Abstract
Here, we provide a detailed description of proteomic, Western blot and RT-PCR analyses performed to examine fat biopsy samples from lean insulin-sensitive and obese insulin-resistant nondiabetic individuals for evidence of endoplasmic reticulum (ER) stress.
Subcutaneous fat biopsies were obtained from the upper thighs of six lean and six obese nondiabetic subjects. Fat homogenates were used for proteomic (two-dimensional gel (2DE) and MALDI-TOF/TOF), Western blot, and RT-PCR analysis.
Proteomic analysis revealed 19 differentially upregulated proteins in fat of obese subjects. Three of these proteins were the ER stress-related unfolded protein response (UPR) proteins calreticulin, protein disulfide-isomerase A3, and glutathione-S-transferase P; Western blotting revealed upregulation of several other UPR stress-related proteins, including calnexin, a membrane-bound chaperone, and phospho c-jun NH2-terminal kinase (JNK)-1, a downstream effector protein of ER stress; RT-PCR analysis revealed upregulation of the spliced form of X-box-binding protein-1s, a potent transcription factor and part of the proximal ER stress sensor inositol-requiring enzyme-1 pathway.
These findings demonstrate of UPR activation in subcutaneous adipose tissue of obese human subjects. As JNK can inhibit insulin action and activate proinflammatory pathways, ER stress activation of JNK may be a link between obesity, insulin resistance, and inflammation.
1. Introduction
Obesity is closely associated with insulin resistance and with a state of low-grade inflammation characterized by elevation of inflammatory cytokines in blood and tissues (Bray, 2004). Insulin resistance and inflammation contribute to the development of several disorders including type 2 diabetes, hypertension, atherogenic dyslipidemia, and abnormalities in blood coagulation and fibrinolysis, all of which are independent risk factors for athero-sclerotic vascular disease such as heart attacks, strokes, and peripheral arterial disease (Bray, 2004). Therefore, it has become important to understand why and how obesity is so tightly linked with insulin resistance and inflammation. On one hand, free fatty acids (FFA) have been established as important links between obesity, insulin resistance, and inflammation. Most obese people have elevated plasma FFA levels (Reaven et al., 1988) and elevated plasma FFA levels have been shown to cause insulin resistance and inflammation (Boden et al., 1994, 2001, 2005; Itani et al., 2002; Santomauro et al., 1999; Yu et al., 2002). On the other hand, not all of these insulin resistant subjects have elevated plasma FFA levels which means that there must be other causes for obesity-related insulin resistance and inflammation. One of these appears to be endoplasmic reticulum (ER) stress (Ozcan et al., 2004). This raises the question as to why there is ER stress in obesity and how obesity-associated ER stress can cause insulin resistance and inflammation. Excessive macronutrient intake is the main cause for obesity and several recent studies in rodents have implicated ER stress as an early sign of nutrient excess and a cause for the development of insulin resistance and inflammation. For instance, in mice fed high fat diets for 3 months, the chronic excessive macronutrient intake caused obesity and ER stress in adipose tissue and liver, whereas overexpression or administration of ER stress reducing chaperone proteins reduced ER stress, insulin resistance, and inflammation (Schroder and Kaufman, 2005). ER stress can lead to the development of insulin resistance and inflammation. Proposed mechanisms include ER stress-induced phosphorylation and activation of C-jun N-terminal kinase (JNK), and activation and nuclear translocation of nuclear factor κB (NFκB), which is a key promoter of inflammation (Zhang and Kaufman, 2008). In addition, ER stress is a major source for the production of reactive oxygen species (ROS) which can produce insulin resistance and inflammation (Schroder and Kaufman, 2005; Zhang and Kaufman, 2008). Thus, it has been proposed that the ER may be a proximal site that senses nutritional excess and translates that into signals producing insulin resistance and inflammatory responses (Zhang and Kaufman, 2008). Until very recently, however, all the evidence linking ER stress with obesity, insulin resistance, and inflammation were based on in vitro and animal data. We have recently shown, however, that there is ER stress in fat of obese people (Boden et al., 2008). In this chapter, details of this study are described.
