01 with an M value (i.e., log2 fold change) < −1, present if they had an M value > −0.8, and considered not to be predictive if neither of these criteria applied.
Statistical analyses
Comparison of survival between groups of mice was done with the Log Rank chi-squared test on the Kaplan-Meier survival data. Comparison of bacterial and viral titers was done using analysis of variance (ANOVA). A p-value of < 0.05 was considered significant for these comparisons. SigmaStat for Windows (SysStat Software, Inc., V 3.11) was utilized for all statistical analyses.
Results
Pneumococcal strain dependent differences in secondary bacterial infections in mice
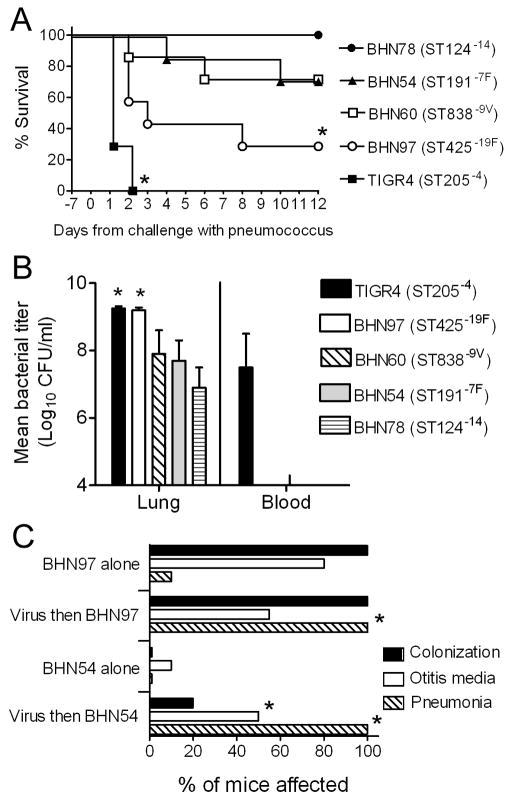
The first hypothesis to be tested was that influenza virus infection differentially affects the expression of disease from different pneumococcal strains. Several clinical S. pneumoniae isolates of different clonal types of known invasive disease potential in humans [7] and mice [8] were administered to influenza-infected mice using a dose that was non-lethal considering the bacteria alone. Considerable differences in mortality were observed that were pneumococcal strain dependent (Figure 1A). The ability to kill mice correlated with bacterial lung load (Figure 1B), and only the highly virulent TIGR4 caused bacteremia.
Figure 1. The impact of influenza virus on disease differs with pneumococcal strain used.
A) Groups of 6 mice were infected intranasally with 30 TCID50 of influenza virus PR8 7 days prior to challenge with 1×105 CFU of S. pneumoniae strains BHN78 (type 14), BHN54 (type 7F), BHN60 (type 9V), BHN97 (type 19F), or TIGR4 (type 4) and followed for mortality. An asterisk (*) indicates a significant difference (p < 0.05) by log-rank test on the Kaplan-Meier survival data vs. the other groups. B) Lung and blood titers were taken 48 hours after secondary challenge from groups of 4 mice infected as above. An asterisk (*) indicates a significant difference (p < 0.05) by ANOVA vs. the BHN78, BHN54, and BHN60 groups. C) Groups of 5 mice were infected intranasally with 30 TCID50 of influenza virus PR8 or were mock-infected with PBS 7 days prior to challenge with 1×105 CFU of S. pneumoniae using versions of BHN54 and BHN97 engineered to express luciferase, and the incidence of rhinitis, otitis media, and pneumonia as determined by bioluminescent imaging were determined over a 14 day period. An asterisk (*) indicates a significant difference (p < 0.05) by Student’s t-test with Bonferroni correction vs. the corresponding bacteria alone group.
In order to assess nasal colonization and site specific disease expression using bioluminescent imaging, we chose two pneumococcal strains which caused intermediate mortality in association with influenza, BHN54 (ST191−7F) and BHN97 (ST425−19F) and engineered them to express luciferase. BHN97 colonized 100% of mice assessed (Figure 1C) and persisted for a median of 33 days in animals infected only with S. pneumoniae, compared to 56 days in animals pre-infected with influenza. By contrast, BHN54 could only be found transiently in influenza infected animals (median 1 day). Both strains could cause otitis media, although this was more common with BHN54 following influenza. Neither strain caused pneumonia with any frequency in the absence of influenza virus, but 100% of mice in both groups pre-infected with influenza virus developed pneumonia (Figure 1C). These data indicate that there are differences in the support that influenza virus is able to provide to different strains of pneumococcus, and that the predominant effect in mice of pre-infection with influenza virus is to increase duration of carriage and enhance bacterial pneumonia in a strain dependent manner.
