Abstract
West Nile virus (WNV) has successfully spread throughout the USA, Canada, Mexico, the Caribbean and parts of Central and South America since its 1999 introduction into North America. Despite infecting a broad range of both mosquito and avian species, the virus remains highly genetically conserved. This lack of evolutionary change over space and time is common with many arboviruses and is frequently attributed to the adaptive constraints resulting from the virus cycling between vertebrate hosts and invertebrate vectors. WNV, like most RNA viruses studied thus far, has been shown in nature to exist as a highly genetically diverse population of genotypes. Few studies have directly evaluated the role of these mutant spectra in viral fitness and adaptation. Using clonal analysis and reverse genetics experiments, this study evaluated genotype diversity and the importance of consensus change in producing the adaptive phenotype of WNV following sequential mosquito cell passage. The results indicated that increases in the replicative ability of WNV in mosquito cells correlate with increases in the size of the mutant spectrum, and that consensus change is not solely responsible for alterations in viral fitness and adaptation of WNV. These data provide evidence of the importance of quasispecies dynamics in the adaptation of a flavivirus to new and changing environments and hosts, with little evidence of significant genetic change.
INTRODUCTION
West Nile virus (WNV) is a mosquito-borne flavivirus that was first detected in North America in 1999 in the New York City area. The virus is maintained in nature in an enzootic cycle in which it is transmitted between ornithophilic mosquitoes and avian hosts. Non-avian vertebrates become infected as a result of feeding by infected vectors, but generally represent ‘dead-end’ hosts due to the low level of viraemia following infection (Kramer & Bernard, 2001). Evolutionary pressures on the virus, therefore, are applied predominately by the mosquito and avian environments. Since its introduction, WNV has spread throughout the USA, Canada, Mexico, the Caribbean and Central and South America (Austin, et al., 2004; Cruz et al., 2005; Dupuis et al., 2003, 2005; Elizondo-Quiroga, 2005; Granwehr et al., 2004; Morales et al., 2006) and has infected a broad range of both mosquito and avian species (Hayes et al., 2005). There have been some recent population level changes in WNV accompanying its expansion in the western hemisphere, but the virus remains highly conserved (Anderson et al., 2001; Beasley et al., 2003; Davis et al., 2005; Ebel et al., 2001, 2004; Lanciotti et al., 2002). This lack of evolutionary change over space and time is common with many arboviruses (Cilnis et al., 1996; Jenkins et al., 2002; Weaver et al., 1992) and is frequently attributed to the adaptive constraints resulting from the virus cycling between vertebrate hosts and invertebrate vectors (Scott et al., 1994; Weaver et al., 1992). This hypothesis is consistent with studies that suggest that WNV and other arboviruses are subject to high levels of purifying selection (Holmes, 2003; Jerzak et al., 2005; Woelk & Holmes, 2002). Despite this genetic stability, WNV has been highly successful in its emergence and establishment in North America (Davis et al., 2005).
WNV, like most RNA viruses studied, has been shown in nature to be a highly genetically diverse population of genotypes around a dominant master sequence (Jerzak et al., 2005). This finding indicates that WNV is most likely perpetuated in nature as a quasispecies. The term quasispecies refers to the diverse mutant spectrum surrounding a master genotype that presumably possesses the highest fitness value (Domingo et al., 1998; Eigen, 1993; Eigen & Biebricher, 1988; Holmes & Moya, 2002). Genetic diversity within virus populations results from the high error rates and lack of proofreading ability of the RNA-dependent RNA polymerase (Holland et al., 1982), together with high rates of virus replication and large population sizes (Drake & Holland, 1999). The quasispecies theory concludes that selection acts on viral populations as a whole, rather than on individual virions (Eigen, 1993; Martinez et al., 1991; Ruiz-Jarabo et al., 2000). Quasispecies have been shown to be important in viral fitness in previous studies (Duarte et al., 1994; Martinez et al., 1991; Ruiz-Jarabo et al., 2002). In studies with vesicular stomatitis virus (VSV), fitness increases have been measured with no changes in consensus sequences (Novella & Ebendick-Corpus, 2004). The existence of genetic diversity within WNV populations therefore suggests that intrapopulation variants may play a large role in viral fitness and replication, and in the ability of WNV to adapt successfully to new and changing environments.
We reported previously that WNV serially passaged in mosquito (C6/36) cell culture displayed large gains in fitness and replicative ability relative to parental virus populations when measured in mosquito cells. However, this did not correspond to similar changes when measured in avian (DF-1) cell culture (Ciota et al., 2007). These cell-specific phenotypic changes were coupled with modest genetic variation in the consensus sequence. Specifically, two non-synonymous changes were identified by full-genome analysis following 39 mosquito cell passages. These included an A→G mutation at position 1712 of the envelope region resulting in a Lys→Arg substitution, and a G→T mutation at position 6687 of the NS4A region resulting in a His→Gln substitution. The envelope change is fairly conservative in nature and, given its position and orientation, could not be clearly implicated in any obvious function such as receptor binding or membrane fusion. With no known role for NS4A in replication, it is also difficult to implicate the change there in any specific function, although the amino acid substitution (positively charged residue to polar uncharged residue) is potentially significant. Follow-up studies are presented here with the same passage series using: (i) molecular cloning analyses to evaluate the extent of non-consensus variation that correlated with adaptation, and (ii) reverse genetics to evaluate whether replicative advantages in C6/36 cells were a result of consensus mutations alone or whether the mutant spectrum had a significant impact in generating the adaptive phenotype. These are important questions, as the role of genotypic diversity in viral adaptation has not been examined directly. We also investigated whether WNV that had been serially passaged in avian cells displayed similar cell-specific replicative advantages and changes in genetic diversity. Understanding quasispecies dynamics and the resultant phenotypes of viruses in cell culture systems provides a basis to evaluate in vivo results and is the first step towards understanding the importance of this phenomenon in viral emergence and evolution.
METHODS
Cells and media
African green monkey kidney cells (Vero; ATCC CCL-81) were grown in minimal essential medium (MEM; Gibco) supplemented with 10 % fetal bovine serum (FBS), 2 mM l-glutamine, 1.5 g sodium bicarbonate l−1, 100 U penicillin ml−1 and 100 µg streptomycin ml−1. Aedes albopictus mosquito cells (C6/36; ATCC CRL-1660) were grown in MEM supplemented with 10 % FBS, 2 mM l-glutamine, 1.5 g sodium bicarbonate l−1, 0.1 mM nonessential amino acids, 100 U penicillin ml−1 and 100 µg streptomycin ml−1. Chicken embryo fibroblast cells (DF-1; ATCC CRL-12203, cell line derived from East Lansing ELL-0 chicken eggs) were grown in Dulbecco’s modified Eagle’s medium (ATCC 30–2002) supplemented with 10 % FBS, 100 U penicillin ml−1 and 100 µg streptomycin ml−1. Vero, C6/36 and DF-1 cells were grown and maintained at the optimum temperature for each cell line: 37, 28 and 39 °C, respectively. Confluent monolayers of each cell line were prepared for infection by seeding six-well plates (Costar) with 6 × 105 cells per well in 3 ml of appropriate medium and incubating at the proper temperature for 3 days. Confluent cell monolayers and virus-infected cells were maintained in the appropriate medium for each cell line, supplemented with 2 % FBS.