2. Study Subjects and Fat Biopsies
2.1. Subjects
Six lean and six obese healthy volunteers were studied. None of the participants had a family history of diabetes or other endocrine disorders or were taking medications. Their body weights were stable for at least 2 months before the biopsies. Compared with the nonobese volunteers, the obese volunteers were heavier (93.4 vs. 77.4 kg; P < 0.03) and had more body fat 40.7 vs. 19.9 kg; P < 0.004) but had the same fat-free mass (57.6 vs. 57.6 kg) and were insulin resistant (1/homeostasis model assessment 0.44 vs. 0.29; P < 0.05). Informed written consent was obtained from all subjects after explanation of the nature, purpose, and potential risks of these studies. The study protocol was approved by the Institutional Review Board of Temple University Hospital.
2.2. Fat biopsies
The subjects were admitted to the Temple University Hospital Clinical Research Center on the day before the studies. At ~8 a.m. on the day after admission, a venous blood sample was obtained and an open fat biopsy was performed by a surgeon. Fat biopsies were obtained from the lateral aspect of the upper thigh (~15 cm above the patella) under local anesthesia, as described (Boden et al., 1994). The excised fat was dropped immediately into isopentane and kept at its freezing point (–160 °C) by liquid nitrogen.
The frozen fat was stored at –80 °C until analyzed.
3. Proteomic Analysis
3.1. Required materials
SYPRO® Ruby (Invitrogen, Carlsbad, California)
Milli-Q System (Millipore, Billerica, MA)
The following is from GE Healthcare, Piscataway, NJ:
IPGphor horizontal electrophoresis apparatus
IPGphor strip holders, IEF sample applicator strip
Immobiline DryPlate 4–7 and/or 3–10
IPG buffers (pH range 4–7 and/or 3–10)
IPG cover fluid
DeStreak reagent
The following is from Sigma–Aldrich, St. Louis, MO:
CHAPS
Urea
Thiourea
DTT
Iodoacetamide
3.2. Protein extraction protocol for two-dimensional electrophoresis
Place the amount of tissue sample to be processed in a precooled (–20 °C), clean mortar.
Freeze the tissue sample thoroughly by adding a small volume (2–5 mL) of liquid nitrogen.
Allow most of the liquid nitrogen to evaporate, leaving 1–2 mL.
Grind the tissue with a precooled (–20 °C) pestle until a fine powder is obtained.
Transfer the powder into a 1.5-mL tube and measure the wet weight. Add 0.5 mL of extraction buffer: 7 M urea,1 2 M thiourea, 4% CHAPS, 43 mM DTT,2 60 mM Tris/HCl (pH 8.8), and 0.1% SDS; additives: 1 × protease inhibitor cocktail2 and 0.2 mM EDTA.
Sonicate sample with five pulses of 10 s each. Add 2 mM MgCl2 and Benzonase (50 U per 100 μl of sample solution) mix and incubate at room temperature (RT) for 15 min with vortexing every 5 min.3
Spin solution at max speed (21,000×g) for 30 min at RT. Discard the pellet.
Mix protein extract (supernatant) with –20 °C cold acetone/0.1% DTT at 1:3 (v/v) ratio and store the mixture at –20 °C overnight.
Spin at 21,000×g for 30 min at 4 °C, discard the supernatant and resuspend the pellet with 1.2 mL 80% (–20 °C) cold acetone/0.2% DTT2 and spin (21,000×g/4 °C) for 5 min and discard the supernatant. Repeat wash.4
Air dry the pellet till no acetone is remaining and redissolved in DeStreak rehydration solution or two-dimensional electrophoresis (2DE) sample buffer: 7 M urea, 2 M thiourea, 4% CHAPS, 1.2% DeStreak, and 0.01% bromophenol blue. Shaking for 3 h at RT to solubilize.