Influenza enhances secondary bacterial disease in ferrets in a pneumococcal strain dependent manner
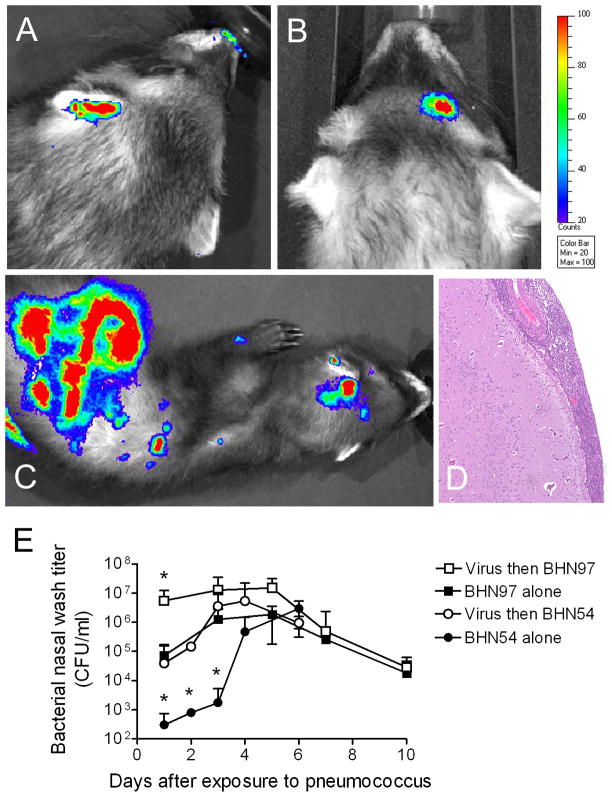
We previously showed that influenza virus can increase pneumococcal nasal titers and increase the incidence of secondary bacterial sinusitis and otitis media in young adult ferrets [9]. To determine whether these effects were pneumococcal strain specific, influenza-infected and influenza-naïve ferrets were challenged with pneumococcal strains BHN97 or BHN54 and assessed daily by nasal wash and bioluminescent imaging. For the first 3 days following infection, BHN97 was recovered from nasal wash at significantly higher titers then BHN54 (Figure 2E), a disparity that was no longer evident after 72 hours. In the absence of influenza virus, 1 of 5 ferrets in each group (20%) developed secondary bacterial infections detectable by bioluminescent imaging. Four out of 5 ferrets infected with BHN97 following influenza virus had secondary infections, including otitis media (Figure 2A), sinusitis (Figure 2B), disseminated disease including meningitis (Figure 2C, D), and meningitis with bacteremia (3 of the 4 had positive blood cultures). By comparison, influenza virus did not enhance the incidence of infections with BHN54, as only 1 out of 5 (20%) of ferrets had a secondary bacterial infection (blood culture negative sinusitis). All influenza infected ferrets were lethargic and were noted to sneeze. Ferrets infected with pneumococcus alone did not sneeze, and did not show overt clinical signs unless they developed meningitis, at which time they became obtunded. Based on these data from mice and ferrets, we conclude that prior influenza virus infection enhances the incidence and severity of pneumococcal disease in a strain dependent manner.
Figure 2. Influenza infection predisposes ferrets to secondary pneumococcal infections.
Ferrets infected with 1×105 TCID50 of influenza virus A/Sydney/5/97 (H3N2) and challenged 3 days later with 1×107 CFU of pneumococcal strain BHN97 were assessed by bioluminescent imaging for foci of bacteria representing sites of secondary bacterial infections. Representative images from ferrets with A) otitis media, B) sinusitis, and C) disseminated disease are pictured. The scale indicates the relative light units per pixel. The ferret in C) had D) meningitis characterized by expansion of the meninges by exuberant infiltrates of neutrophils with admixed macrophages, lymphocytes and necrotic cellular debris. Inflammation was confined to the meninges; however there was a mild response on the superficial brain surface consisting of edema and gliosis. E) Groups of 5 ferrets were infected with influenza or were mock-infected with PBS 3 days prior to challenge with pneumococcal strains BHN54 or BHN97. An asterisk (*) indicates a significant difference by ANOVA in nasal wash titer compared to all other groups at that timepoint (p < 0.05).
