Abstract
Evidence supporting an early origin of prostate cancer is growing. We demonstrated previously that brief exposure of neonatal rats to estradiol or bisphenol A elevated their risk of developing precancerous lesions in the prostate upon androgen-supported treatment with estradiol as adults. Epigenetic reprogramming may be a mechanism underlying this inductive event in early life, because we observed overexpression of phosphodiesterase 4D variant 4 (Pde4d4) through induction of hypomethylation of its promoter. This epigenetic mark was invisible in early life (postnatal d 10), becoming apparent only after sexual maturation. Here, we asked whether other estrogen-reprogrammable epigenetic marks have similar or different patterns in gene methylation changes throughout life. We found that hypomethylation of the promoter of nucleosome binding protein-1 (Nsbp1), unlike Pde4d4, is an early and permanent epigenetic mark of neonatal exposure to estradiol/bisphenol A that persists throughout life, unaffected by events during adulthood. In contrast, hippocalcin-like 1 (Hpcal1) is a highly plastic epigenetic mark whose hypermethylation depends on both type of early-life exposure and adult-life events. Four of the eight genes involved in DNA methylation/demethylation showed early and persistent overexpression that was not a function of DNA methylation at their promoters, including genes encoding de novo DNA methyltransferases (Dnmt3a/b) and methyl-CpG binding domain proteins (Mbd2/4) that have demethylating activities. Their lifelong aberrant expression implicates them in early-life reprogramming and prostate carcinogenesis during adulthood. We speculate that the distinctly different fate of early-life epigenetic marks during adulthood reflects the complex nature of lifelong editing of early-life epigenetic reprogramming.
The factors contributing to prostate cancer (PCa) risk are not fully understood. Family history, ethnicity, and advancing age are the only risk factors known with certainty (1–3). However, migration and twin studies strongly argue for the environment as a major determinant of PCa risk (4–6). New evidence now supports the action of environmental factors early in life (7–9). Like most human organs, the prostate begins development early in gestation but reaches full maturation only after puberty (10, 11). Before the organ is fully matured, it is developmentally plastic and can respond to a variety of endogenous or external cues to modify its developmental course, resulting in a phenotype with presumed adaptive values for adult life (12). However, if the phenotype established in early life significantly contradicts the needs later in life or has no apparent adaptive value, the risk of developing prostate disease can increase (7, 9, 12). A prime example is exposure of the fetal gland to higher levels of maternal hormones such as testosterone (T), estrogens, and IGF-I and IGF-II, resulting in elevated risk for PCa and/or the aggressive form of the cancer (13–18).
One mechanism by which maternal hormones may predispose the prostate to disease development is by epigenetic reprogramming of transcriptional programs (transcriptome remodeling) and hence the phenotype of the adult gland. Epigenetics allows a cell/tissue to “remember” its experiences, such as a developmental sequence or various environmental stimuli, and modify how the genome is “read” (19). These epigenetic memories are represented as methylation of specific CpG dinucleotides (DNA methylation), unique histone modifications, changes in microRNA profiles, alterations of nucleosome position, and other changes in chromatin structure. Epigenetic changes are replicated into daughter cells (mitotically heritable) (20) and may pass to subsequent generations (21, 22). Because they do not involve changes in primary DNA sequence or in chromosomal organization, these changes are potentially reversible, but evidence is still sparse.