Virus strains
WNV NY003356 is a primary isolate from the kidney tissue of an American crow that was collected in 2000 in Staten Island, NY, USA, and was prepared by one round of amplification in Vero cells (Ebel et al., 2001). Biological clones of this strain (3356.1.1.1) were generated by three rounds of plaque purification on Vero cells in order to establish a homogeneous population for all studies. The biological clone was propagated for use in passaging and growth studies by two rounds of Vero cell amplification and was quantified by plaque assay on Vero cells (Payne et al., 2006). A full-length WNV infectious clone (FL-WNV) was constructed as described previously (Shi et al., 2002b) based on the sequence of the same isolate. Multiple aliquots of all viral stocks were stored at −80 °C.
Serial passage of virus
The biological clone was passaged 40 (C6/36) or 20 (DF-1) times sequentially in cell culture, using an m.o.i. of 0.1 p.f.u. per cell (approx. 5 log10 p.f.u. virus), based on Vero cell titres, for each passage as described previously (Ciota et al., 2007). The C6/36 passage series used in this study was the same series used in the Ciota et al. (2007) study. Briefly, virus was adsorbed to confluent cell monolayers in six-well plates for 1 h at 28 °C (C6/36) or 39 °C (DF-1), with frequent rocking. Following adsorption, the inoculum was removed, cells were washed with MEM, 3 ml maintenance medium was added and plates were incubated at appropriate temperatures. Medium from each well was harvested at 72 h post-inoculation (p.i.) and multiple aliquots were stored at −80 °C. Following each passage, the viral harvest was quantified by plaque assay on Vero cells and diluted to produce an m.o.i. of 0.1 p.f.u. per cell for each subsequent passage. Virus titres varied during passage, but at no time was there greater than a 0.5 log10 difference between C6/36- and DF-1-derived virus. Alternate-passaged WNV was derived from 39 serial passages in C6/36 cells followed by a single passage in DF-1 cells.
Viral growth curve analysis
Six-well plates containing confluent monolayers of Vero, C6/36 or DF-1 cells were infected with virus, in duplicate, at an m.o.i. of 0.01 p.f.u. per cell, based on Vero cell titres. At the end of a 1 h absorption period at 37 °C (Vero), 28 °C (C6/36) or 39 °C (DF-1), the inoculum was removed, wells were washed and 3 ml of maintenance medium were added to each well. Samples, consisting of 100 µl media, were taken in duplicate at 12, 24, 36, 48, 72, 96 and 120 h p.i., diluted 1 : 10 in BA-1 medium containing 20 % FBS and stored at −80 °C. All samples were titrated in duplicate by plaque assay on Vero cells and growth curves were constructed using the mean titre for each time point.
Plasmid construction
Standard site-specific mutagenesis by overlap extension was used for the introduction of mutations into a modified full-length WNV cDNA clone (pFLWNV; Shi et al., 2002a). Primers 5′-ACCACACGCCACGAGGCAGTCTGTGATAG-3′, 5′-CTATCACAGACTGCCTCGTGGCGTGTGGTTC-3′, 5′-CATACCAACAGCCGCTGTCAGTT-3′ and 5′-CAAGATTCCGAATACCGCAAG-3′ were used to make the single-nucleotide mutant A1712G. The modified pFLWNV vector and the final PCR product were digested with ClaI and StuI. The 1.5 kb fragment was inserted into the digested vector. Primers 5′-TCCTCCTCATGCATCGGAAGGGCATTG-3′, 5′-CTCTATGACTTTCGGCATGTAGG-3′, 5′-AATGCCCTTCCGATGCATGAGGAG-3′ and 5′-GGTTTGGTTTGTGCCTAGTG-3′ were used to make the single-nucleotide mutant G6687T. The PCR product was digested with KpnI and the 2.4 kb fragment was inserted into the digested vector. The full-length WNV cDNA vector containing the G6687T mutation was digested with ClaI and StuI and ligated with the 1.5 kb fragment above to make the double mutant. Before ligation, the 1.5 and 2.4 kb fragments were sequenced to confirm the mutations. Sequencing of the plasmids of pFLWNV containing mutations indicated that no other changes were present.
In vitro transcription of RNA and transfection
Transcription and transfection of RNAs were performed as described previously (Shi et al., 2002a). Briefly, plasmids were purified using a MaxiPrep kit (Qiagen) and digested with XbaI. The linearized plasmids were extracted and resuspended in RNase-free water. An mMessage mMachine T7 kit (Ambion) was used to transcribe full-length WNV RNA in vitro and DNA template was removed by DNase I digestion. RNA was quantified by spectrophotometer and 10 µg of wild-type or mutant RNA was electroporated into 8 × 106 BHK-21 cells. Transfected cells were seeded into T-150 flasks. Cell supernatants were collected on days 1 to 5 post-transfection and WNV infectious particles were quantified by plaque assay on Vero cells.
Sequencing of mutant strains
RNA was extracted from WNV using RNeasy spin columns (Qiagen) according to manufacturer’s protocol. Primers for WNV were designed from GenBank accession no. AF260967. One-step RT-PCR (Qiagen) was conducted using primers to generate overlapping PCR products. Reverse transcription reactions were carried out at 50 °C for 30 min, followed by inactivation of the transcriptase at 95 °C for 15 min. Amplification was then carried out for 40 cycles of 94 °C for 20 s, 55 °C for 30 s and 72 °C for 2 min, with a final elongation at 72 °C for 10 min. PCR products were visualized on a 1.5 % agarose gel and bands were allowed to run through 1 % Nusieve GTG low melting point agarose (Cambrex BioScience). Sequencing was performed using ABI 3700 automated sequencers (Applied Biosystems) using overlapping primers to give a minimum of twofold redundancy. Sequences were compiled and edited by using the dnastar software package.
High-fidelity RT-PCR, cloning and sequencing
Production and analysis of clones was performed as described previously (Jerzak et al., 2005). RNA was extracted from infected specimens using RNeasy spin columns (Qiagen) and RT-PCR was conducted using primers designed to amplify the 3′ 1159 nt of the WNV envelope (E) coding region and the 5′ 779 nt of the WNV non-structural protein 1 (NS1) coding region (forward primer WNV1311: 5′-ATGCGCCAAATTTGCCTGCTCTAC-3′; reverse primer WNV3248: 5′-ATGGGCCCTGGTTTTGTGTCTTGT-3′). Reverse transcription of 5 µl RNA was performed with Sensiscript reverse transcriptase (Qiagen) at 45 °C for 40 min. Reverse transcriptase reactions were followed by heat inactivation at 95 °C for 5 min. The resulting cDNA was used as template for PCR amplification. WNV cDNA was then amplified with a ‘high-fidelity’ protocol using PfuUltra (Stratagene), according to the manufacturer’s specifications. Amplification was carried out for 40 cycles of 94 °C for 30 s, 50 °C for 30 s and 72 °C for 4 min, with final extension at 72 °C for 10 min. PCR products were visualized on a 1.5 % agarose gel and DNA was recovered using a MinElute gel extraction kit (Qiagen) as specified by the manufacturer. The recovered DNA was ligated into the cloning vector pCR-Script Amp SK(+) and transformed into XL10-Gold ultracompetent cells (Stratagene) according to the manufacturer’s protocol. The blue–white colour-screening method was used to select transformed colonies. White colonies were screened by direct PCR using primers specific for the desired insert. Plasmid DNA was purified by using a QIAprep Spin Miniprep kit (Qiagen) as specified by the manufacturer. Sequencing was carried out using five pairs of overlapping primers and the T3 reverse primer. Sequencing was performed at the Wadsworth Center Molecular Genetics Core using ABI 3700 and 3100 automated sequencers (Applied Biosystems). Between 15 and 20 clones were sequenced per sample.