Determine the protein concentration using 2D Quant Kit as per manufacturer's instructions (Amersham).
3.3. First dimension of 2DE
-
12
Mix sample to the final volume with DeStreak rehydration buffer supplemented with the corresponding immobilized pH gradient (IPG) buffer.
-
13
Rehydrate the IPG strips overnight at 22 °C in rehydration buffer: 7 M urea, 2 M thiourea, 4% CHAPS, 5% glycerol, 15% 2-propanol, 1.2% DeStreak reagent2, 0.5% IPG buffer 7–112, and 0.2% methylcellulose mixed with an appropriate sample volume.5
-
14Use Ethan IPGphor system (GE Health system) to perform isoelectric focusing (IEF). Program the settings to temperature of 20 °C and use the following settings.
Step T (°C) Start volt (V) End volt (V) Duration (h) Total (Vh) IEF protocol (7 cm) 1 20 0 250 0.5 2 20 250 1500 1 3 20 1500 3000 1 4 20 3000 8000 3 11,000
3.4. Second dimension of 2DE
Dissolve 100 mg of DTT in 10 mL of equilibration buffer 6 M urea, 30% (w/v) glycerol, and 2% (w/v) SDS in 0.05 M Tris–HCl buffer (pH 8.8). Take out the focused IPG gel strips from the freezer and place them into individual test tubes. Add 10 mL of equilibration buffer I and 50 μL of the bromophenol blue (0.25%, w/v) solution. Seal the test tubes with Parafilm, rock them 15 min on a shaker, and then pour off equilibration buffer I.
Dissolve 400 mg of iodoacetamide in 10 mL of equilibration buffer. Add equilibration buffer II and 50 μL of bromophenol blue solution to the test tube as above and equilibrate for another 15 min on a rocker.
After the second equilibration, rinse the IPG gel strip with deionized water for a second and place it on a piece of filter paper at one edge for a few minutes to drain off excess equilibration buffer.
Place IPG gel strip on top of the vertical SDS gel.
Fill the electrophoresis chamber with electrode buffer and turn on the SDS gel cassettes in a vertical position to facilitate the application of the first dimension IPG strips.
Equilibrate the IPG gel strips as described above for first dimension and immerse them in electrode buffer for a few seconds.
Place the IPG gel strip on top of an SDS gel. Carefully press the IPG strip with a spatula onto the surface of the SDS gel to achieve complete contact. Repeat this procedure for the remaining IPG strips.
Insert the gel cassettes in the electrophoresis apparatus and start electrophoresis.6
Run the SDS–PAGE gels in Bio-Rad Mini-PROTEAN® 3 System (Hercules, CA) at 200 V for 45 min.
Terminate the run when the bromophenol blue tracking dye has migrated off the lower end of the gel.
Open the cassettes carefully with a spatula. Peel the gel off the glass plate carefully, lifting it by the lower edge, and place it in a tray containing fixing solution or transfer buffer, respectively. Then continue with fixing, protein staining, or blotting.
3.5. Fluorescent staining with SYPRO® Ruby
After electrophoresis, place the gel into a clean container with 100 mL of fixing solution (50% methanol, 7% acetic acid) and agitate on an orbital shaker for 30 min. Repeat once more with fresh fixing solution. Pour off the used fixing solution.7 After fixing, perform three washes in ultrapure water for 10 min each, before proceeding to the staining step.
Add 60 mL of SYPRO® Ruby gel stain. Agitate on an orbital shaker overnight.
Transfer the gel to a clean container and wash in 100 mL of wash solution (10% methanol, 7% acetic acid) for 30 min. The transfer step helps minimize background staining irregularities and stain speckles on the gel. Before imaging rinse the gel in ultrapure water a minimum of two times for 5 min to prevent possible corrosive damage to the imager.
SYPRO® Ruby protein gel stain has two excitation maxima, one at ~280 nm and one at ~450 nm, and has an emission maximum near 610 nm. Proteins stained with the dye can be visualized using a 300 nm UV transilluminator, a blue-light transilluminator, or a laser scanner. The stain has exceptional photostability allowing long exposure times for maximum sensitivity.