Apropos to early-life transcriptome remodeling through epigenetics in the prostate, we provided the first evidence that brief exposure of neonatal rats to 17β-estradiol-3-benzoate (EB) or the environmental estrogen mimic bisphenol A (BPA) induced hypomethylation of the promoter of phosphodiesterase 4D variant 4 (Pde4d4) and its overexpression in their prostates during adulthood (23). Coincidentally, the risk of developing precancerous prostate lesions under the oncogenic effect of protracted stimulation by 17β-estradiol (E2) supported by T (T+E2) was significantly elevated if the rats were treated neonatally with EB/BPA. Interestingly, in this “two-hit” PCa-induction model, hypomethylation of the Pde4d4 promoter and gene overexpression were undetectable at postnatal d 10 and appeared only after puberty. A scenario was similar for hypomethylation of the promoter of nucleosome binding protein-1 (Nsbp1) [or high-mobility group nucleosome binding domain 5 (Hmgn5)] induced by brief treatment of mouse neonates with diethylstilbestrol (DES) or genistein (GEN). The epigenetic change remained dormant in the immature uteri of the treated mice but emerged in their uteri after sexual maturation (24). The appearance of this epigenetic alteration and associated aberrant gene expression was associated with a heightened incidence of uterine cancer at old age in mice treated neonatally. Noteworthy is the finding that both hypomethylation of the Nsbp1 promoter and uterine carcinogenesis induced by neonatal DES/GEN treatment was effectively obliterated by ovariectomy. The findings from these two studies support the postulate that early-life epigenetic memories may require later-life events such as sexual maturation to bring them to the surface.
We further elucidated the roles of adult-life events on activating/modifying early-life epigenetic memories by 1) characterizing the temporal changes in promoter methylation and gene expression of hippocalcin-like 1 (Hpcal1) [or visinin-like protein 3 (Nvp-3)] and Nsbp1 in the prostates of animals treated neonatally with EB or BPA and that received or did not receive the adult-life oncogenic stimulus (T+E2 treatment), comparing results with those published for Pde4d4 (23); and 2) determining the life-time changes in expression of genes encoding DNA methylation-regulating proteins, including DNA methyltransferases and methyl-CpG binding domain proteins, findings that may offer a mechanistic explanation for early-life epigenetic reprogramming by estrogens.
In our previous study on Pde4d4 (23), we identified Hpcal1, a member of the visinin-like subfamily of neural calcium sensor and a putative tumor suppressor (25–28), as a putative candidate but did not select it for validation until now. In the current investigation, we also selected to study Nsbp1 because it was identified originally as an early-life estrogen (DES/GEN)-reprogrammable gene in the mouse uterus (24), and the human gene was recently shown to be a putative oncogene in glioma (29) and cancers of the bladder (30) and prostate (31, 32). Here, we demonstrated that both Hpcal1 and Nsbp1, like Pde4d4, were reprogrammable by neonatal exposure of EB/BPA in the prostate but differed significantly in their time of appearance during development and responses to adult-life modulations. Furthermore, early and persistent alterations in expression of DNA methyltransferase-3a (Dnmt3a), Dnmt3b, and methyl-CpG binding domain protein 2 (Mbd2) may explain in part epigenetic reprogramming by early-life exposures to estrogens.
Materials and Methods
Animals and tissue preparation
Animal housing and treatments were approved by the Animal Use Committee at the University of Illinois at Chicago and carried out as previously described (23). Pregnant Sprague Dawley rats (Zivic-Miller Laboratories, Pittsburgh, PA) and male offspring were maintained under the following conditions to minimize BPA and estrogenic exposures: housing in new polysulfone cages, supplying deionized water from glass bottles, and feeding with a single feed lot of soy-free, phytoestrogen-reduced diet (Zeigler Reduced Rodent Diet 2; Ziegler Brothers, Inc., Gardners, PA) with 12 ppm phytoestrogens as determined by high-performance liquid chromatography. Pregnant dams were monitored, and the day of birth was designated postnatal d 0. The pups were sexed by anogenital distance, and each litter was culled to 10 pups by removing or adding female pups as necessary. Newborn male pups were assigned to one of four neonatal treatment groups with 30–35 pups/group: 1) controls given tocopherol-stripped corn oil vehicle alone (ICN Biomedicals, Aurora, OH), 2) high-dose EB (HEB) [2500 μg/kg body weight (BW)], 3) low-dose EB (LEB) (0.1 μg/kg BW), or 4) BPA (10 μg/kg BW). All steroids were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO). Litter effects were avoided by randomly assigning male pups within each litter to a treatment and toe clipping for permanent identification. Treatments were given on d 1, 3, and 5 of life through sc injections in the nape of the neck, which produced serum BPA levels less than 2 ng/ml during the first hour after treatment (33). Male rats from each group were killed on d 10 (n = 10/group), d 90 (n = 8/group), and d 200 (n = 8/group). An additional cohort of neonatally treated rats (n = 8/group) were implanted with T- and E2-filled SILASTIC capsules (Dow Corning Corp., Midland, MI) on d 90 and killed on d 200. The T- and E2-capsule lengths were designed to elevate serum E2 levels to approximately 75 pg/ml and maintain serum T levels close to the physiological level at approximately 3 ng/ml (34). The T+E2 regimen was reported to produce approximately 40% prostatic intraepithelial neoplasia in the dorsal lobe (DP) of controls, 100% in HEB-treated rats, 50% in LEB-treated rats, and 100% in BPA-treated animals (23). RNA and DNA were isolated from rat DP with TRIzol (Invitrogen, Carlsbad, CA) and DNeasy kit (QIAGEN, Valencia, CA), respectively.