Sequence analysis of populations
Sequences were compiled and edited using the seqman module of the dnastar software package, and a minimum of twofold redundancy throughout each clone was required for sequence data to be considered complete. Between 15 and 20 clones from each individual sample were aligned using megalign within dnastar. The consensus sequence for each sample was determined and the sequence of each clone was compared with the consensus. Consensus sequences attained by averaging cloning products agreed with those attained by direct PCR sequencing. The percentage of nucleotide mutations (total number of mutations divided by total number of bases sequenced), amino acid mutations (total number of amino acid changes divided by total number of amino acids sequenced) and the sequence diversity (percentage of clones with at least one difference from the consensus) were used as indicators of genetic diversity. Normalized Shannon entropy (Sn) was calculated based on the frequency of genotypes in populations as follows: Shannon entropy (Sn)=Σ−i Pi lnPi/lnN, where Pi is the frequency of an individual genotype and N is the number of clones sequenced. Sn values range from 0 (completely homogeneous) to 1 (completely heterogeneous). In order to assess selective pressures on populations, dN/dS ratios were calculated using dnasp (Rozas & Rozas, 1999) and Microsoft excel, as described previously (Jerzak et al., 2005). Statistical analyses were performed using both Microsoft excel 2003 and graphpad prism version 4.00.
RESULTS
Genetic diversity of WNV populations following in vitro passage
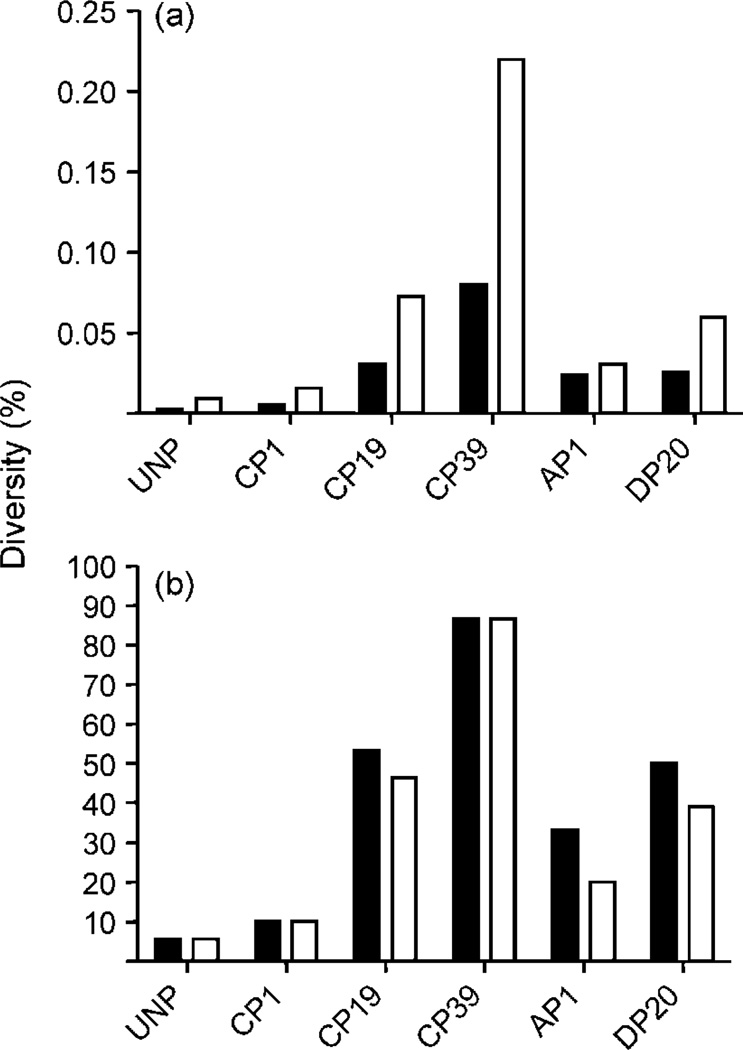
Genetic diversity of WNV was assessed by clonal analysis of nt 1311–3248 (E/NS1 regions) for the following virus populations: unpassaged WNV 3356.1.1.1 (UNP), C6/36 passage 1 (CP1), C6/36 passage 19 (CP19), C6/36 passage 39 (CP39), DF-1 passage 20 (DP20) and alternate passaged, i.e. C6/36 passage 39–DF-1 passage 1 (AP1). The level of nucleotide diversity for WNV UNP was 0.003 % (Fig. 1a). This represented just one mutation in 18 clones analysed (Table 1). A low level of homogeneity was anticipated for this population of clones, as the virus strain was itself a biological clone. This result confirmed the high fidelity of the PfuUltra enzyme used for RT-PCRs and allowed us to have confidence in the low levels of misincorporations in subsequent reactions. Overall, the genetic diversity increased in a reasonably linear fashion with serial passage in C6/36 cell culture (Fig. 1). The level of nucleotide diversity identified for CP19 (0.031 %) was significantly higher than that measured for UNP (Fisher’s exact test, P=0.012) and continued to increase significantly from CP19 to CP39 (P=0.022). Fourteen different genotypes in the cloned region were identified as resulting from C6/36 passage (Table 1). Levels of entropy (Sn) within populations based on the frequency of new genotypes also increased with C6/36 passage, progressing from 0.07 for UNP (highly homogeneous) to 0.70 for CP39 (highly heterogeneous). This represented a total of eight genotypes identified for CP39 with analysis of just 15 clones. As indicated by values for amino acid diversity (Fig. 1b), the majority of mutations were non-synonymous. For the entire region analysed, the dN/dS ratio for CP39 was 0.94, which is very close to the 1.0 predicted by random accumulation of mutations without significant positive or purifying selection (Holmes, 2003). For the E coding region alone, however, 100 % of mutations identified (6/6) were non-synonymous. Furthermore, the two synonymous changes in the NS1 region were only identified in 2/15 clones, whereas the non-synonymous changes found in the E region were identified in 13/15 clones (Table 1). The majority of clones (57 %) differed from the consensus sequence in at least one position by passage 19. Despite this, the single most common genotype (master sequence) remained the consensus sequence (genotype C). By passage 39, 87 % of clones differed from the consensus sequence, and the master sequence (genotype I) contained a nonconservative amino acid change in the envelope protein (F432S) not identified by consensus analysis.
Fig. 1.