3.6. 2DE image analysis
Perform 2DE analysis by PDQuest software (Bio-Rad), version 8.0.
Enumerate and analyze each gel from obese and lean fat samples for spot detection, background subtraction, and protein spot volume quantification.
Make manual corrections to validate the matches automatically generated by the software.
Normalize spot volume values in each gel by dividing the raw quantity of each spot by the total volume of all spots included in the same gel.
Determine SD for each protein spot and the average spot volume values.
Perform Student's t-test on the match spots in order to determine the spots that were differentially expressed.
Choose spots that show a statistically significant difference with a confidence level of 0.05.
To test for inter gel reproducibility, perform 2DE analysis in triplicate using one representative sample.
3.7. In-gel digestion
Wearing gloves and sleeve protectors, wipe down ALL surfaces in the hood with methanol/water-moistened lint-free cloth, including the outside of all the tubes (make sure to not wipe off the labeling), the outside and inside of the Speed Vac and centrifuge, tube racks, bottles, etc. Wipe razor blades with methanol-soaked lint-free cloth.
Prepare the following solutions: 25 mM NH4HCO3 (100 mg/50 mL) 25 mM NH4HCO3 in 50% ACN 50% ACN/5% formic acid (may substitute TFA or acetic acid) 12.5 ng/μL trypsin (Promega) in 25 mM NH4HCO3 (freshly diluted)
Dice each gel slice into small pieces (1 mm2) and place into 0.65-mL siliconized tubes (PGC Scientific).
Add ~100 μL (or enough to cover) of 25 mM NH4HCO3/50% ACN and vortex for 10 min.
Using gel loading pipet tip, extract the supernatant and discard.
Repeat steps 3 and 4 once or twice.
Speed Vac the gel pieces to complete dryness (~20 min).
Prepare fresh solutions: 10 mM DTT in 25 mM NH4HCO3 (1.5 mg/mL) 55 mM iodoacetamide in 25 mM NH4HCO3 (10 mg/mL)
Add 25 μL (or enough to cover) 10 mM DTT in 25 mM NH4HCO3 to dried gels. Vortex and spin briefly. Allow reaction to proceed at 56 °C for 1 h.
Remove supernatant, add 25 μL 55 mM iodoacetamide to the gel pieces. Vortex and spin briefly. Allow reaction to proceed in the dark for 45 min at RT.
Remove supernatant (discard). Wash gels with ~100 μL NH4HCO3, vortex 10 min, spin.
Remove supernatant (discard). Dehydrate gels with ~100 μL (or enough to cover) of 25 mM NH4HCO3 in 50% ACN, vortex 5 min, spin. Repeat one time.
Speed Vac the gel pieces to complete dryness (~20 min). Proceed with trypsin digest.
Add trypsin solution (step 2) to just barely cover the gel pieces. Estimate the gel volume and add about 3× volume of trypsin solution. This volume will vary from sample to sample, but on average ~5–25 μL is sufficient.
Rehydrate the gel pieces on ice or at 4 °C for 10 min Spin. Add 25 mM NH4HCO3 as needed to cover the gel pieces.
Spin briefly and incubate at 37 °C for 4 h—overnight.
3.8. Extraction of peptides
Transfer the digest solution (aqueous extraction) into a clean 0.65-mL siliconized tube.
To the gel pieces, add 30 μL (enough to cover) of 50% ACN/5% formic acid, vortex 20–30 min, spin, sonicate 5 min. Repeat.
Vortex the extracted digests, spin and Speed Vac to reduce volume to 10 μL.
Either proceed with C18 ZipTip (Millipore) cleanup or analyze with LC–MS. Add 2–5 μL of 5% formic acid. When analyzing low levels of protein, concentrate the peptides by eluting from ZipTips using 3 μL of elution solution, into a clean 0.65-mL siliconized tube.