Influenza virus enhances transmission of S. pneumoniae in a pneumococcal strain dependent manner
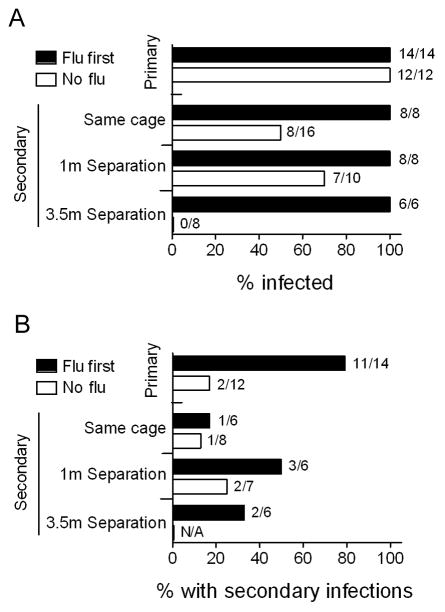
The second major question we wished to address was whether influenza virus altered transmission of S. pneumoniae. Transmission was studied using pairs of infected and uninfected ferrets as described in the methods. All directly inoculated donor ferrets were infected with S. pneumoniae, and pneumococcus was able to transmit from infected to contact ferrets in the same cage or up to a distance of 1 meter (Table 1). This is the first report of a small animal model of natural transmission of S. pneumoniae. Infection with influenza of the donors increased the incidence of pneumococcal acquisition in contact ferrets from 25% to 75% in same cage contacts and from 50% to 83% in ferrets 1 meter away, but did not affect the absence of transmission to ferrets 3.5 meters away. Infection with influenza of the contact ferrets had a more robust effect, as 100% of contacts including those 3.5 meters away were infected. As summarized in Figure 3A, primary infection occurred in every instance when pneumococci were introduced directly into the nose of anesthetized ferrets. Transmission to close contacts, either in the same cage or a close, adjacent cage, occurred 50–70% of the time if the contacts had not been previously infected with influenza virus. Influenza rendered contact ferrets extremely susceptible to acquisition of pneumococcus as 100% of ferrets, even those 3.5 meters away, were infected.
Table 1.
Transmission of S. pneumoniae between ferrets
| Pneumococcal Strain | Experimental Group | Donors - #Shedding/#Exposed (%) | Donors - #Secondary infections/#Assessed (%) | Degree of Contact | Contacts - #Shedding/#Exposed (%) | Contacts - #Secondary infections/#Assessed |
|---|---|---|---|---|---|---|
| BHN97 | Pneumo onlya | 8/8 (100) | 2/8 (25) | Same cage | 2/8 (25) | 1/4 (25) |
| 3 feet separation | 2/4 (50) | 1/4 (25) | ||||
| 10 feet separation | 0/4 (0) | 0/4 (0) | ||||
| Donors flub | 12/12 (100) | 10/12 (83) | Same cage | 6/8 (75) | 0/4 (0) | |
| 3 feet separation | 5/6 (83) | 1/4 (25) | ||||
| 10 feet separation | 0/4 (0) | 0/2 (0) | ||||
| Contacts fluc | 4/4 (100) | 0/4 (0) | Same cage | 4/4 (100) | 0/4 (0) | |
| 3 feet separation | 4/4 (100) | 2/4 (50) | ||||
| 10 feet separation | 4/4 (100) | 1/4 (25) | ||||
| Both flud | 4/4 (100) | 1/2 (50) | Same cage | 4/4 (100) | 1/2 (50) | |
| 3 feet separation | 4/4 (100) | 1/2 (50) | ||||
| 10 feet separation | 2/2 (100) | 1/2 (50) | ||||
| BHN54 | Pneumo only | 4/4 (100) | 0/4 (0) | Same cage | 0/4 (0) | 0/4 (0) |
| 3 feet separation | 0/4 (0) | 0/4 (0) | ||||
| Donors flu | 4/4 (100) | 1/4 (25) | Same cage | 0/4 (0) | 0/4 (0) | |
| 3 feet separation | 0/4 (0) | 0/4 (0) | ||||
| Contacts flu | 4/4 (100) | 0/4 (0) | Same cage | 0/4 (0) | 0/4 (0) | |
| 3 feet separation | NDe | ND | ||||
| Both flu | 7/7 (100) | 2/7 (24) | Same cage | 0/4 (0) | 0/4 (0) | |
| 3 feet separation | 0/4 (0) | 0/4 (0) |
Pneumo only – The donor ferrets were infected with S. pneumoniae while the contact ferrets were naïve.