Genetic diversity of WNV generated by in vitro passage in C6/36 cells (CP), DF-1 cells (DP) and in C6/36 cells followed by DF-1 cells (AP1). Diversity values (%) were determined by clonal analyses of the region containing nt 1311–3248 (E/NS1). (a) Nucleotide diversity and amino acid diversity represent the percentage of changes per sequenced units. (b) Sequence diversities represent the percentage of genotypes different from the consensus sequence. Filled bars, nucleotide diversity; empty bars, amino acid diversity.
Table 1. Genotypes identified in populations of unpassaged (UNP), C6/36-passaged (CP), DF-1-passaged (DP) and alternate-passaged (AP) WNV for nt 1311–3248.
Values for Sn represent calculations of normalized Shannon entropy based on frequency of variable genotypes. Consensus sequences are highlighted in grey. Only changes relative to consensus sequences of each population are shown for non-consensus variants, and genotypes are listed based on frequency in the population.
| Population (Sn) | Genotype | Frequency | WNV nucleotide coordinate | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E | NSI | |||||||||||||||||
| 1391 | 1444 | 1712 | 1714 | 1780 | 1788 | 1844 | 1949 | 1975 | 1993 | 2063 | 2367 | 2586 | 2699 | 2704 | 3012 | |||
| UNP (0.07) | A | 17/18 | T | G | A | C | G | T | A | A | A | G | C | T | A | T | T | G |
| B | 1/18 | – | – | – | – | – | – | – | – | G⋆ | – | – | – | – | – | – | – | |
| CP1 (0.13) | A | 18/20 | T | G | A | C | G | T | A | A | A | G | C | T | A | T | T | G |
| C | 1/20 | – | – | G⋆ | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| D | 1/20 | – | – | – | – | – | – | – | – | – | – | – | – | – | C⋆ | – | – | |
| CP19 (0.54) | C | 6/15 | T | G | G | C | G | T | A | A | A | G | C | T | A | T | – | – |
| A | 5/15 | – | – | A⋆ | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| E | 1/15 | – | – | A⋆ | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| F | 1/15 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| G | 1/15 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| H | 1/15 | – | – | – | – | C⋆ | – | – | – | – | – | – | – | – | – | – | – | |
| CP39 (0.70) | I | 5/15 | C⋆ | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| C | 2/15 | T | G | G | C | G | T | A | A | A | G | C | T | A | T | T | G | |
| J | 2/15 | – | – | A⋆ | G⋆ | – | – | G⋆ | – | – | – | – | – | – | – | – | – | |
| K | 2/15 | C⋆ | – | – | – | – | – | G⋆ | – | – | – | – | – | – | – | – | – | |
| L | 1/15 | – | – | – | – | – | – | G⋆ | – | – | – | – | – | – | – | – | – | |
| M | 1/15 | – | – | A⋆ | G⋆ | – | – | – | – | – | – | – | – | – | – | – | – | |
| N | 1/15 | – | – | – | – | – | – | – | T⋆ | – | A⋆ | – | – | – | – | – | A | |
| O | 1/15 | – | – | – | – | – | – | G⋆ | – | – | – | – | – | – | – | – | – | |
| AP1 (0.36) | C | 10/15 | T | G | G | C | G | T | A | A | A | G | C | T | A | T | T | G |
| K | 2/15 | C⋆ | – | – | – | – | – | G⋆ | – | – | – | – | – | – | – | – | – | |
| L | 2/15 | – | – | – | – | – | – | G⋆ | – | – | – | – | – | – | – | – | – | |
| P | 1/15 | – | – | – | – | – | – | – | T⋆ | – | – | – | – | – | – | – | – | |
| DP20 (0.47) | A | 12/18 | T | G | G | C | G | T | A | A | A | G | C | T | A | T | T | G |
| Q | 3/18 | – | A⋆ | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| R | 3/18 | – | – | – | – | – | – | – | – | – | – | A⋆ | – | – | – | – | – | |
| S | 2/18 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| T | 1/18 | – | – | – | – | – | – | – | G⋆ | – | – | – | – | – | – | – | – | |
Genotypes shared among populations are represented by repeat letters and amino acid changes are indicated by ⋆.
CP39 lost a substantial amount of genetic variability after a single passage in DF-1 cells (AP1) (Fig. 1). Comparisons of overall nucleotide diversity, amino acid diversity, nucleotide sequence diversity and amino acid sequence diversity all indicated statistically significant decreases from CP39 to AP1 (Fisher’s exact test, P<0.05). Nucleotide diversity decreased from 0.08 to 0.02 % and amino acid diversity decreased from 0.22 to 0.03 % (Fig. 1a). The percentage of clones with consensus genotypes increased from 13 % for CP39 to 67 % for AP1 (Fig. 1b) and levels of entropy decreased from 0.70 to 0.36 (Table 1). These values indicated a significant increase in genotypic homogeneity from CP39 after just 48 h of replication in DF-1 cells. Genotypes C (consensus), K and L were identified in both CP39 and AP1. No additional mutations were identified with AP1.
Although measurements of diversity on all levels (nucleotide, amino acid and sequences) for DP20 were slightly lower than those measured for CP19 (Fig. 1), none were statistically significant (Fisher’s exact test). Nine out of 18 clones analysed for DP20 represented the consensus genotype, while 6/15 for CP19 represented the consensus (Table 1). Calculations of entropy for DP20 (0.47) also indicated a slightly lower genotypic heterogeneity compared with CP19 (0.54).
Viral growth patterns of WNV following cell-culture passage
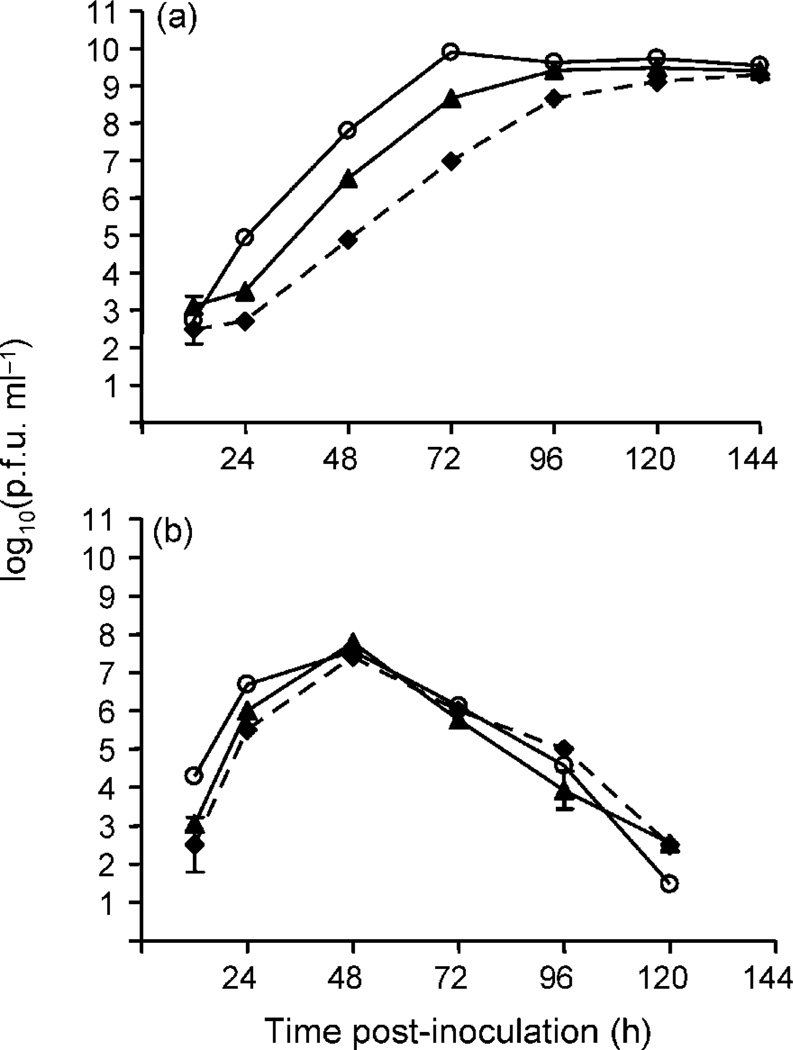
The replicative advantage measured by growth analyses in C6/36 for WNV CP40 was similar to the advantage reported previously following 39 passages (Fig. 2a; Ciota et al., 2007). However, a single passage in DF-1 cells following 39 passages in C6/36 (AP1), significantly decreased the WNV titre compared with CP40 at 24, 48 and 72 h p.i. in C6/36 cells (Fig. 2a; paired t-test, P<0.05), but not in DF-1 cells (Fig. 2b). There was a decrease in the rate of replication of AP1 in C6/36 cells compared with CP40 up to 96 h p.i. However, AP1 titres in C6/36 cells did remain significantly higher than those of UNP up to 120 h p.i (P<0.05).