Use 1 μL of the unseparated digests for analysis by Matrix Assisted Laser Desorption Ionization/Time of Flight (MALDI/TOF).
3.9. Preparation of MALDI matrix
Dissolve the contents of a 10-mg tube of α-cyano-4-hydroxycinammic acid in 1 mL of the 50% acetonitrile in 0.05% triflouroacetic acid solution. For best performance, once in solution, the matrix should be stored in the dark and used within 1 week.
Some residual crystals may be visible in the matrix solution. The acetonitrile concentration can be adjusted to suit individual preferences. A mixture of 70% ACN and 30% of the 0.1% TFA solution can also be used.
3.10. Sample plate preparation and MALDI-TOF/TOF analysis
Transfer 10 μL of the matrix solution to a small tube.
Add 1 μL of the standard/sample to the tube containing the matrix and vortex.
Apply 1.0 μL from the mixture to wells of an AnchorChip™ sample target plate used for the Bruker Auto-flex MALDI-TOF/TOF.
Once the liquid has evaporated (2 h), the target is ready for analysis.
Obtain peptide mass fingerprints using the reflective and positive ion mode of Autoflex MALDI TOF/TOF mass spectrometer (Bruker, Daltonics Inc., Billerica, MA)
Use FlexAnalysis™ software with signal-to-noise ratio of 2:1 to collect mass spectra from 100 to 400 laser shots.
Generate mono-isotopic peaks by mass peak value calculation.
Use two trypsin auto-digestion peptides with M + H values 842.509 and 2211.104 as internal standards.
Indentify protein by matching the calibrated peptide mass values within NCBInr protein databases using an in-house version of Mascot Server 2.2 imbedded in Bruker's Biotool software.
Select Homo sapiens species for taxonomy.
Use the following match variances: mass tolerance of 40 ppm, one missed trypsin cleavage, fixed modification of carbamidomethyl cysteine, and variable modification of methionine oxidation.
3.11. Results of proteomic analysis
Gels with isoelectric focusing ranges of PI 4–7 and 6–10 produced a total of ~900 protein spots in each gel (Fig. 4.1). The comparison of all spots visualized yielded 24 spots that were significantly different in lean versus obese volunteers. Three of these spots represented multiple isoforms of vimentin. Three vimentin isoforms were considered as one protein (24 – 2 = 22). Two other spots could not be identified (22 – 2 = 20). Of 20 remaining differentially expressed proteins, 10 were upregulated and 1 (α-enolase) was downregulated in obese versus lean volunteers (Table 4.1). The differentially expressed proteins were grouped into the following categories: (1) UPR and stress (seven proteins), (2) energy and FFA metabolism (five proteins), (3) structural proteins (four proteins), and (4) protein transport and signaling (four proteins).
Figure 4.1.
Expression of the UPR proteins CRT and PDI in adipose tissue of one lean and one obese subject (upper panels). Differentially expressed proteins in fat homogenates from one lean and one obese subject. Proteins were separated by isoelectric focusing and molecular weight (2DE) as described in Section 3. The subproteom from each sample was assessed using PI ranges of 4–7 (middle panels) and 6–10 (lower panels). The proteins were stained with SPYRO® Ruby and images compared by PD Quest software. The numbers correspond to the spot numbers in Table 4.1. The arrows indicate upregulated proteins in the fat of obese subjects (from Boden et al., 2008 with permission).
Table 4.1.