Bisulfite genomic sequencing
Genomic DNA (100–200 ng) from DP was bisulfite modified with use of the EZ Methylation Modification kit (Zymo Research, Irvine, CA) before PCR. In silico analyses and detailed database searches were used to predict the 5′-CpG island(s) (CGI) in each gene (www.urogene.org/methprimer/). Primers were designed to amplify fragments encompassing the 5′-CGI of candidate genes: Nsbp1, Hpcal1, Dnmt3b, and Mbd2 from bisulfite-modified DNA (Table 1). Amplicons were generated from DP of individual animals and subcloned into the pCR2.1 vector (Invitrogen) as previously described (23). Six or more clones were picked and sequenced for each sample (Macrogen, Rockville, MD).
Table 1.
Primers used for bisulfite genomic sequencing, MSPCR, and real-time PCR
| Primers for BSPCR | ||
| rBS-NSBP1 | Forward | 5′-GTTTTTTTGTTAATTGGGAAAGATT-3′ |
| Reverse | 5′-AAAAAAATCTCTACCTAACTACTCC-3′ | |
| rBS-PDE4D4 | Forward | 5′-TTATTGTTGTGAAGAGTAGATTTTGTG-3′ |
| Reverse | 5′-ATCCTAAATTTCTTCAAACCTAACC-3′ | |
| rBS-HPCAL1 | Forward | 5′-GGAGTGATTAGGTTGTAGTTGAGG-3′ |
| Reverse | 5′-TCCTACCCTCAAATTCCTAAACTAC-3′ | |
| rBS-DNMT3b | Forward | 5′-AGGTGGAGAAGTTTGGGTATTATTT-3′ |
| Reverse | 5′-CAAAAACCTCCAAAAAACCTATC-3′ | |
| rBS-MBD2 | Forward | 5′-GGTTATAGTTAATTTTATTTGTAATAATGT-3′ |
| Reverse | 5′-CCAACACTAAACCCTAATTTTC-3′ | |
| Primers for MSPCR | ||
| rMS-NSBP1 | Forward | 5′-CGGGTTACGTTTTTCGAATC-3′ |
| Reverse | 5′-ATCTCTACCTAACTACTCCGCGAA-3′ | |
| rUS-NSBP1 | Forward | 5′-GTGGGTTATGTTTTTTGAATTGG-3′ |
| Reverse | 5′-AAAAAAATCTCTACCTAACTACTCCACAA-3′ | |
| rMS-HPCAL1 | Forward | 5′-AGTGGGTTTGTAGTTGATCGTAGAC-3′ |
| Reverse | 5′-CCCCCGTAAAAAAAATATAAACG-3′ | |
| rUS-HPCAL1 | Forward | 5′-GTAGTGGGTTTGTAGTTGATTGTAGAT-3′ |
| Reverse | 5′-CCCCCATAAAAAAAATATAAACACC-3′ | |
| Primers for real-time PCR | ||
| rNSBP1 | Forward | 5′-CTATGCCTGTGCCCTTTACAC-3′ |
| Reverse | 5′-TTTTGGCTTCTCCATTTTCAG-3′ | |
| rPDE4D4 | Forward | 5′-ACGAGCAGCACCACCAGTA-3′ |
| Reverse | 5′-AAAGACGAGGGCCAGGACAT-3′ | |
| rHPCAL1 | Forward | 5′-GAGATTGTGCAGGCCATCTAC-3′ |
| Reverse | 5′-TGCCATCATTGTTTGTGTCC-3′ | |
| rRPL19 | Forward | 5′-GCATATGGGCATAGGGAAGA-3′ |
| Reverse | 5′-CCATGAGAATCCGCTTGTTT-3′ | |
| rDNMT1 | Forward | 5′-GAGGTGGGCGACTGCGTCTC-3′ |
| Reverse | 5′-TGTGGATGTAGGAAAGTTGCA-3′ | |
| rDNMT3a | Forward | 5′-CAGAATAGCCAAGTTCAGCAAAGTGA-3′ |