Fig. 2.
Growth of unpassaged (◆), C6/36-passage 40 (○) and alternate-passages (▲) WNV in C6/36 and DF-1 cell cultures. AP1 refers to WNV passaged 39 times in C6/36 cells followed by a single passage in DF-1 cells. The m.o.i. for all growth curves was 0.01 p.f.u. per cell. Results are presented as the means ± sd of duplicate assays. (a) WNV growth in C6/36 cell culture. (b) WNV growth in DF-1 cell culture.
Growth analyses of WNV passaged 10 and 20 times in DF-1 cells (DP10 and DP20, respectively) indicated no significant differences in peak virus titre in either C6/36 (Fig. 3a) or DF-1 cell culture (Fig. 3b) compared with DF-1 passage and UNP (paired t-test). This finding differed from the substantial cell-specific increases in peak growth titre observed following passage of WNV in C6/36 cells (Fig. 2a). However, both DP10 and DP20 reached their peak titre a day earlier than UNP in DF-1 cell culture, and virus titres were significantly higher at 48 h p.i. for DP10 and at both 36 and 48 h p.i. for DP20 relative to UNP (paired t-test, P<0.01). Virus titres of DP20 at 72 h p.i. and DP10 at 96 h p.i. were significantly lower than UNP (P<0.01). Although statistically significant, the biological significance of these differences is uncertain. Only one titre difference (DP20 vs UNP at 36 h p.i.) was greater than 1 log10 p.f.u. ml−1. Nevertheless, these data suggest a slightly increased rate of growth in DF-1 cell culture following passage in DF-1.
Fig. 3.
Growth of unpassaged (◆) and DF-1-passaged (□, DP10; △, DP20) WNV in C6/36 and DF-1 cell culture. The m.o.i. for all growth curves was 0.01 p.f.u. per cell. Results are presented as the means ± sd of duplicate assays. (a) WNV growth in C6/36 cell culture. (b) WNV growth in DF-1 cell culture.
Viral growth patterns of mutant virus strains
Growth analyses of the single mutants WNV A1712G and WNV G6687T and the double mutant WNV E/NS4 were performed to evaluate the importance of consensus mutations in producing the replicative advantages of C6/36-passaged WNV in C6/36 cell culture. Sequencing of mutant strains confirmed that no additional mutations resulted from insertion of desired changes. Growth of both single mutants and the double mutant in C6/36 and Vero cell cultures were similar to the wild-type strain (Fig. 4). Although virus titres were slightly higher for WNV A1712G in C6/36 cells, these differences were only statistically significant (paired t-test, P<0.05) at 12 h p.i. Significantly lower titres (paired t-test, P<0.05) for both WNV A1712G and WNV E/NS4 were measured in DF-1 cell culture when compared with wild-type strain at 72 h p.i. to 120 h p.i. (Fig. 4). No differences in growth were measured between WNV G6687T in DF-1 cell culture and the wild-type strain. All trends were verified in replicate growth analyses (data not shown).
Fig. 4.
Growth of full-length WNV infectious clone (–––) and WNV mutants (- - -) in C6/36, DF-1 and Vero cell cultures. Single mutants are WNV A1712G and WNV G6687T. WNV E/NS4 contains the mutations A1712G and G6687T. The m.o.i. for all growth curves was 0.01 p.f.u. per cell. Results are presented as the means ± sd of duplicate assays.
DISCUSSION
WNV has perpetuated itself in nature with relatively few genetic changes (Anderson et al., 2001; Beasley et al., 2003; Davis et al., 2005; Ebel et al., 2001, 2004; Lanciotti et al., 2002), even as it has expanded its range in the naive environment of North America (Davis et al., 2005). Worldwide, WNV has infected over 75 species of mosquitoes (Higgs et al., 2004) and over 300 species of birds (Marra et al., 2003). Although it is clear that WNV, like all RNA viruses, has the capacity for rapid accumulation of mutations (Drake & Holland, 1999), the impact of nonconsensus genetic variants and quasispecies populations on WNV adaptation has not been studied previously.
We reported previously that WNV is capable of cell-specific adaptation with minimal genetic alterations in consensus sequences following passage in mosquito cell culture (Ciota et al., 2007). This cell-specific adaptation results in a significant replicative advantage in mosquito cells (Fig. 2). Here, we have shown that despite minimal consensus change, multiple intrapopulation genotype variants were generated with mosquito cell passage (Table 1) and that the size of the mutant spectrum increased with adaptation to mosquito cells (Fig. 1, Table 1). The level of variation was such that the consensus genotype was no longer the dominant genotype by passage 39. In fact, as the clones sequenced here represent only a portion of the genome (2 kb), it seems likely that a complete genome identical to the consensus sequence would be quite rare. As population diversity increases, it becomes more likely that the consensus sequence may not represent the most common genotype (master sequence). Given that the complete set of sequences and not individual mutations represent the unit of selection, it is very important to address this when studying viral adaptation and evolution. This is particularly true with WNV, where unlike reports for some other flaviviruses, no evidence of recombination has been noted (Holmes et al., 1999; Twiddy & Holmes, 2003). The most common sequence in the C6/36 passage 39 population that we identified was genotype I (Table 1). This genotype includes a T→C change at position 1391 of the E coding region, which translates into a potentially significant phenylalanine to serine change in almost 50 % of the population. These data demonstrated that attempting to identify important mutations using consensus change alone can be misleading.
