Abstract
Quantitative trait locus (QTL) analysis of genetic crosses has proven to be a useful tool for identifying loci associated with specific phenotypes and for dissecting genetic components of complex traits. Inclusion of a mutation that interacts epistatically with QTLs in genetic crosses is a unique and potentially powerful method of revealing the function of novel genes and pathways. Although we know that a mutation within the novel tub gene leads to obesity and cochlear and retinal degeneration, the biological function of the gene and the mechanism by which it induces its phenotypes are not known. In the current study, a QTL analysis for auditory brainstem response (ABR) thresholds, which indicates hearing ability, was performed in tubby mice from F2 intercrosses between C57BL/6J-tub/tub and AKR/J-+/+ F1, hybrids (AKR intercross) and between C57BL/6J-tub/tub and CAST/Ei.B6-tub/tub F1 hybrids (CAST intercross). A major QTL, designated as modifier of tubby hearing1 (moth1), was identified on chromosome 2 with a LOD score of 33.4 (P <; 10−33) in the AKR intercross (181 mice) and of 6.0 (P <; 10−6) in the CAST intercross (46 mice). This QTL is responsible for 57 and 43% of ABR threshold variance, respectively, in each strain combination. In addition, a C57BL/6J congenic line carrying a 129/Ola segment encompassing the described QTL region when made homozygous for tubby also exhibits normal hearing ability. We hypothesize that C57BL/6J carries a recessive mutation of the moth1 gene which interacts with the tub mutation to cause hearing loss in tub/tub mice. A moth1 allele from either AKR/J, CAST/Ei or 129/Ola is sufficient to protect C57BL/6J-tub/tub mice from hearing loss.
INTRODUCTION
Tubby mice have an autosomal recessive mutation which results in maturity onset obesity associated with insulin resistance and retinal and cochlear degeneration. The biological function of tub and the mechanisms by which it induces these phenotypes are still unclear. While obesity is not observed until 12–18 weeks of age, abnormal electroretinogram and auditory brainstem response (ABR) are observed by 3 weeks of age (1). Histological features of the sensorineural alterations are well studied (1,2). The retinal degeneration is characterized by progressive apoptotic loss of photoreceptor cells within the outer nuclear layer and the cochlear degeneration by loss of outer hair cells and afferent neurons (1 – 3).
Since the identification of the mutation as a splicing defect in a novel gene (4,5), we have determined that tubby is a member of a new gene family (6,7). The N-terminus of the four genes within this family (tubby and the tubby-like proteins Tulp1, Tulp2 and Tulp3) is divergent whereas the C-terminus is highly conserved, suggesting that the C-terminus confers a common function to the family members. These genes, however, must have also evolutionarily acquired some unique functions and lost a part of some common function because Tulp1, Tulp2 or Tulp3 are unable to compensate for the tub mutation in tubby mice. In part, this divergence may have resulted in cell-specific expression (3). In addition to the mutation in the mouse tub gene, mutations within the human TULP1 gene (8,9) have also been implicated in retinal degeneration, suggesting that these genes may share a similar function necessary for retinal cell survival. To understand the causes and the variability observed in the mutant phenotypes, it is important to investigate the function of the tubby gene family members.