| Reverse | 5′-CTTTGCCCTGCTTTATGGAG-3′ | |
| rDNMT3b | Forward | 5′-GTTAAAGAAAGTACAGACAATAACCAC-3′ |
| Reverse | 5′-TCTGATGACTGGCACACTCC-3′ | |
| rMeCP2 | Forward | 5′-GTCGCTCTGCTGGAAAGTAT-3′ |
| Reverse | 5′-TGGGCTTCTTAGGTGGTTTC-3′ | |
| rMBD1 | Forward | 5′-CAGCAGTCACAACCTTCCTG-3′ |
| Reverse | 5′-GGTGCCAATCCCTCCTATCT-3′ | |
| rMBD2 | Forward | 5′-GTCGGCCCAGGTAGTAATGAT-3′ |
| Reverse | 5′-GACTCGCTCTTCCTGTTTCCT-3′ | |
| rMBD3 | Forward | 5′-CTGAACACTGCACTGCCTGTA-3′ |
| Reverse | 5′-GTTTCTTCTCCCAGAAAAGCTG-3′ | |
| rMBD4 | Forward | 5′-CCTACCGGATCTTTTGTGTCA-3′ |
| Reverse | 5′-GATTTTCCCAAAGCCAGTCAT-3′ | |
Treatment of NbE-1 and AIT cells with 5-aza-deoxycytidine (5-AZA-dC)
Two rat cell lines, the immortalized normal prostatic epithelial cell line NbE-1 (35) and the tumorigenic cell line AIT (36), were maintained in DMEM/F12 medium (Invitrogen) supplemented with 5% fetal bovine serum (HyClone, Logan, UT), 1× insulin-transferrin-selenium, 1 mm sodium pyruvate, and 100 mm nonessential amino acids (Invitrogen). Cells were treated with 0.5 or 1.0 μm 5-AZA-dC (Tocris, Ellisville, MO) or with dimethylsulfoxide (DMSO) (0.1%) as control every 2 d for up to 8 d. RNA was isolated for real-time RT-PCR analyses of steady-state transcript levels, and DNA was isolated for methylation-specific PCR (MSPCR) as previously described (23, 24).
Real-time PCR
As previously described (23, 24), total RNA was isolated from cells or tissues reverse transcribed to cDNA; and expression levels of Nsbp1, Hpcal1, ribosomal protein L19 (Rpl19), Dnmt1, Dnmt3a, Dnmt3b, methyl CpG binding protein 2 (Mecp2), Mbd1, Mbd2, Mbd3, and Mbd4 determined by a SYBR Green-based real-time PCR method on the ABI PRISM 7500 FAST System (Applied Biosystems, Foster City, CA). The expression level of each gene was normalized to the expression of Rpl19. Rpl19 was chosen as the house-keeping gene because its expression was not changed in response to EB and BPA throughout life (23). Primers specific for every gene were designed in the exon/exon spanning region or unique regions (Table 1). The threshold cycle (Ct) number for each sample was determined in triplicate, and the experiments were repeated with three individual sets of samples. The relative expression of the real-time PCR products was determined by the ΔΔCt method (23).