The generation of quasispecies populations in other RNA viruses such as human immunodeficiency virus and Hepatitis C virus has been attributed to the avoidance of host immune responses (Farci et al., 2000, 2002; Frost et al., 2005; Geffin et al., 2003; Weiner et al., 1995). Studies with Foot-and-mouth disease virus (FMDV) have demonstrated that molecular memory is a function of quasispecies structure (Arias et al., 2004; Ruiz-Jarabo et al., 2000). These studies point to the idea that the accumulation of diversity results from variable selective pressures, which, in turn, allow viruses to succeed in dynamic environments. Results presented here clearly indicate that such factors are not a prerequisite for the accumulation of diverse genotypes within populations. This result is consistent with findings from another study with FMDV, which identified genetically diverse populations following sequential cell culture passage (Arias et al., 2001). Diverse genotypes within in vitro-passaged viruses could represent tolerance for neutral mutations or even somewhat deleterious mutations, due to the low levels of selection and lack of bottlenecks that occur with this type of large population passage. Given the well-documented high mutation rate of RNA viruses (approx. 1 mutation per 104 bp replicated), it is easy to see how such high levels of diversity could accumulate in the absence of selection (Drake & Holland, 1999). A previous study with a high-fitness variant of VSV obtained from cell-culture passage showed high levels of phenotypic heterogeneity within the population, with the majority of subclones possessing fitness values lower than that of the parental population (Duarte et al., 1994). In our study, the dN/dS ratio for mutations generated with C6/36 passage (0.94) was not significantly different from what would be predicted by accumulation of random mutations in the absence of high levels of purifying or positive selection (Holmes, 2003). However, for CP39, the majority of the change was found in the E coding region (Table 1) and all of the mutations in this region were non-synonymous. This indicates that positive selection is probably acting on the envelope region, where as purifying selection is acting on the NS1 region, and that selective pressures can vary drastically between regions for WNV. One possible explanation for the apparent contradicting results of high diversity together with selection is that the mutant spectrum somehow provides a replicative advantage and that the unit of selection, as predicted by the concept of quasispecies, is the entire population (Eigen, 1993). In a study by Jerzak et al. (2005), which analysed the same region, no significant bias was identified for mutation in the envelope region in natural isolates. These discrepant results can probably be attributed to the fact that, in the present study, we measured changes in an adapting population. It is not surprising that changes in the E protein are important in creating this adaptive phenotype given this region’s well-defined role in binding and replication (Chambers et al., 1990; Scherret et al., 2001), yet full-genome analysis of large numbers of clones would be necessary to characterize fully the selective pressures on the quasispecies population.
The finding of less cell-specific adaptation (Figs 2a, 3a) with avian cell-passaged WNV relative to mosquito cell-passaged WNV suggests that these disparate cell lines represent different environments for WNV replication and adaptation. Jerzak et al. (2005) demonstrated that mosquito-derived WNV is more heterogeneous than avian-derived WNV in field isolates. The trend we observed towards less variability for WNV in avian cells provides further evidence that the size of the mutant spectrum for WNV may at least be partially dependent on the host cell. Studies with the plant viruses Tobacco mosaic virus and Cucumber mosaic virus have demonstrated host-dependent effects of population diversity (Schneider & Roossinck, 2001). Inherent differences in selective pressures or replicase fidelity between different environments may explain host cell-dependent effects. Further studies with larger sample sizes of WNV populations are necessary for an accurate assessment of this phenomenon and its potential effects on adaptation.
Previous studies have also attempted to measure the level of evolutionary constraint that is imposed by alternating environments through assessment of changes in consensus sequence alone (Chen et al., 2003; Greene et al., 2005; Novella et al., 1999; Weaver et al., 1999). These studies have drawn conclusions using small numbers of mutations without consideration of potentially significant change within the mutant spectra. Here, we have shown that the levels of genetic diversity generated by sequential passage in mosquito cell culture are significantly decreased with a single passage in the ‘bypassed’ alternate cell line (DF-1) (Fig. 1). Furthermore, the phenotypic advantage measured in mosquito cell culture was significantly decreased after a single passage in avian cell culture (Fig. 2). Although the complexity of in vivo systems and differences in replicative strategy between hosts are critical factors to consider when assessing the factors affecting viral evolution, the data presented here support the idea that differential selection in alternating cellular environments constrains the level of WNV mutation maintained in the virus population and, therefore, the rate of genetic change.
Many previous studies have identified consensus mutations associated with alterations in viral fitness, but none has evaluated directly the importance of such changes in generating these phenotypes (Chen et al., 2003; Greene et al., 2005; Novella et al., 1999; Weaver et al., 1999). Our results clearly indicate that the cell-specific replicative advantage acquired with passaging of WNV is not solely the result of consensus change in amino acid sequences (Fig. 4), and, therefore, implicate minority mutations in the adaptive phenotype of the passaged population. Neither the single nor double mutants displayed a significant replicative advantage in any cell line studied. In fact, in avian cells, the A1712G mutation seemed to be less stable in response to the shifting pH seen in the media at later time points (Fig. 4). Given that this difference was not observed with replication of the heterogeneous CP40 population, this result also implicates non-consensus variants in the adaptability of WNV to disparate environments. The phenotypic importance of the mutant spectrum has been noted in previous reports with other viruses (Farci et al., 2002; Sauder et al., 2006). In our study, adaptation must be conferred either exclusively from individual, non-consensus variants within the population possessing high fitness levels or as a result of complementation between genotypes that increases the overall fitness of the population. A recent study with Dengue virus suggests that a defective genome can be maintained within the mutant spectrum by complementation of functional genomes (Aaskov et al., 2006). A separate study with Poliovirus provides evidence for the role of complementation and cooperative interactions in pathogenesis (Vignuzzi et al., 2006). The fitness level conferred by the combination of variants of the adapted WNV population studied here could potentially be greater than that of any individual variant, yet evaluation of the fitness of multiple variants within the population is necessary before the source of adaptation can be characterized accurately. These data provide us with evidence of the importance of the mutant spectrum in generating phenotypes in WNV and point to quasispecies dynamics as an explanation of how this virus has succeeded in changing environments and hosts with little evidence of significant genetic change.
ACKNOWLEDGEMENTS
The authors thank the Wadsworth Center Molecular Genetics Core for sequencing and the Wadsworth Center Media and Tissue Culture Facility for providing cells and media for this work. We thank G. D. Ebel for assistance with data analysis and interpretation. We appreciate the efforts of the Wadsworth Center Arbovirus Laboratories personnel in assisting with cell-culture work. We also thank William Reisen for funding support. This work was supported partially by federal funds from the National Institute of Allergy and Infectious Disease, National Institutes of Health (contract number NO1-AI-25490) and, National Institutes of Health (grant numbers RO1-AI-47855 and RO1-AI-50758).