Donors flu – The donor ferrets were infected with influenza virus then 3 days later superinfected with S. pneumoniae while the contact ferrets were naïve.
Contacts flu – The contact ferrets were infected with influenza virus 3 days prior to being exposed to donor ferrets who had been infected with S. pneumoniae.
Both flu – The donor ferrets were infected with influenza virus then 3 days later superinfected with S. pneumoniae while the contact ferrets were infected with influenza virus 3 days prior to being exposed to donor ferrets.
ND = Not done.
Figure 3. Influenza virus enhances transmission between ferrets.
A summary of several experiments (from Table 1) is presented demonstrating the percentage of ferrets A) infected or B) which developed secondary bacterial infections with S. pneumoniae strain BHN97 (considering only those which were shedding), stratified by whether the ferrets were directly infected by inoculation under anesthesia (primary) or infected naturally by transmission (secondary), by whether they were previously infected with influenza virus (flu first) or were naïve to influenza virus at the time of infection or exposure (no flu), and, for contact ferrets, by physical proximity to the donor ferrets (same cage, 1 meter of separation, 3.5 meters of separation). N/A indicates no ferrets were shedding.
Due to space constraints, the experimental design employed here involved ferrets housed together in pairs for all experiments. Thus, it was theoretically possible for one contact ferret of a pair to acquire pneumococcus and then transmit it to the other ferret in the pair, which could alter the interpretation of these outcomes. Of the 21 ferrets which were infected at a distance, 17 were positive at the initial assessment within 24 hours of exposure, while 4 were positive on the second day (Table 2) with a mean time to positivity of 1.2 days post-exposure. Ferrets exposed by direct contact had similar or slightly longer times to positivity, with 9 of 16 positive on the first day, 5 positive on the second day, and 2 positive on the third day with a mean time to positivity of 1.6 days post-exposure. Examining pairs of ferrets, discordant results where one ferret shed and the other did not only occurred once. From these data we conclude that some secondary transmission within pairs of contact ferrets likely occurred in the same cage contact ferrets in some cases. While we do not believe this alters the conclusions reached on the effect of influenza on transmission, it does mean that the true efficiency of transmission in the model is likely overestimated by looking at the raw percentages, and the length of exposure to infected animals (16 hours exposure to donor ferrets in this study) required to transmit cannot be estimated.
Table 2.
Characterization of transmission of BHN97 between ferrets
| Time to positivity (TTP)a | Mean (d) | |||
|---|---|---|---|---|
| 24 h | 48 h | 72 h | ||
| Same Cage (# ferrets) | 9 | 5 | 2 | 1.6 |
| Distant Cage (# ferrets) | 17 | 4 | 0 | 1.2 |
| Difference in TTP between ferrets | ||||
| Same day | 24 h | 48 h | ||
| Same Cage (# ferrets) | 10 | 5 | 1 | |
| Distant Cage (# ferrets)b | 19 | 1 | 0 | |
Indicates time between start of exposure and detection of pneumococcus in nasal washes (positivity)
Complete discordance where 1 ferret was positive and 1 negative was only seen in a single pair of ferrets
An examination of bacterial titers from primary (direct inoculation) compared to secondary (via transmission) infections showed no differences in nasal washes between virus infected ferrets (Figure 4A). However, secondary acquisition of pneumococcus in virus naïve animals resulted in lower titers and more rapid clearance of bacteria compared to direct inoculation of a high titer of bacteria. Viral titers were different 24 hours after primary inoculation compared to secondary acquisition, but could not be distinguished beginning 2 days after infection or exposure (Figure 4B). Secondary bacterial infections were detected by bioluminescent imaging more commonly in virus infected ferrets than in virus naïve animals following primary inoculation, but the incidence of secondary disease was lower following natural acquisition than with direct inoculation (Figure 3B and Table 1). Donor ferrets which were naïve to influenza virus but exposed to virus-infected contacts all developed influenza virus infections. Viral titers in these ferrets, which contracted pneumococcal infection prior to influenza, did not differ from animals primarily infected with influenza (data not shown), and none of these ferrets developed otitis media, sinusitis, pneumonia, or invasive disease.