Identification of genetic modifiers to reveal novel gene interactions is a well-established and powerful methodology in bacteria, yeast, nematodes and Drosophila research. Since the establishment of quantitative trait locus (QTL) analysis (10), the identification of such modifiers in mice has become practical and shown to be an extremely useful tool (discussed in ref. 11). In addition, with the development of genetic resources such as single-strand length polymorphism (SSLP) markers, expressed sequence tags (ESTs) and large insert genomic libraries, identification of genes which are responsible for QTLs is feasible. In the current study, QTL analysis is used to map genes affecting hearing in tub/tub mice. These genes are necessary for the generation of the tub phenotype and. therefore, may mediate its normal function. Identification of such genes will help to elucidate the pathways through which TUB functions.
RESULTS
Auditory brainstem response (ABR) analysis was performed to estimate the hearing ability of 6- to 8-week-old mice.
AKR intercross
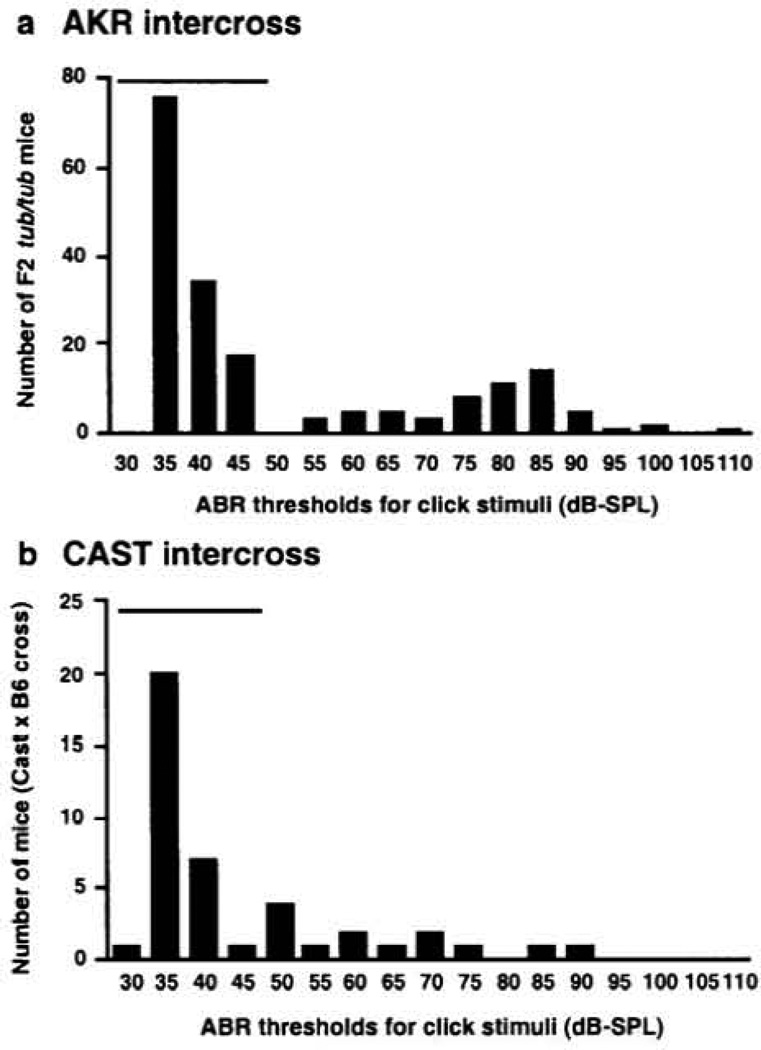
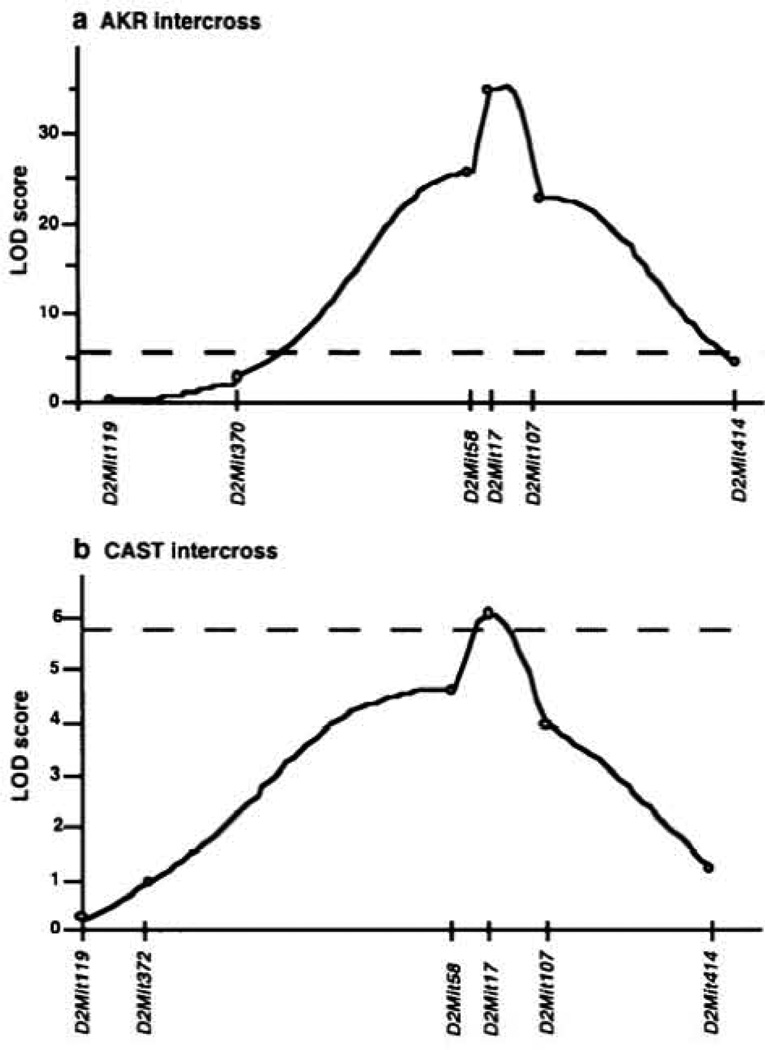
A total of 181 F2 mice homozygous for tubby were tested in the AKR/J × C57BL/6J-tub/tub intercross (AKR intercross). A bimodal distribution was observed for ABR thresholds for all four test stimuli; results for the broadband click stimulus are shown in Figure 1. Thirty-two percent of tub/tub mice showed abnormal click ABR thresholds (55 db SPL and higher), indicating the existence of one major recessive gene, designated as moth, which affected hearing ability in tubby mice (25% would be expected for a single recessive gene). In contrast, 7-week-old Fl tub/+ and F2 +/+ mice did not show an elevation or a large variability in ABR thresholds (36.3 ± 3 and 36.7 ± 3 dB SPL, respectively, ranging from 35 to 40 dB SPL) in comparison with the values observed for inbred AKR/J, C57BL/6J and CAST/Ei mice at 22–33 weeks (42 ± 3, 39 ± 4 and 30 ± 0 dB SPL, respectively) (12). The large variation in ABR thresholds is observed only in mice that arc homozygous for the tubby mutation, suggesting the existence of genetic factors specifically interacting with the tubby mutation. All mice that were phenotype by ABR were also genotyped using 57 markers which were distributed across the entire genome. Additional markers were used to define the region encompassing the significantly linked QTL on chromosome 2. The results of the linkage analyses are shown in Table 1. In the AKR intercross, marker D2Mit17 on chromosome 2 was highly associated with click ABR thresholds (P < 4.22 × 10−34, LOD = 33.4) (Fig. 2a and Table 1). The AKR/J allele of this QTL normalized ABR thresholds in tubby mice in a dominant fashion; a single copy prevented hearing loss (Table 1). This QTL is responsible for 57% of the click threshold variance observed in this intercross. Significant linkage to chromosome 2 was also observed with the other stimuli tested (8, 16 and 32 kHz). However, the LOD scores were lower than observed for the click stimulus (Table 1).
Figure 1.
Frequency distribution of click ABR threshold values from F2 mice homozygous for tubby. (a) F2 mice from the AKR/J × C57BL/6J-tub/tub intercross. (b) F2 mice from the CAST.B6-tub/tub × C57BL/6J-tub/tub intercross. The horizontal bar indicates the range of click ABR thresholds observed in mice from the parental strains C57BL/6J, AKR/J and CAST/Ei tested at the same age.
Table 1.