Proteins differentially expressed in 2DGs
| Spot no. | Protein identification | Swiss protein accession no. | Mouse score | Peptides matched | Normalized spot volumes ± SD (lean; n = 6) | Normalized spot volumes ± SD (lean; n = 6) | P |
|---|---|---|---|---|---|---|---|
| UPR and stress | |||||||
| 3 | CRT | P27797 | 66 | 7 | 0 | 1019 ± 236 | < 0.001 |
| 23 | PDI | P30101 | 61 | 6 | 421 ± 328 | 1170 ± 63 | < 0.01 |
| 20 | 20 kDa HSPβ-6 | 014558 | 76 | 5 | 2534 ± 1157 | 7530 ± 248 | 0.001 |
| 21 | 27 kDa HSPβ-1 | P04792 | 78 | 8 | 4242 ± 1438 | 8140 ± 1643 | 0.001 |
| 25 | HSPβ-5 | P02511 | 74 | 9 | 4162 ± 558 | 8967 ± 2918 | 0.049 |
| 16 | 60 kDa HSP | P10809 | 69 | 6 | 600 ± 558 | 1868 ± 56 | < 0.001 |
| 19 | Glutathione-S-transferase P | P09211 | 107 | 8 | 2231 ± 698 | 3936 ± 390 | 0.013 |
| Energy and FFA metabolism | |||||||
| 12 | ATP synthase subunit-β | P06576 | 138 | 11 | 2413 ± 801 | 5499 ± 1251 | 0.01 |
| 15 | Perilipin | 060240 | 57 | 6 | 1602 ± 381 | 3960 ± 1019 | 0.007 |
| 22 | Aldehyde dehydrogenase | P05091 | 87 | 9 | 1341 ± 161 | 4088 ± 799 | 0.001 |
| 24 | α-Enolase | P06733 | 65 | 7 | 803 ± 214 | 293 ± 74 | 0.018 |
| 26 | Carbonic anhydrase-1 | P00915 | 198 | 11 | 2463 ± 774 | 4846 ± 523 | 0.012 |
| Structural proteins | |||||||
| 1 | Myosin light-chain polypeptide-6 | P60660 | 60 | 6 | 1792 ± 528 | 3837 ± 284 | 0.002 |
| 2 | Tropomyosin β-chain | P07951 | 58 | 6 | 1141 ± 443 | 4072 ± 531 | 0.03 |
| 4 | Tropomyosin α4-chain | P67936 | 94 | 8 | 770 ± 355 | 3429 ± 779 | 0.002 |
| 7–9 | Vimentin | P08670 | 204 | 19 | 3136 ± 1418 | 11,005 ± 3353 | 0.007 |
| Protein transport and signaling | |||||||
| 6 | γ-Synuclein | 076070 | 66 | 4 | 488 ± 465 | 1714 ± 670 | 0.034 |
| 11 | pGDP dissociation inhibitor-1 | P52565 | 75 | 5 | 2163 ± 612 | 3168 ± 286 | 0.049 |
| 5 | 14-3-3 protein-γ | P61981 | 78 | 6 | 875 ± 525 | 3619 ± 1005 | 0.005 |
| 10 | Galectin-1 | P09382 | 113 | 10 | 9030 ± 1387 | 25,049 ± 4175 | 0.001 |
Data are means ± S.D.
UPR and stress
Levels of expression of the following UPR proteins were overexpressed in 2DEs in adipose tissue from obese volunteers: CRT, a protein chaperone, increased from undetectable to 1019 ± 236 arbitrary units; PDI, a protein foldase, increased approximately threefold; and glutathione-S-transferase P, an antioxidant protein belonging to a UPR-upregulated pathway increased ~1.8-fold.
Several cytosolic small heat-shock proteins (HSPs) (20 and 27 kDa HSP) and one mitochondrial HSP (60 kDa HSP) were also overexpressed in the adipose tissue of obese volunteers, suggesting the presence of cytosolic and mitochondrial stress in addition to ER stress (Table 4.1).
4. Western Blot Analysis
4.1. Required materials
Blotting chamber (Bio-Rad)
Protein disulfide isomerase A3 (Santa Cruz, CA, SC-20132)
Calreticulin (Santa Cruz, CA, SC-11398)
Calnexin (Santa Cruz, CA, SC-11397)
Mouse anti-rabbit horseradish peroxidase-conjugated secondary antibody (Biomeda, Foster City, CA)
4.2. Expression analysis
Separate the proteins (30–80 μg) from the same adipose tissue lysates as used for the 2DEs by 10–14% gradient SDS-PAGE.