Figure 4. Bacterial and viral titers in nasal washes are similar in singly infected compared to co-infected ferrets.
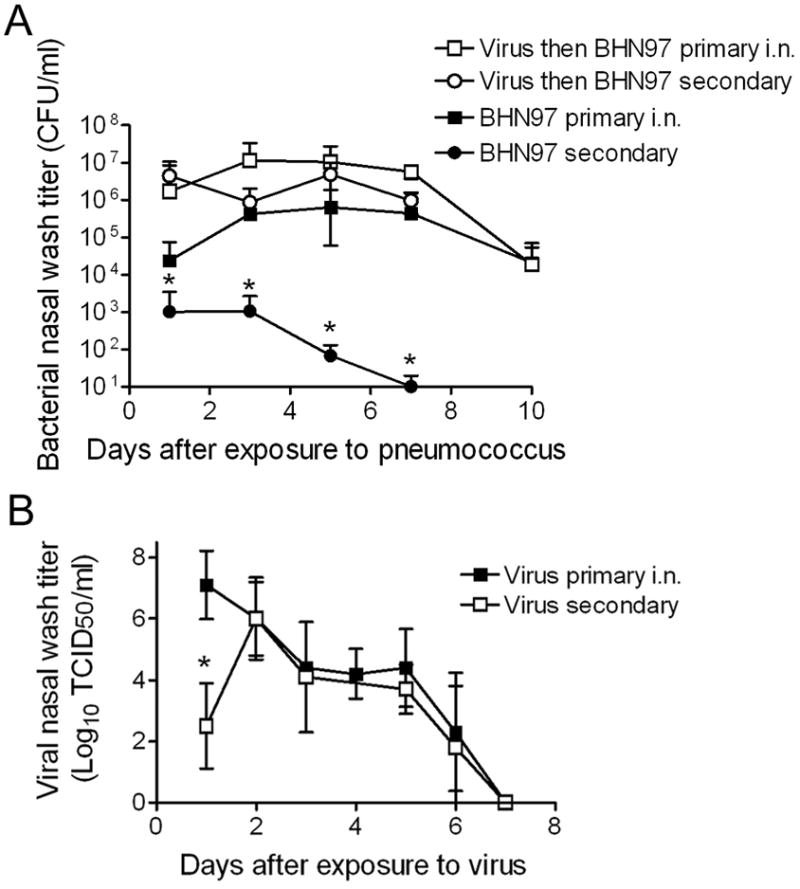
A summary of several experiments is presented demonstrating A) mean bacterial titers in nasal washes from ferrets directly infected with S. pneumoniae strain BHN97 by inoculation under anesthesia (primary i.n.; n = 8 per group) or infected naturally by transmission (secondary; n = 15 per group) stratified by whether they were previously infected with influenza virus or were naive to influenza virus at the time of infection or exposure, and B) mean viral titers in ferrets directly infected with influenza virus by inoculation under anesthesia (primary i.n.; n = 13 per group) or infected naturally by transmission (secondary; n = 4 per group). An asterisk (*) indicates a significant difference (p < 0.05) in titer compared to all other groups at that time point.
Pattern of accessory regions and gene composition differs between BHN97 and BHN54
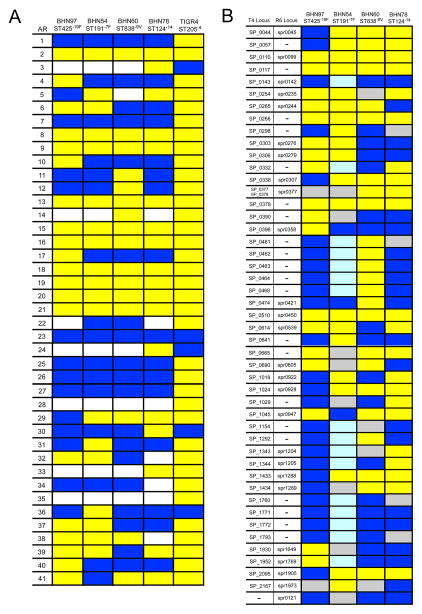
Since BHN97 and BHN54 differed substantially in their ability to colonize both mice and ferrets and transmit between ferrets, we sought to determine genetic differences between the strains using a whole genome microarray approach [10]. Examination of the accessory regions (ARs) present in the genomes of these 2 strains (Figure 5A) revealed several instances where an AR was present in BHN97 and absent in BHN54 or vice versa. ARs 4, 10, 40, and 41 (present in BHN97) and 29 (present in BHN54) have been associated with clonal complexes and serotypes that have a high invasive disease potential [10]. Of note, AR6, which is found in nearly all isolates from serotypes with the highest invasive disease potential, is present in both strains, while AR11, which encodes invasive pili that enhance adherence and colonization of the 19F isolate of ST162 [8], was absent in both. The differences in AR loci should provide target genes for understanding the colonization and transmission phenotypes of these strains.