A major QTL and genotypic effect at D2Mit17 in the AKR intercross
| Sound stimuli | Genotype at locusa | ABR threshold (db SPL)b | LOD score | Variance explained (%) |
|---|---|---|---|---|
| Click | AA | 41.2 ± 2.15b (47) | 33.4 | 57 |
| AB | 41.1 ± 1.19 (89) | |||
| BB | 76.2 ± 2.35 (45) | |||
| 8 kHz | AA | 27.3 ± 1.98 (47) | 23.7 | 45 |
| AB | 26.2 ± 1.00 (89) | |||
| BB | 50.9 ± 2.06 (45) | |||
| 16 kHz | AA | 25.2 ± 2.40 (47) | 17.8 | 36 |
| AB | 24.0 ± 1.64 (89) | |||
| BB | 50.6 ± 2.08 (45) | |||
| 32 kHz | AA | 48.2 ± 1.98 (47) | 24.4 | 46 |
| AB | 46.2 ± 1.20 (89) | |||
| BB | 73.4 ± 2.09 (45) |
AA, homozygous for AKR/J; AB, heterozygous; BB, homozygous for C57BL/6J alleles at D2Mit17.
Mean ± SEM (number of mice in parentheses).
Figure 2.
LOD score distributions for associations of click ABR threshhold with loci on chromosome 2 in the AKR intercross (a) and in the CAST intercross (b). The dotted line represents highly significant linkage as assessed by permutation testing (P < 0.001).
CAST intercross
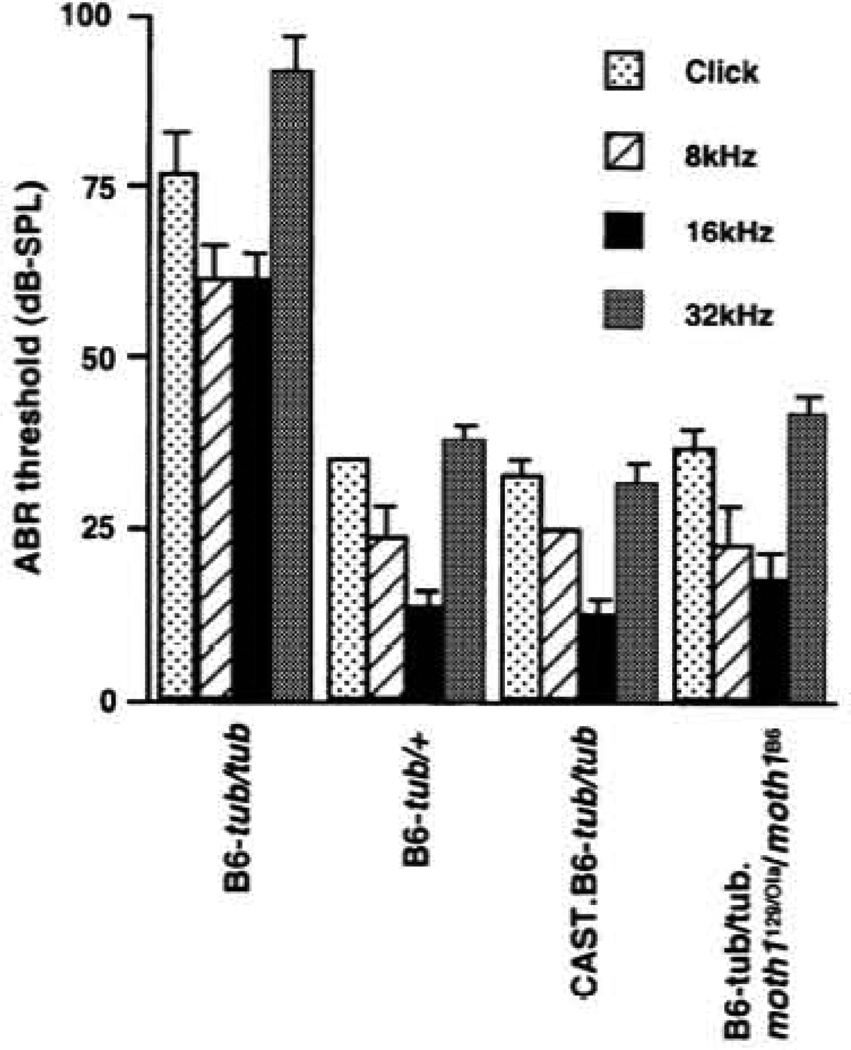
Congenic CAST/Ei.B6-tub/tub mice, generated in our laboratory, showed normal ABR thresholds (Fig. 3), suggesting that CAST/Ei, like AKR/J, had alleles of genes which protect tubby mice from hearing loss. To test whether CAST/Ei carried a protective allele of the moth1 gene on chromosome 2, QTL analysis for ABR thresholds was performed using 46 F2 mice homozygous for tub from a CAST/Ei.B6-tub/tub × C57BL/6J-tub/tub F1 hybrid intercross (CAST intercross). A similar pattern of segregation was observed in the CAST intercross as in the AKR intercross (Fig. I b). Although a smaller number of mice was tested, a highly significant LOD score of 6.0 at D2Mit17 was observed for click thresholds (Fig. 2b and Table 2). The LOD score distribution for click ABR thresholds in the CAST intercross is shown in Figure 2b. This QTL is responsible for 47% of click threshold variance (Table 2). A similar stimulus-specific effect was also observed in the CAST intercross as seen in the AKR intercross (Table 2).
Figure 3.
The ABR thresholds in C57BL/6J-tub/tub.moth1129/Ola/moth1B6 and CAST/Ei.B6-tub/tub mice. The mean ± SD of the sound pressure threshold is represented by a bar and shown for each genotype and stimulus as indicated. Five to six mice at 6–8 weeks of age were tested for each column. C57BL/6J-tub/tub and C57BL/6J-tub/+ strains were used as an abnormal and normal control, respectively. C57BL/6J-tub/tub.moth1129/Ola/moth1B6 and CAST/Ei.B6-tub/tub mice were generated as described in Materials and Methods.