REFERENCES
- Aaskov J, Buzacott K, Thu HM, Lowry K, Holmes EC. Long-term transmission of defective RNA viruses in humans and Aedes mosquitoes. Science. 2006;311:236–238. doi: 10.1126/science.1115030. [DOI] [PubMed] [Google Scholar]
- Anderson JF, Vossbrinck CR, Andreadis TG, Iton A, Beckwith WH, III, Mayo DR. Characterization of West Nile virus from five species of mosquitoes, nine species of birds, and one mammal. Ann N Y Acad Sci. 2001;951:328–331. doi: 10.1111/j.1749-6632.2001.tb02709.x. [DOI] [PubMed] [Google Scholar]
- Arias A, Lazaro E, Escarmis C, Domingo E. Molecular intermediates of fitness gain of an RNA virus: characterization of a mutant spectrum by biological and molecular cloning. J Gen Virol. 2001;82:1049–1060. doi: 10.1099/0022-1317-82-5-1049. [DOI] [PubMed] [Google Scholar]
- Arias A, Ruiz-Jarabo CM, Escarmis C, Domingo E. Fitness increase of memory genomes in a viral quasispecies. J Mol Biol. 2004;339:405–412. doi: 10.1016/j.jmb.2004.03.061. [DOI] [PubMed] [Google Scholar]
- Austin RJ, Whiting TL, Anderson RA, Drebot MA. An outbreak of West Nile virus-associated disease in domestic geese (Anser anser domesticus) upon initial introduction to a geographic region, with evidence of bird to bird transmission. Can Vet J. 2004;45:117–123. [PMC free article] [PubMed] [Google Scholar]
- Beasley DW, Davis CT, Guzman H, Vanlandingham DL, Travassos da Rosa APA, Parsons RE, Higgs S, Tesh RB, Barrett ADT. Limited evolution of West Nile virus has occurred during its southwesterly spread in the United States. Virology. 2003;309:190–195. doi: 10.1016/s0042-6822(03)00150-8. [DOI] [PubMed] [Google Scholar]
- Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Chen W-J, Wu H-R, Chiou S-S. E/NS1 modifications of dengue 2 virus after serial passages in mammalian and/or mosquito cells. Intervirology. 2003;46:289–295. doi: 10.1159/000073208. [DOI] [PubMed] [Google Scholar]
- Cilnis MJ, Kang W, Weaver SC. Genetic conservation of Highlands J viruses. Virology. 1996;218:343–351. doi: 10.1006/viro.1996.0203. [DOI] [PubMed] [Google Scholar]
- Ciota AT, Lovelace AO, Ngo KA, Le AN, Maffei JG, Franke MA, Payne AF, Jones SA, Kauffman EB, Kramer LD. Cell-specific adaptation of two flaviviruses following serial passage in mosquito cell culture. Virology. 2007;357:165–174. doi: 10.1016/j.virol.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz L, Cardenas VM, Abarca M, Rodriguez T, Reyna RF, Serpas MV, Fontaine RE, Beasley DW, Da Rosa AP. Short report: serological evidence of West Nile virus activity in El Salvador. Am J Trop Med Hyg. 2005;72:612–615. [PubMed] [Google Scholar]
- Davis CT, Ebel GD, Lanciotti RS, Brault AC, Guzman H, Siirin M, Lambert A, Parsons RE, Beasley DW. Phylogenetic analysis of North American West Nile virus isolates, 2001–2004: evidence for the emergence of a dominant genotype. Virology. 2005;342:252–265. doi: 10.1016/j.virol.2005.07.022. [DOI] [PubMed] [Google Scholar]
- Domingo E, Escarmis C, Sevilla N, Baranowski E. Population dynamics in the evolution of RNA viruses. Adv Exp Med Biol. 1998;440:721–727. doi: 10.1007/978-1-4615-5331-1_93. [DOI] [PubMed] [Google Scholar]
- Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc Natl Acad Sci U S A. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte EA, Novella IS, Ledesma S, Clarke DK, Moya A, Elena SF, Domingo E, Holland JJ. Subclonal components of consensus fitness in an RNA virus clone. J Virol. 1994;68:4295–4301. doi: 10.1128/jvi.68.7.4295-4301.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis AP, II, Marra PP, Kramer LD. Serologic evidence of West Nile virus transmission, Jamaica, West Indies. Emerg Infect Dis. 2003;9:860–863. doi: 10.3201/eid0907.030249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis AP, II, Marra PP, Reitsma R, Jones MJ, Louie KL, Kramer LD. Serologic evidence for West Nile virus transmission in Puerto Rico and Cuba. Am J Trop Med Hyg. 2005;73:474–476. [PubMed] [Google Scholar]
- Ebel GD, Dupuis AP, II, Ngo KA, Nicholas D, Kauffman E, Jones SA, Young D, Maffei J, Shi P-Y, et al. Partial genetic characterization of West Nile virus strains, New York State. Emerg Infect Dis. 2001;7:650–653. doi: 10.3201/eid0704.010408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel GD, Carricaburu J, Young D, Bernard KA, Kramer LD. Genetic and phenotypic variation of West Nile virus in New York, 2000–2003. Am J Trop Med Hyg. 2004;71:493–500. [PubMed] [Google Scholar]
- Eigen M. Viral quasispecies. Sci Am. 1993;269:42–49. doi: 10.1038/scientificamerican0793-42. [DOI] [PubMed] [Google Scholar]
- Eigen M, Biebricher DK. Sequence space and quasispecies distribution. In: Domingo E, Holland JJ, Ahlquist P, editors. RNA Genetics. Boca Raton, FL: CRC Press; 1988. pp. 211–245. [Google Scholar]
- Elizondo-Quiroga D, Davis CT, Fernandez-Salas I, Escobar-Lopez R, Olmos DV, Gastalum LCS, Acosta MA, Elizondo-Quiroga A, Gonzalez-Rojas JI, et al. West Nile virus isolation in human and mosquitoes, Mexico. Emerg Infect Dis. 2005;11:1449–1452. doi: 10.3201/eid1109.050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, Strazzera A, Chien DY, Munoz SJ, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- Farci P, Strazzera R, Alter HJ, Farci S, Degioannis D, Coiana A, Peddis G, Usai F, Serra G, et al. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc Natl Acad Sci U S A. 2002;99:3081–3086. doi: 10.1073/pnas.052712599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost SDW, Wrin T, Smith DM, Kosakovsky Pond SL, Liu Y, Paxinos E, Chappey C, Galovich J, Beauchaine J, et al. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci U S A. 2005;102:18514–18519. doi: 10.1073/pnas.0504658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geffin R, Hutto C, Andrew C, Scott GB. A longitudinal assessment of autologous neutralizing antibodies in children perinatally infected with human immunodeficiency virus type 1. Virology. 2003;310:207–215. doi: 10.1016/s0042-6822(03)00137-5. [DOI] [PubMed] [Google Scholar]
- Granwehr BP, Lillibridge KM, Higgs S, Mason PW, Aronson JF, Campbell GA, Barrett ADT. West Nile virus: where are we now? Lancet Infect Dis. 2004;4:547–556. doi: 10.1016/S1473-3099(04)01128-4. [DOI] [PubMed] [Google Scholar]