Transfer the separated proteins to a nitrocellulose membrane in a semi-dry blotting chamber according to the manufacturer's protocol.
Block the blots with 5% milk in Tris-buffered saline solution (pH 7.6) containing 0.05% Tween-20.
Probe with the following rabbit anti-human antibodies from Santa Cruz Biotechnology (Santa Cruz, CA) at a concentration of 0.4 μg/mL: protein disulfide isomerase A3 (PDI), calreticulin (CRT), and calnexin (CNX).
Use a rabbit antiserum that detects phosphor c-jun NH2-terminal kinase (JNK)-1, -2, and -3 (Cell Signaling Technology, Danvers, MA) and a rabbit antiserum that detects total JNK.
Incubate the blots with primary antibody overnight at 4 °C at with gentle shaking and then incubate with a mouse anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:10,000) (Biomeda) for 1 h at RT.
Expose the blots using a chemiluminescent detection method (Enhanced ECL Detection System; GE Healthcare BioSciences).
4.3. Results of Western blot analysis
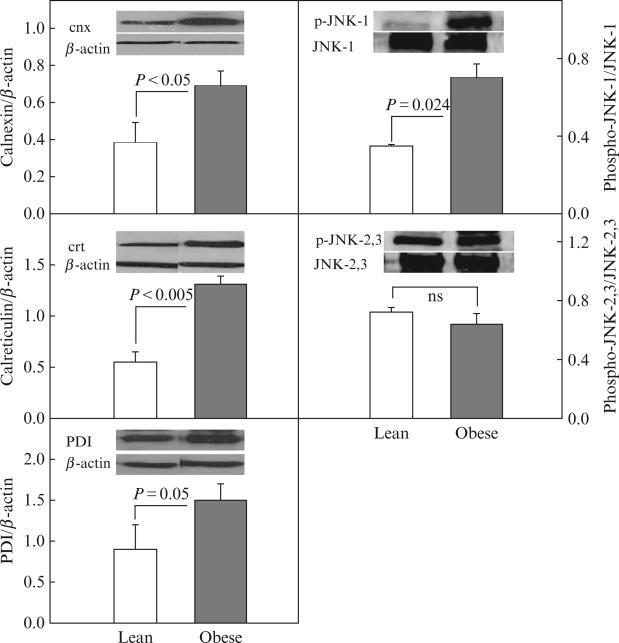
Western blotting confirmed upregulation of CRT and PDI and revealed upregulation of CNX (~1.8-fold), a membrane bound chaperone and phospho-JNK-1 ~2.0-fold, a downstream effector protein of the UPR. Phospho-JNK 2/3, however, was unchanged (Fig. 4.2).
Figure 4.2.
Protein abundance (by Western blots) of CNX/β-actin, CRT/β-actin, PDI, phospho-JNK-1/JNK-1, and phospho-JNK-2,3/JNK-2,3 in subcutaneous adipose tissue from four lean (insufficient fat was available for Western analysis from two of six lean subjects) and six obese nondiabetic subjects. Inserts show representative Western blots (from Boden et al., 2008 with permission).
5. RT-PCR Analysis
Total RNAs were isolated from frozen adipose tissues, and real-time RT-PCR was performed with a SYBR Green One-Step qRT-PCR kit (Invitrogen) and a Light-Cycler (Roche, Indianapolis, IN). Primers for X-box-binding protein (XBP)-1s (NM-005080) were sense: TTGAGAACCAGGAGTTAAG and antisense CCTGCACCTGCTGCGGACT.
5.1. Results of RT-PCR analysis
Upregulation of the spliced form of XPP-1s, a part of the IRE-1/XBP-1 proximal ER stress sensor, and of tumor necrosis factor-α, a proinflammatory cytokine, were documented with RT-PCR, whereas there were no differences in interleukin-1β and -6 mRNA in adipose tissue of lean and obese subjects (Fig. 4.3).
Figure 4.3.