Table 2.
A major QTL and genotypic effect at D2Mit17 in the CAST intercross
| Sound stimuli | Genotype at locusa | ABR threshold (db SPL)b | LOD score | Variance explained (%) |
|---|---|---|---|---|
| Click | CC | 35.4 ± 0.89 (13) | 6.0 | 43 |
| CB | 38.8 ± 2.08 (20) | |||
| BB | 59.2 ± 4.90 (13) | |||
| 8 kHz | CC | 25.0 ± 1.70 (13) | 3.5 | 26 |
| CB | 24.8 ± 1.90 (20) | |||
| BB | 37.3 ± 3.23 (13) | |||
| 16 kHz | CC | 16.7 ± 3.06 (13) | 2.0 | 14 |
| CB | 16.8 ± 2.03 (20) | |||
| BB | 34.3 ± 7.75 (13) | |||
| 32 kHz | CC | 40.0 ± 1.96 (13) | 5.1 | 37 |
| CB | 42.3 ± 2.42 (20) | |||
| BB | 61.9 ± 4.50 (13) |
CC, homozygous for CAST/Ei; CB, heterozygous; BB, homozygous for C57BL/6J alleles at D2Mil17.
Mean ± SEM (number of mice in parentheses).
129/Ola allele of moth1
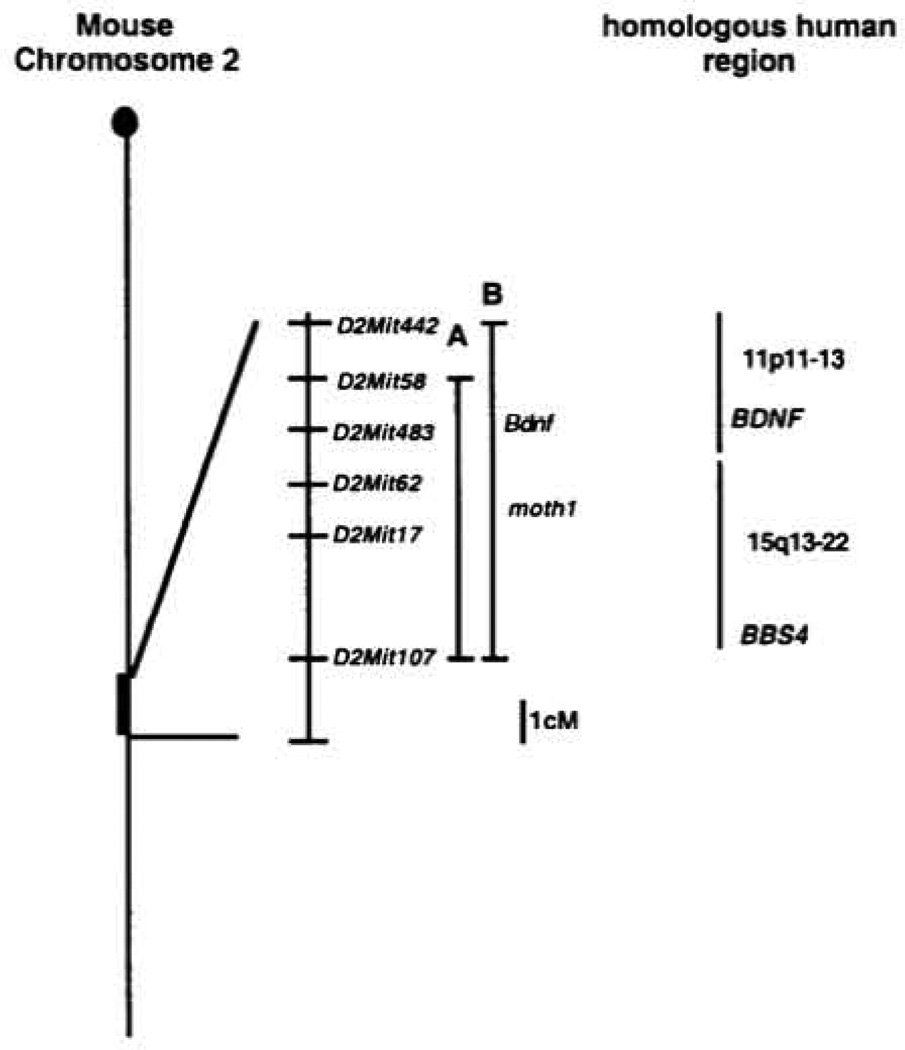
To investigate other strain-specific alleles of moth1 mice from a C57BL/6J stock (C57BL/6J.129-B2mTml/unc congenic for a 129/Ola-derived donor segment including moth1 were crossed to C57BL/6J-tub/tub to produce F1 hybrids. Mice homozygous for tubby and heterozygous for moth1129/Ola from an intercross of the F1 mice were phenotyped by ABR analysis. The 129/Ola congenic segment on chromosome 2 is ~11 cM in length, extending from D2Mit442 to D2Mit107 (Fig. 4). This region covers the majority of the 95% confidence interval identified for the click QTL in the AKR intercross (Fig. 4). The C57BL/6J-tub/tub.moth1129/Ola /moth1B6 mice showed normal ABR thresholds for all sound stimulations: click and 8, 16 and 32 kHz (Fig. 3).
Figure 4.
Map position of the moth1 gene and the syntenic human genetic regions. Bar A represents the confidence intervel including the moth1 gene. Bar B represents the 129/Ola segment which the C57BL/6J.129/Ola congenic line carries.