Figure 5. Presence or absence of accessory regions (ARs) and virulence genes in strains used in this study.
A) Presence or absence of ARs were determined for strains utilized in the study including TIGR4. Yellow, AR is present. White, some genes from the AR are present and some are absent. Dark blue, AR is absent. ARs are defined in [10]. B) Presence or absence of specific genes linked to invasion by signature tagged mutagenesis [10] were determined for strains used in the study compared to known loci in TIGR4 and R6. Yellow, gene is present. Light blue, gene is likely to be absent but the p value is not significant. Dark blue, gene is absent (p < 0.01). Gray, there are no data or the data are unclear from this analysis.
Analysis of the isolates for the presence or absence of genes associated with invasion and virulence in signature tagged mutagenesis (STM) studies [10–13] delineated a number of genes that could be associated with the enhanced virulence and invasive capacity of TIGR4 (Figure 1) compared to the other isolates studied (Figure 5B). Interestingly, BHN97, which was more virulent in association with influenza virus pre-infection than BHN54, has considerably fewer of these genes than the other isolates. Only 2 genes from this analysis, SP_0396 (mtlF, a putative mannitol-specific enzyme involved in carbohydrate transport) and SP_1045 (a hypothetical protein), are clearly present in BHN97 and absent in BHN54 and thus represent candidates to explain the enhanced relative virulence of this strain in influenza virus infected mice. However, since the published STM studies used invasiveness in mice that are not infected with influenza to identify these genes, it is likely that there are other potential genes that are important in the context of prior influenza infection but that do not fall out in screens in the absence of influenza. Thus, further work to identify virulence genes in S. pneumoniae that are context specific (i.e., important in the post-influenzal host) is required.
Discussion
The influence of respiratory viruses on pneumococcal transmission has been studied in humans in only a cursory fashion. Gwaltney et al. demonstrated in a longitudinal study of families that in 56% of cases where a defined episode of transmission could be documented, the donor had symptoms of an upper respiratory tract infection [6]. They suggested that increased pneumococcal titers in the nasopharynx mediated by viral co-infection, modification of the site of colonization by viruses from the nasopharynx to the anterior nares, or increased production and dissemination of respiratory virus secretions due to intercurrent viral illness was responsible for this phenomenon. A study of prevalence in adults showed that carriage could be detected in adults twice as often when they had an upper respiratory tract infection, implying acquisition of pneumococci was favored during viral co-infections [14]. In this report utilizing the ferret model, both transmission from the donor and acquisition by contacts was enhanced by prior influenza virus infection. The effect on the contacts seemed stronger, since only when the contacts were virally infected was transmission possible over 3.5 meters, and enhanced nasal titers as a sole mechanism seems unlikely since influenza virus enhanced nasal titers of BHN54 to levels similar to that of BHN97, yet transmission of BHN54 could not be demonstrated. Similar effects by other respiratory viruses should be assessed.
We did not assess the theory espoused by Gwaltney et al. [6] that influenza facilitates a transition from the posterior nasopharynx to the anterior nares – simultaneous culturing of both sites in this model would be revealing. We also did not assess the mode of transmission. It is likely that aerosol transmission was required for acquisition by contact ferrets placed 3.5 meters away, as large droplets are unlikely to be capable of crossing this space, particularly in a room with such a high rate of air exchange. Sneezing by influenza infected ferrets may have contributed either to formation of aerosols or dissemination across that distance. However, influenza infected contacts were capable of acquiring pneumococcus from donors 3.5 meters away who were not infected with influenza and were not sneezing. Changes in respiratory secretions and increased dissemination could be one of the mechanisms by which influenza virus infection enhances transmission, however, since influenza caused significant sneezing and increased the quantity and thickness of secretions. This is more likely to have contributed to direct contact within the same cage and to transmission across short spaces where large droplet transmission appears possible.