Methylation-specific PCR
Bisulfite-modified genomic DNA was subjected to MSPCR. Sets of primers were designed to amplify the region of the 5′ promoter showing differential methylation in response to E2/BPA. The M primer set amplified the methylated target sequence; the U primer set amplified only the unmethylated allele (Table 1). Thirty-five-cycle PCR were performed with Platinum Taq polymerase (Invitrogen) under the following conditions: denaturation at 94 C for 30 sec, annealing at 56 C (Nsbp1) or 58 C (Hpcal1) for 1 min, and extension at 72 C for 1 min, followed by a 12-min final extension. PCR products were separated on 2% agarose gel and visualized with ethidium bromide.
RNA interference [small interfering RNA (siRNA)]-mediated gene silencing
To determine whether the differentially methylated genes can regulate cellular functions in rat prostatic epithelial cells, we used siRNA to knockdown gene expression in NbE-1 or AIT cells and studied the effects on cell viability. Transfection protocols were optimized to achieve maximal gene silencing in preliminary experiments. Final protocols used 0.2 μm Accell siRNA SMARTpool against Nsbp1–3′untranslated region, Hpcal-3′ untranslated region, Pde4d4-open reading frame, or a nontargeting control (Thermo Scientific, Pittsburgh, PA) and Lipofectamine 2000 (Invitrogen) to assist siRNA delivery. After a 3-d incubation, alamarBlue (Invitrogen) was added to the cells, and cell viability was determined for all treatment groups by measuring the absorbance at 570/600 nm. RNA was isolated for real-time PCR analysis of target-gene expression (23, 24).
Statistical analysis
Data were expressed as mean ± sd. Bonferroni post hoc test (correction test) was performed after ANOVA (Prism version 4.0; GraphPad, San Diego, CA) for multiple comparisons among all data groups to determine the statistical significance. A statistically significant difference between groups was accepted at P < 0.05.
Results
Aberrant Nsbp1 promoter demethylation and transcriptional overexpression in the prostates of rats neonatally exposed to EB or BPA persist throughout life and are insensitive to adult-life modification
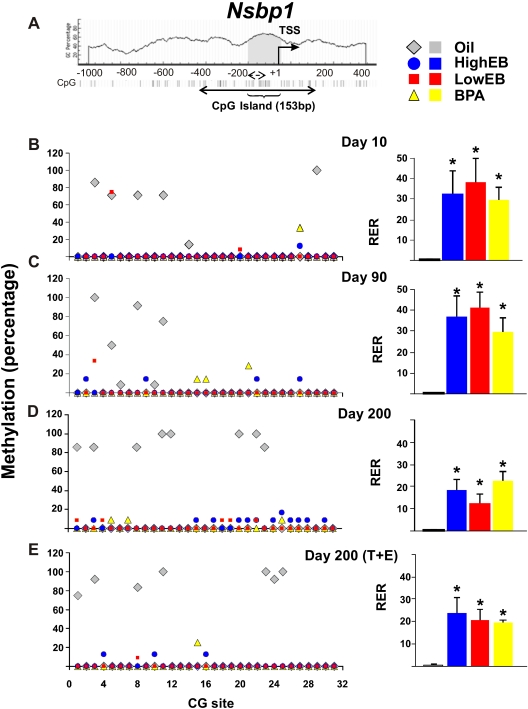
In silico analysis located a 153-bp CGI in the Nsbp1 5′ flanking region (Fig. 1A). A total of 31 CG sites along the Nsbp1 promoter were analyzed by bisulfite genomic sequencing. CG sites 3, 5, 8, and 11 were methylated (80–100%) in control (oil treated) animals at d 10 (Fig. 1B, left panel). Promoter methylation persisted at CG sites 3, 8, and 11 in oil-treated animals during life, and methylation of the promoter became more widespread with aging (Fig. 1, C–E, left panels). Expression of Nsbp1 in oil-treated animals was low (Fig. 1, B–E, right panels), in concordance with the hypermethylation status of the Nsbp1 promoter. Animals neonatally exposed to EB or BPA exhibited obvious demethylation of the promoter as compared with controls at d 10. This early epigenetic reprogramming persisted after sexual maturation (d 90) and throughout adult life (d 200), remaining unaltered even under the protracted action of a cancer-promoting treatment with T+E2 (Fig. 1, B–E, left panel). In concordance with promoter methylation status, the prostates of animals exposed to EB/BPA during early-life showed significant overexpression of Nsbp1 (∼30-fold at d 10, ∼30- to 40-fold at d 90, ∼15- to 25-fold at d 200, and ∼20-fold at d 200 under T+E2-treatment as compared with controls) (P < 0.05) (Fig. 1, B–E, left panels). These data suggest that the Nsbp1 is an estrogen-reprogrammable gene whose methylation status, once altered, persists throughout life.