Immunohistochemistry
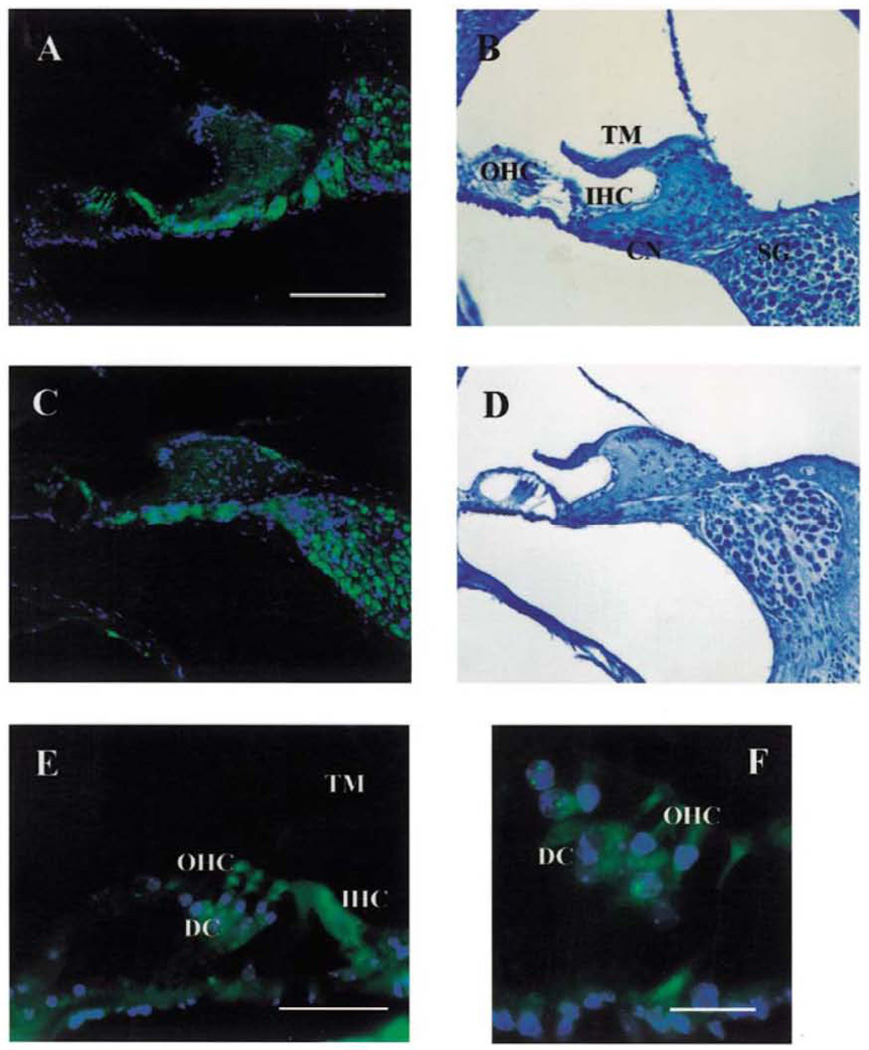
To determine whether the TUB protein is indeed expressed in the ear and to identify the primary site of action of the tubby gene, immunohistochemical studies were performed. A polyclonal tubby antibody that specifically recognizes the N-terminal end of both normal and mutant TUB protein was used (3). TUB is localized in the outer and inner hair cells and spiral ganglion cells, as well as in the supporting cells of Deiters adjacent to the outer hair cells. A characteristic subcellular staining pattern could be detected, with a predominant cytoplasmic staining in outer and inner hair cells and in spiral ganglion cells and an intense nucleolar staining in the external phalangeal cells of Deiters (Fig. 5). Localization patterns of anti-TUB antibodies were the same in both C57BL6/J and CS7BL/6J-tub/rub mice. We could not find any significant morphological differences between the inner ear of normal and tub/tub mice at 8–10 weeks of age.
Figure 5.
Distribution of tubby immunoreactivity (A, C, E and F) and light microscopic morphology (B and D) of the cochlea of C57BL/6J (wt) and C57BL/6J-tub/tub (tub) mice at 10 weeks of age. The TUB protein is strongly expressed (green) in the outer and inner hair cells (OHC/IHC), the adjacent external phalangeal cells of Deiters (DC), cochlear nerve fibers (CN) and in the spiral ganglion cells (SC). Note that there are no obvious differences in distribution or expression of the tubby protein between normal and mutant mice (A and C). At this 8–10 weeks time point, no histopathological changes in the organ of Corti could be detected (B and D). Higher magnifications of representative sections of the Corti organ of C57BL/6J mice reveal the subcellular distribution of TUB. A cytoplasmic localization pattern can be observed in the outer and inner hair cells, depicted by arrowheads (E and F), whereas there is a distinct nucleolar immunoreactivity detectable in the external phalangeal cells (marked by arrows in F). TM, tectorial membrane, Scale bars: (A–D) 100 µm; (E) 5 µm; (F) 20 µm.
DISCUSSION
Genetic identification of moth1 on chromosome 2, a major gene affecting hearing loss in tubby mice
In both the AKR and the CAST intercrosses, the same chromosomal locus was found to be linked to ABR thresholds by QTL analysis. The AKR/J and CAST/Ei alleles at this locus were able to protect tub mice from hearing loss. In addition, by congenic strain analysis we show that the 129/Ola al1ele of this locus also acts as a dominant protective allele. This gene, named moth1 for modifier of tub hearing, may be directly involved in the hearing impairment cascade induced by the tub mutation. It is known that cochlear neuroepithelial degeneration, characterized by loss of hair and ganglion cells, is one possible cause of hearing loss in tubby mice (1,2). Although the function of TUB is still unknown, the tubby mutation is known to induce cell death in sensory neurons (1,3). Because a single moth1 allele from the genetically unrelated strains AKR/J, CAST/Ei and 129/Ola is able to protect C57BL/6J-tub/tub mice against hearing loss, it is likely that B6 carries a recessive mutation at the moth1 locus which is necessary to mediate the effects of the tub mutation on hearing. Because homozygosity for the moth1 allele itself cannot induce bearing impairment in C57BL/6J mice, the allelic difference in the moth1 gene between C57BL/6J and the other inbred strains used in this study may not be critical for maintaining the hair and spiral ganglion cells, but becomes significant only when the tubby gene is mutated and unable to function properly. A similar epistatic gene interaction has been described for the dfw and mdfw genes (13).