Secondary bacterial disease in this model manifests as increased mucosal disease (otitis media and sinusitis), as well as increased pneumonia and invasive disease. Interestingly, this finding was pneumococcal strain specific. We have confirmed through whole genome microarray analysis that several ARs differ between the strains, with 4 of the 5 regions previously implicated in virulence [8,10] present in BHN97, which was superior in colonization and transmission compared to BHN54. These data should provide targets to explore the mechanisms underlying these differences in pathogenesis. Analysis of specific genes identified in STM studies [11–13] also revealed differences in the strains studied that could be related to relative disparities in virulence. However, these analyses are limited because there are likely virulence factors that are specifically important in the context of post-influenza pneumonia that cannot be identified through screens in the absence of influenza.
A final, interesting question is that suggested by studies from Gray et al. [15,16], does pneumococcal disease typically occur shortly after acquisition of a new strain, prior to development of type-specific immunity? Or can disease in colonized individuals occur at any time, independent of duration of carriage? Henderson et al. argued that timing of acquisition did not matter for subsequent development of otitis media, but excluded all cases where an intercurrent viral infection was suspected [17]. Syrjanen et al. examined this issue in more detail and found that acute otitis media was most common shortly after acquisition of a new serotype, particularly if a viral infection was present at the time of presentation [18]. If respiratory viruses such as influenza virus enhance acquisition of new strains, and also increase the chances of disease, then understanding the interaction between viruses and S. pneumoniae, particularly understanding serotype-specific differences in the ability to benefit by viral co-infection, may be critical information for prevention of disease. If the competing concept is true, that invasion and disease can occur at any time following acquisition, perhaps aided by a super-imposed viral infection, then length of carriage is a more important concept. In these studies, it is interesting to note that otitis media does occur in mice colonized with pneumococcus which are subsequently challenged with influenza virus [19], but pneumonia or invasive disease are not seen in either mice or ferrets in this scenario. These data favor the Gray hypothesis and argue that point prevalence studies of carriage, although easier to perform, are not as useful as longitudinal studies that can determine timing of acquisition.
Acknowledgments
The work described here was supported by PHS grant AI-66349 and ALSAC (JAM) and the Swedish Research council and Torsten and Ragnar Söderbergs foundation (BHN).
Footnotes
The authors declare no conflicts of interest exist.
References
- 1.Bridy-Pappas AE, Margolis MB, Center KJ, Isaacman DJ. Streptococcus pneumoniae: description of the pathogen, disease epidemiology, treatment, and prevention. Pharmacotherapy. 2005;25(9):1193–212. doi: 10.1592/phco.2005.25.9.1193. [DOI] [PubMed] [Google Scholar]
- 2.De Wals P, Erickson L, Poirier B, Pepin J, Pichichero ME. How to compare the efficacy of conjugate vaccines to prevent acute otitis media? Vaccine. 2009;27(21):2877–83. doi: 10.1016/j.vaccine.2009.02.102. [DOI] [PubMed] [Google Scholar]
- 3.McCullers JA. Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev. 2006;19(3):571–82. doi: 10.1128/CMR.00058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peltola VT, McCullers JA. Respiratory viruses predisposing to bacterial infections: role of neuraminidase. Pediatr Infect Dis J. 2004;23(1 Suppl):S87–S97. doi: 10.1097/01.inf.0000108197.81270.35. [DOI] [PubMed] [Google Scholar]
- 5.Finland M. Recent advances in the epidemiology of pneumococcal infections. Medicine (Baltimore) 1942;21(3):307–44. [Google Scholar]
- 6.Gwaltney JMJ, Sande MA, Austrian R, Hendley JO. Spread of Streptococcus pneumoniae in families. II. Relation of transfer of S. pneumoniae to incidence of colds and serum antibody. J Infect Dis. 1975;132(1):62–8. doi: 10.1093/infdis/132.1.62. [DOI] [PubMed] [Google Scholar]