Messenger RNA (mRNA) corrected for 18S ribosomal RNA (18S rRNA) of sXBP-1, tumor necrosis factor-α, interleukin-1β, and interleukin-6 in six lean and six obese nondiabetic subjects (from Boden et al., 2008 with permission).
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01-DK58895, a grant from the American Diabetes Association (1-10-CT-06), a grant from the Department of Health, Commonwealth of Pennsylvania, and a grant from the Groff Foundation (all to G. B.) and R01-A1064017 (to S. M.).
Footnotes
Deionize urea stirring it during 10 min with w/v ~0.6% mixed-ion exchanger (e.g., Amberlite MB-1).
These reagents should be added fresh.
In order to prevent carbamylation modifications, avoid incubation at 37 °C. Since benzonase is a protein it may appear on the 2D map.
To increase protein recovery, sample can be incubated at –20 °C/1 h during the first wash prior spin.
Rehydration and sample volumes depend on pH range, separation distance, and detection method.
Remove the IPG gel strips from the surface of the vertical SDS gel once the proteins have migrated out of the IPG gel strip.
For IEF gels, place the gel into a clean container with 100 mL of IEF fix solution and agitate on an orbital shaker for 3 h.
REFERENCES
- Boden G, Chen X, Ruiz J, Rossetti L. Mechanisms of fatty acid induced inhibition of glucose uptake. J. Clin. Invest. 1994;93:2438–2446. doi: 10.1172/JCI117252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden G, Lebed B, Schatz M, Homko C, Lemieux S. Effects of acute changes on plasma FFA on intramyocellular fat content and insulin resistance in healthy subjects. Diabetes. 2001;50:1612–1617. doi: 10.2337/diabetes.50.7.1612. [DOI] [PubMed] [Google Scholar]
- Boden G, She P, Mozzoli M, Gumireddy K, Reddy P, Xiang X, Luo Z, Ruderman N. Free fatty acids produce insulin resistance and activate the proinflammatory NFκB pathway in rat liver. Diabetes. 2005;54:3458–3465. doi: 10.2337/diabetes.54.12.3458. [DOI] [PubMed] [Google Scholar]
- Boden G, Duan X, Homko C, Molina EJ, Song W, Perez O, Cheung P, Merali S. Increase in endoplasmic reticulum stress-related proteins and genes in adipose tissue of obese insulin-resistant individuals. Diabetes. 2008;57:2438–2444. doi: 10.2337/db08-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray GA. Medical consequences of obesity. J. Clin. Endocrinol. Metab. 2004;89:2583–2589. doi: 10.1210/jc.2004-0535. [DOI] [PubMed] [Google Scholar]
- Itani SI, Ruderman NB, Schmieder F, Boden G. Lipid-induced insulin resistance in human muscle is associated with changes in diacylglycerol, protein kinase C, and IκB-α. Diabetes. 2002;51:2005–2011. doi: 10.2337/diabetes.51.7.2005. [DOI] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- Reaven GM, Hollenbeck C, Jeng C-Y, Wu MS, Chen YD. Measurement of plasma glucose, free fatty acid, lactate and insulin for 24 h in patients with NIDDM. Diabetes. 1988;37:1020–1024. doi: 10.2337/diab.37.8.1020. [DOI] [PubMed] [Google Scholar]
- Santomauro AT, Boden G, Silva M, Rocha DM, Santos RF, Ursich MJ, Strassmann PG, Wajchenberg BL. Overnight lowering of free fatty acids with acipimox improves insulin resistance and glucose tolerance in obese diabetic and nondiabetic subjects. Diabetes. 1999;48:1836–1841. doi: 10.2337/diabetes.48.9.1836. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Yu C, Chen Y, Cline GW, et al. Mechanism by which fatty acids inhibit activation of insulin receptor substrate-1 (IRS-1)-associated phosphatidylinositol 3-kinase activity in muscle. J. Biol. Chem. 2002;277:50230–50236. doi: 10.1074/jbc.M200958200. [DOI] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]