C57BL/6J mice carry the susceptibility allele (ahl) for late onset of hearing loss, which has been mapped to chromosome 10 (14). In the present study, all mice were tested at 6–8 weeks of age, well before the late onset of hearing loss attributable to ahl. It appears that the tub mutation is not interacting with the 66 ahl allele to accelerate the ahl-induced hearing loss, because we did not observe a QTL affecting hearing ability on chromosome 10. Thus, the abnormality in hearing observed in both crosses is not due to ahl and can be explained by the effect of the tubby mutation without the protective allele of the moth1 gene.
Although in the AKR intercross the most significant linkage for A6R thresholds was observed on chromosome 2 over all sound stimulations (click and 8, 16 and 32 kHz), a difference in the level of significance between each stimulus was observed. A similar pattern was observed in the CAST intercross as well. The lowest LOD scores were obtained with thresholds evoked from the 16 kHz stimulus; mice are most sensitive to frequencies of 12–24 kHz (15). It is also known that different strains exhibit different sensitivities to these particular stimuli (12), suggesting that there might be a genetic basis for the frequency-dependent sensitivities.
TUB protein is expressed in the cochlea
TUB protein was found to be expressed in the outer and inner hair cells and the spiral ganglion cells, regions which degenerate in tubby mice (1,2). In tubby mice, retinal degeneration has been shown to be caused by apoptosis (1,3). It is possible that the delayed cochlear degeneration may also occur via the same mechanism. However, while tubby mice show broadband hearing impairment at 3 weeks of age (1), cochlear degeneration is not observed (2). In this study, we also did not observe significant degeneration of the cells in the organ of Corti at 8–10 weeks of age. The cells may, therefore, lose their function prior to showing signs of degeneration.
A candidate gene for modifying tubby hearing
Genes expressed in neurons and associated with cell survival could be candidates for moth1. Brain-derived neurotrophic factor (BDNF), a gene known to play a role in neural cell survival (reviewed in ref. 16), is located on chromosome 2 at 62 cM (MGD), well within the 7.86 ± 1.39 cM region identified for moth1. Furthermore, it is known that BDNF and its high affinity receptor trkB are highly expressed in exactly the same subset of cells positive for the TUB protein (reviewed in ref. 17). This information, together with the fact that mice homozygous for a null allele for BDNF show sensory neural defects (18), suggested it as a good candidate gene for moth1. Sequence comparison of the BDNF coding region, however, showed no differences between C57BL6-tub/tub and AKR/J, thus making it less likely that BDNF is the moth1 gene. Potential expression level differences will have to be explored using congenic lines that are currently being generated.
Chromosomal localization of moth1
The phenotypes of tubby mice resemble human diseases such as Alström syndrome and Bardet–Biedl syndromes, which are characterized by obesity, blindness and deafness. Interestingly, moth1 maps to the region of mouse chromosome 2 which is homologous to the human chromosome 15q13–22, where one of the Bardet–Biedl genes (BBS4) maps (human chromosome 15q22.3–23) (19,20). Although only the hearing phenotype was characterized in the experiments described here, it would be interesting to see how the moth1 gene affects the other tubby phenotypes, such as retinal degeneration and obesity.
Further studies are ongoing to identify the moth1 gene. Identification of a gene which interacts with tubby may allow us to elucidate the function of tubby and, thereby, identify pathways that significantly correlate with the tubby phenotypes ill vivo.
MATERIALS AND METHODS
Congenic mice and crosses
All mice used in this study were originally obtained from the Jackson Laboratory. To generate the congenic strain CAST/Ei.B6-tub/tub, tub/+ mice originally in the C57BL/6J background were sequentially backcrossed to strain CAST/Ei for 10 generations and intercrossed. The congenic strain B6.129-B2mTMl/Unc/Dcr (N11) has been previously described (21). In its ~ 11 cM congenic 129/Ola-derived segment, it carries a null allele of the gene β2-microglobulin (B2m) (which only expresses a detectable physiological phenotype when homozygous) and 129/Ola-derived markers including D2Mit58 and D2Mit107. These mice were out-crossed to C57BL/6J-tub/tub and progeny were intercrossed to produce F2 mice homozygous for tub but heterozygous for the 129/Ola congenic segment. An allele-specific PCR assay (22) was used to identify tub/tub mice and markers D2Mit58 and D2Mit107 were used to distinguish between the 129/Ola and C57BL/6J alleles of moth1. For the AKR intercross, Fl progeny from a breeding pair of C57BL/6J-tub/tub and AKR/J-+/+ mice were intercrossed to produce F2, progeny. The F2 mice homozygous for tub were selected by the allele-specific PCR assay as described above. For the CAST intercross, F1 progeny from a breeding pair of C57BL/6J-tub/tub and CAST/Ei.B6-tub/tub mice were intercrossed to produce F2 progeny. Phenotyping and the genome-wide scan of these F2 progeny were carried out as described below.
ABR phenotyping
A computer-aided evoked potential system (Intelligent Hearing System, IHS; Miami, FL) was used to test mice for ABR thresholds as previously described (12). Mice were anesthetized with tribromoethanol (5.3 mg/10 g body wt i.p.). Subdermal needle electrodes were inserted at the vertex (active) and ventrolaterally to the right ear (reference) and to the left ear (ground). Specific acoustic stimuli were delivered binaurally through 1 cm plastic tubes channeled from high frequency transducers. Mice were tested with click stimuli and also with 8, 16 and 32 kHz tone pips at varying intensity, from low to high (10–90 dB SPL). An ABR threshold was determined for each stimulus frequency by identifying the lowest intensity which produced a recognizable ABR pattern (at least two consistent peaks) (12).