- 7.Sandgren A, Sjostrom K, Olsson-Liljequist B, et al. Effect of clonal and serotype-specific properties on the invasive capacity of Streptococcus pneumoniae. J Infect Dis. 2004;189(5):785–96. doi: 10.1086/381686. [DOI] [PubMed] [Google Scholar]
- 8.Sandgren A, Albiger B, Orihuela CJ, Tuomanen E, Normark S, Henriques-Normark B. Virulence in mice of pneumococcal clonal types with known invasive disease potential in humans. J Infect Dis. 2005;192(5):791–800. doi: 10.1086/432513. [DOI] [PubMed] [Google Scholar]
- 9.Peltola VT, Boyd KL, McAuley JL, Rehg JE, McCullers JA. Bacterial sinusitis and otitis media following influenza virus infection in ferrets. Infect Immun. 2006;74(5):2562–7. doi: 10.1128/IAI.74.5.2562-2567.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blomberg C, Dagerhamn J, Dahlberg S, et al. Pattern of accessory regions and invasive disease potential in Streptococcus pneumoniae. J Infect Dis. 2009;199(7):1032–42. doi: 10.1086/597205. [DOI] [PubMed] [Google Scholar]
- 11.Hava DL, Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol. 2002;45(5):1389–406. [PMC free article] [PubMed] [Google Scholar]
- 12.Lau GW, Haataja S, Lonetto M, et al. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol Microbiol. 2001;40(3):555–71. doi: 10.1046/j.1365-2958.2001.02335.x. [DOI] [PubMed] [Google Scholar]
- 13.Polissi A, Pontiggia A, Feger G, et al. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect Immun. 1998;66(12):5620–9. doi: 10.1128/iai.66.12.5620-5629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regev-Yochay G, Raz M, Dagan R, et al. Nasopharyngeal carriage of Streptococcus pneumoniae by adults and children in community and family settings. Clin Infect Dis. 2004;38(5):632–9. doi: 10.1086/381547. [DOI] [PubMed] [Google Scholar]
- 15.Gray BM, Converse GM, 3d, Dillon HC., Jr Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J Infect Dis. 1980;142(6):923–33. doi: 10.1093/infdis/142.6.923. [DOI] [PubMed] [Google Scholar]
- 16.Gray BM, Dillon HCJ. Clinical and epidemiologic studies of pneumococcal infection in children. Pediatr Infect Dis. 1986;5(2):201–7. doi: 10.1097/00006454-198603000-00009. [DOI] [PubMed] [Google Scholar]
- 17.Henderson FW, Collier AM, Sanyal MA, et al. A longitudinal study of respiratory viruses and bacteria in the etiology of acute otitis media with effusion. N Engl J Med. 1982;306(23):1377–83. doi: 10.1056/NEJM198206103062301. [DOI] [PubMed] [Google Scholar]
- 18.Syrjanen RK, Auranen KJ, Leino TM, Kilpi TM, Makela PH. Pneumococcal acute otitis media in relation to pneumococcal nasopharyngeal carriage. Pediatr Infect Dis J. 2005;24(9):801–6. doi: 10.1097/01.inf.0000178072.83531.4f. [DOI] [PubMed] [Google Scholar]
- 19.McCullers JA, Karlstrom A, Iverson AR, Loeffler JM, Fischetti VA. Novel strategy to prevent otitis media caused by colonizing Streptococcus pneumoniae. PLoS Pathog. 2007;3(3):e28. doi: 10.1371/journal.ppat.0030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith MW, Schmidt JE, Rehg JE, Orihuela C, McCullers JA. Induction of pro- and anti-inflammatory molecules in a mouse model of pneumococcal pneumonia following influenza. Comp Med. 2007;57(1):82–9. [PMC free article] [PubMed] [Google Scholar]
- 21.Francis KP, Yu J, Bellinger-Kawahara C, et al. Visualizing pneumococcal infections in the lungs of live mice using bioluminescent Streptococcus pneumoniae transformed with a novel gram-positive lux transposon. Infect Immun. 2001;69(5):3350–8. doi: 10.1128/IAI.69.5.3350-3358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McAuley JL, Hornung F, Boyd KL, et al. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe. 2007;2(4):240–9. doi: 10.1016/j.chom.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCullers JA, Rehg JE. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J Infect Dis. 2002;186(3):341–50. doi: 10.1086/341462. [DOI] [PubMed] [Google Scholar]
- 24.McCullers JA, Bartmess KC. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J Infect Dis. 2003;187(6):1000–9. doi: 10.1086/368163. [DOI] [PubMed] [Google Scholar]
- 25.Sjostrom K, Blomberg C, Fernebro J, et al. Clonal success of piliated penicillin nonsusceptible pneumococci. Proc Natl Acad Sci U S A. 2007;104(31):12907–12. doi: 10.1073/pnas.0705589104. [DOI] [PMC free article] [PubMed] [Google Scholar]