- Greene IP, Wang E, Deardorff ER, Milleron R, Domingo E, Weaver SC. Effect of alternating passage on adaptation of Sindbis virus to vertebrate and invertebrate cells. J Virol. 2005;79:14253–14260. doi: 10.1128/JVI.79.22.14253-14260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes EB, Komar N, Nasci RS, Montgomery SP, O’Leary DR, Campbell GL. Epidemiology and transmission dynamics of West Nile virus disease. Emerg Infect Dis. 2005;11:1167–1173. doi: 10.3201/eid1108.050289a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs S, Snow K, Gould EA. The potential for West Nile virus to establish outside of its natural range: a consideration of potential mosquito vectors in the United Kingdom. Trans R Soc Trop Med Hyg. 2004;98:82–87. doi: 10.1016/s0035-9203(03)00004-x. [DOI] [PubMed] [Google Scholar]
- Holland JJ, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. Rapid evolution of RNA genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Holmes EC. Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J Virol. 2003;77:11296–11298. doi: 10.1128/JVI.77.20.11296-11298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC, Moya A. Is the quasispecies concept relevant to RNA viruses? J Virol. 2002;76:460–465. doi: 10.1128/JVI.76.1.460-462.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC, Worobey M, Rambaut A. Phylogenetic evidence for recombination in dengue virus. Mol Biol Evol. 1999;16:405–409. doi: 10.1093/oxfordjournals.molbev.a026121. [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Rambaut A, Pybus OG, Holmes EC. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol. 2002;54:156–165. doi: 10.1007/s00239-001-0064-3. [DOI] [PubMed] [Google Scholar]
- Jerzak G, Bernard KA, Kramer LD, Ebel GD. Genetic variation in West Nile virus from naturally infected mosquitoes and birds suggests quasispecies structure and strong purifying selection. J Gen Virol. 2005;86:2175–2183. doi: 10.1099/vir.0.81015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer LD, Bernard KA. West Nile virus in the western hemisphere. Curr Opin Infect Dis. 2001;14:519–525. doi: 10.1097/00001432-200110000-00004. [DOI] [PubMed] [Google Scholar]
- Lanciotti RS, Ebel GD, Deubel V, Kerst AJ, Murri S, Meyer R, Bowen M, McKinney N, Morrill WE, et al. Complete genome sequences and phylogenetic analysis of West Nile virus strains isolated from the United States, Europe, and the Middle East. Virology. 2002;298:96–105. doi: 10.1006/viro.2002.1449. [DOI] [PubMed] [Google Scholar]
- Marra PP, Griffing SM, McLean RG. West Nile virus and wildlife health. Emerg Infect Dis. 2003;9:898–899. doi: 10.3201/eid0907.030277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MA, Carrillo C, Gonzalez-Candelas F, Moya A, Domingo E, Sobrino F. Fitness alteration of foot-and-mouth disease virus mutants: measurement of adaptability of viral quasispecies. J Virol. 1991;65:3954–3957. doi: 10.1128/jvi.65.7.3954-3957.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales MA, Barrandeguy M, Fabbri C, Garcia JB, Vissani A, Trono K, Gutierrez G, Pigretti S, Menchaca H, et al. West Nile virus isolation from equines in Argentina, 2006. Emerg Infect Dis. 2006;12:1559–1561. doi: 10.3201/eid1210.060852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novella IS, Ebendick-Corpus BE. Molecular basis of fitness loss and fitness recovery in vesicular stomatitis virus. J Mol Biol. 2004;342:1423–1430. doi: 10.1016/j.jmb.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Novella IS, Hershey CL, Escarmis C, Domingo E, Holland JJ. Lack of evolutionary stasis during alternating replication of an arbovirus in insect and mammalian cells. J Mol Biol. 1999;287:459–465. doi: 10.1006/jmbi.1999.2635. [DOI] [PubMed] [Google Scholar]
- Payne AF, Binduga-Gajewska I, Kauffman EB, Kramer LD. Quantitation of flaviviruses by fluorescent focus assay. J Virol Methods. 2006;134:183–187. doi: 10.1016/j.jviromet.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Ruiz-Jarabo CM, Arias A, Baranowski E, Escarmis C, Domingo E. Memory in viral quasispecies. J Virol. 2000;74:3543–3547. doi: 10.1128/jvi.74.8.3543-3547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Jarabo CM, Arias A, Molina-Paris C, Briones C, Baranowski E, Escarmis C, Domingo E. Duration and fitness dependence of quasispecies memory. J Mol Biol. 2002;315:285–296. doi: 10.1006/jmbi.2001.5232. [DOI] [PubMed] [Google Scholar]
- Sauder CJ, Vandenburgh KM, Iskow RC, Malik T, Carbone KM, Rubin SA. Changes in mumps virus neurovirulence phenotype associated with quasispecies heterogeneity. Virology. 2006;350:48–57. doi: 10.1016/j.virol.2006.01.035. [DOI] [PubMed] [Google Scholar]
- Scherret JH, MacKenzie JS, Khromykh AA, Hall RA. Biological significance of glycosylation of the envelope protein of Kunjin virus. Ann N Y Acad Sci. 2001;951:361–363. doi: 10.1111/j.1749-6632.2001.tb02719.x. [DOI] [PubMed] [Google Scholar]
- Schneider WL, Roossinck MJ. Genetic diversity in RNA virus quasispecies is controlled by host–virus interactions. J Virol. 2001;75:6566–6571. doi: 10.1128/JVI.75.14.6566-6571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott TW, Weaver SC, Mallampalli VL. Evolution of mosquito-borne viruses. In: Morse SS, editor. The Evolutionary Biology of Viruses. New York: Raven Press Ltd; 1994. pp. 293–324. [Google Scholar]
- Shi P--Y, Tilgner M, Lo MK. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology. 2002a;296:219–233. doi: 10.1006/viro.2002.1453. [DOI] [PubMed] [Google Scholar]
- Shi P-Y, Tilgner M, Lo MK, Kent KA, Bernard KA. Infectious cDNA clone of the epidemic West Nile virus from New York City. J Virol. 2002b;76:5847–5856. doi: 10.1128/JVI.76.12.5847-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiddy SS, Holmes EC. The extent of recombination in members of the genus Flavivirus. J Gen Virol. 2003;84:429–440. doi: 10.1099/vir.0.18660-0. [DOI] [PubMed] [Google Scholar]
- Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Rico-Hesse R, Scott TW. Genetic diversity and slow rates of evolution in New World alphaviruses. Curr Top Microbiol Immunol. 1992;176:99–117. doi: 10.1007/978-3-642-77011-1_7. [DOI] [PubMed] [Google Scholar]
- Weaver SC, Brault AC, Kang W, Holland JJ. Genetic and fitness changes accompanying adaptation of an arbovirus to vertebrate and invertebrate cells. J Virol. 1999;73:4316–4326. doi: 10.1128/jvi.73.5.4316-4326.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Erickson AL, Kansopon J, Crawford K, Muchmore E, Hughes AL, Houghton M, Walker CM. Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a Cytotoxic T Lymphocyte escape variant. Proc Natl Acad Sci U S A. 1995;92:2755–2759. doi: 10.1073/pnas.92.7.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelk CH, Holmes EC. Reduced positive selection in vector-borne RNA viruses. Mol Biol Evol. 2002;19:2333–2336. doi: 10.1093/oxfordjournals.molbev.a004059. [DOI] [PubMed] [Google Scholar]