Genotyping
Tail DNA was isolated according to Buffone (23). SSLP markers polymorphic between strains AKR/J and C57BL/6J or CAST/Ei and C57BL/6J were selected at ~35 cM intervals across the mouse genome. A total of 57 SSLP markers were tested for the initial genome-wide screen. Additional markers were tested around loci that showed significant or suggestive linkage. For PCR amplification, 100 ng of DNA were used in a 10 µl volume containing 50 mM KCl, 10 mM Tris–HCl, pH 8.3, 2.5 mM MgCl2, 0.2 mM oligonucleotides, 200 µM dNTP and 0.02 U AmpliTaq DNA polymerase. The reactions were subjected to the following temperature cycling program: initial denaturation for 2 min at 95°C; 20 s at 94°C, 30 s at 50°C, 40 s at 72°C for 49 cycles: followed by a 7 min extension at 72°C. PCR products were separated by electrophoresis on a 4% MetaPhor (FMC, Rockland, ME) agarose gel and visualized under UV light after staining with ethidium bromide.
Light microscopy and immunofluorescence analysis
C57BL/6J (n = 4) and C57BL/6J-tub/tub (n = 4) mice at 8–10 weeks of age were deeply anesthetized and transcardially perfused with phosphate-buffered saline (PBS) followed by 4% parafonnaldehyde (PFA) in PBS. The cochlea was removed and stored in 4% PFA for 4 h. For decalcification, the cochlea was incubated in 7% EDTA–PBS for 4 days before embedding in paraffin. Serial sections (7 µm) of the cochlea were stained with Mayer’s hematoxylin using standard procedures. Adjacent sect ions were subjected to immunohistochemical analysis using rabbit polyclonal antibody specific for an N-terminal portion of the tubby protein (3). Briefly, after blocking with 2% goat serum in PBS, sections were incubated with the primary antibody (N2, 1:1000) at 4°C overnight. Binding was detected using biotinylated goat anti-rabbit IgG (1:200; Vector. Burlingame, CA) followed by FITC–avidin D (1:200; Vector) or Cy3-conjugated donkey anti-rabbit IgG (1:150; Jackson Immunoresearch, West Grove, CA). A nuclear counterstain was performed with 4,6-diamidine 2-phenylindoldihydrochloride (DAPI) at a final concentration of 5 µg/ml. Images were collected on a Leica DMRXE fluorescent microscope equipped with a SPOT CCD camera using appropriate bandpass filters for each fluorochrome.
QTL and statistical analysis
QTL analysis was performed using MapManager QT (v.3.0b26; Manly). LOD scores were calculated by dividing the F scores by 4.6 (24). To evaluate the significance of the results, a permutation test (25) with 1000 replications was used.
The data generated from the entire set of F2, progeny were also subjected to analysis of variance (ANOVA) between groups defined by genotype for selected genetic loci. Analysis of variance was performed using the Statview v4.5 program for the Macintosh Computer (Abacus Concepts, Berkeley, CA).
ACKNOWLEDGEMENTS
We thank Cindy S. Avery and Gloria York for mouse colony management and Gayle B. Collin for technical assistance. We are also grateful to Drs Wayne N. Frankel, Greg A. COX and Barbara B. Knowles for careful review of the manuscript. This work was supported by a grant from the Foundation for Fighting Blindness and from AXYS Pharmaceuticals Inc. Institutional shared services arc supported by National Cancer Institute Support grant CA-34196.
Contributor Information
Akihiro Ikeda, The Jackson Laboratory, 600 Main Street, Bar Harbor, ME 04609, USA.
Qing Yin Zheng, The Jackson Laboratory, 600 Main Street, Bar Harbor, ME 04609, USA.
Philip Rosenstiel, The Jackson Laboratory, 600 Main Street, Bar Harbor, ME 04609, USA.
Terry Maddatu, The Jackson Laboratory, 600 Main Street, Bar Harbor, ME 04609, USA.
Aamir R. Zuberi, The Jackson Laboratory, 600 Main Street, Bar Harbor, ME 04609, USA.
Derry C. Roopenian, The Jackson Laboratory, 600 Main Street, Bar Harbor, ME 04609, USA
Michael A. North, AXYS Pharmaceuticals Inc., La Jolla, CA 92037, USA
Jürgen K. Naggert, The Jackson Laboratory, 600 Main Street, Bar Harbor, ME 04609, USA
Kenneth R. Johnson, The Jackson Laboratory, 600 Main Street, Bar Harbor, ME 04609, USA
Patsy M. Nishina, The Jackson Laboratory, 600 Main Street, Bar Harbor, ME 04609, USA.
REFERENCES
- 1.Heckenlively JR, Chang B, Erway LC, Peng C, Hawes NL, Hageman Gs, Roderick TH. Mouse model for Usher syndrome: linkage mapping suggests homology to Usher type I reported at human chromosome 11 p15. Proc. Natl Acad. Sci. USA. 1995;92:11100–11104. doi: 10.1073/pnas.92.24.11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohlemiller KK, Hughes RM, Lett JM, Ogilvie JM, Speck JD, Wright JS, Faddis BT. Progression of cochlear and retinal degeneration in the tubby (rd5) mouse. Audiol. Neuro-Otol. 1997;2:175–185. doi: 10.1159/000259242. [DOI] [PubMed] [Google Scholar]
- 3.Ikeda S, He W, Ikeda A, Naggert JK, North MA, Nishina PM. Cell specific expression of tubby gene family members in the retina. Invest. Ophthalmol. Vis. Sci. 1999 in press. [PubMed] [Google Scholar]
- 4.Noben-Trauth K, Naggert JK, North MA, Nishina PM. A candidate gene for the mouse mutation tubby. Nature. 1996;380:534–538. doi: 10.1038/380534a0. [DOI] [PubMed] [Google Scholar]
- 5.Kleyn PW, Fan W, Kovats SG, Lee JJ, Pulido JC, Wu Y, Berkemeier LR, Misumi DJ, Holmgren L, Charlat O, Woolf EA, Tayber O, Brody T, Shu P, Hawkins F, Kennedy B, Baldini L, Ebeling C, Alperin GD, Deeds J, Lakey ND, Culpepper J, Chen H, Glücksmann-Kuis MA, Carlson GA, Duyk GM, Moore KJ. Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell. 1996;85:281–290. doi: 10.1016/s0092-8674(00)81104-6. [DOI] [PubMed] [Google Scholar]
- 6.North MA, Naggert JK, Yan Y, Noben-Trauth K, Nishina PM. Molecular characterization of TUB, TULP1, and TULP2, members of the novel tubby gene family and their possible relation to ocular diseases. Proc. Natl Acad. Sci. USA. 1997;94:3128–3133. doi: 10.1073/pnas.94.7.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishina PM, North MA, Ikeda A, Yan Y, Naggert JK. Molecular characterization of a novel tubby gene family member, TULP3, in mouse and human. Genomics. 1998;54:215–220. doi: 10.1006/geno.1998.5567. [DOI] [PubMed] [Google Scholar]
- 8.Hagstrom S, North M, Nishina PM, Berson E, Dryja T. Recessive mutations in the gene encoding the tubby-like protein, TULP1, in patients with retinitis pigmentosa. Nature Genet. 1998;18:174–176. doi: 10.1038/ng0298-174. [DOI] [PubMed] [Google Scholar]
- 9.Banerjee P, Kleyn PW, Knowls JA, Lewis CA, Ross BM, Parano E, Kovats SG, Lee JJ, Penchaszadeh GK, Ott J, Jacobson SG, Gilliam TC. TULP1 mutation in two extended Dominican kindreds with autosomal recessive retinitis pigmentosa. Nature Genet. 1998;18:177–179. doi: 10.1038/ng0298-177. [DOI] [PubMed] [Google Scholar]
- 10.Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dietrich WF, Lander ES, Smith JS, Moser AR, Gould KA, Luongo C, Borenstein N, Dove W. Genetic identification of Mom-1, a major modifier locus affecting Min-induced intestinal neoplasia in the mouse. Cell. 1993;75:631–639. doi: 10.1016/0092-8674(93)90484-8. [DOI] [PubMed] [Google Scholar]
- 12.Zheng QY, Johnson KR, Erway LC. Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hearing Res. 1999;130:94–107. doi: 10.1016/s0378-5955(99)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noben-Trauth K, Zheng QY, Johnson KR, Nishina PM. mdfw: a deafness susceptibility locus that interacts with deaf waddler (dfw) Genomics. 1997;44:266–272. doi: 10.1006/geno.1997.4869. [DOI] [PubMed] [Google Scholar]
- 14.Johnson KR, Erway LC, Cook SA, Willott JF, Zheng QY. A major gene affecting age-related hearing loss in C57BL/6J mice. Hearing Res. 1997;114:83–92. doi: 10.1016/s0378-5955(97)00155-x. [DOI] [PubMed] [Google Scholar]
- 15.Ehret G. Psychoacoustics. In: Willot JF, editor. Auditory Psychobiology of the Mouse. Springfield, IL: Charles C. Thomas; 1983. pp. 13–53. [Google Scholar]
- 16.Sinder WD, Johnson EM. Neurotrophic molecules. Ann. Neurol. 1989;26:489–506. doi: 10.1002/ana.410260402. [DOI] [PubMed] [Google Scholar]
- 17.Fritzsch B, Silos-Santiago I, Bianchi LM, Farinas I. The role of neurotrophic factors in regulating the development of inner ear innervation. Trends Neurosci. 1997;20:159–164. doi: 10.1016/s0166-2236(96)01007-7. [DOI] [PubMed] [Google Scholar]
- 18.Ernfors P, Lee K-F, Jaenish R. Mice lacking brain-derived neurotrophic factor develop with sensory deficits. Nature. 1994;368:147–150. doi: 10.1038/368147a0. [DOI] [PubMed] [Google Scholar]
- 19.Bruford EA, Riise R, Teague PW, Porter K, Thomson KL, Moore AT, Jay M, Warburg M, Schinzel A, Tommerup N, Tornqvist K, Rosenberg T, Patton M, Mansfield DC, Wright AF. Link-age mapping in 29 Bardet-Biedl syndrome families confirms loci in chromosomal regions 11ql3, 15q22.3–q23, and 16q21. Genomics. 1997;41:93–99. doi: 10.1006/geno.1997.4613. [DOI] [PubMed] [Google Scholar]
- 20.Carmi R, Rokhlina T, Kwitek-Black AE, Elbedour K, Nishimura D, Stone EM, Sheffield VC. Use of a DNA pooling strategy to identify a human obesity syndrome locus on chromosome 15. Hum. Mol. Genet. 1995;4:9–13. doi: 10.1093/hmg/4.1.9. [DOI] [PubMed] [Google Scholar]
- 21.Christianson GJ, Vekasi S, Niles J, Roopenian SL, Roths JB, Roopenian DC. b2 microglobulin-deficient mice show abnormal Ig homeostasis and are protected from lupus-like autoimmunity. J. Immunol. 1997;159:4780–4792. [PubMed] [Google Scholar]
- 22.Maddatu T, Naggert JK. Allele-specific PCR assays for the tub and cpe-fat mutations. Mamm. Genome. 1997;8:857–858. doi: 10.1007/s003359900594. [DOI] [PubMed] [Google Scholar]
- 23.Buffone GJ. Isolation of DNA from biological specimens without extraction with phenol. Clin. Chem. 1985;31:164–165. [PubMed] [Google Scholar]
- 24.Lander ES, Kruglyack L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nature Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 25.Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]