Fig. 1.
Schematic diagram of CpG dinucleotides content (%) in the 5′ flanking region of the Nsbp1 gene. A, A CGI of 153 bp (gray) was identified between −190 to −38 upstream of the transcriptional start site (TSS). The CGI has 12 CG sites. Here, we studied an expanded region encompassing the CGI. It consists of 517 bp containing a total of 31 CG sites (solid double-headed arrow). Individual CG sites are represented as vertical lines. The methylation status of the region was determined by bisulfite genomic sequencing (solid double-headed arrow) and by MSPCR (broken double-headed arrow, see Fig. 3 for results), respectively. B–E, left panel, DNA isolated from the dorsal prostate of animals treated with HEB (blue circles), LEB (red squares), BPA (yellow triangles), or oil (gray diamonds) at postnatal (PN) d 10, 90, 200, or 200 (T+E2) was subjected to bisulfite genomic sequencing analysis. Methylation status of each CG site of the Nsbp1 promoter is indicated as average percentage methylation. B–E, right panel, RNA was isolated from the dorsal prostate of animals treated with HEB (blue bar), LEB (red bar), BPA (yellow bar), or oil (gray bar) at PN d 10, 90, or 200 (±T+E2). Relative levels of Nsbp1 transcript were determined by real-time PCR and normalized to Rpl19 transcript in the same sample. The relative level of Nsbp1 transcript [relative expression ratio (RER)] at PN d 10 in neonatal oil-treated samples was arbitrarily assigned a value of 1.0, and values from all treatment groups at various life stages were normalized to the mean value of this group. Statistical significant differences are indicated (*, P = 0.05).
Aberrant Hpcal1 promoter hypermethylation and transcriptional suppression in the prostates of rats neonatally exposed to EB or BPA are dependent on the nature of early-life exposure and are susceptible to adult-life modification
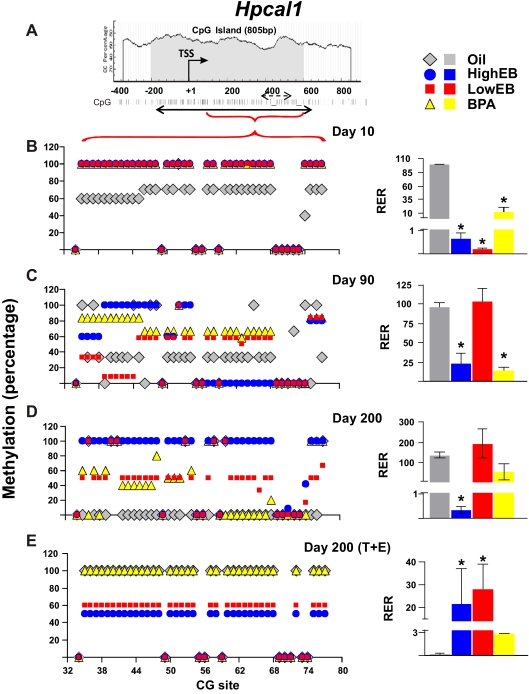
In this study, in silico analysis revealed the presence of an 805-bp CGI with 77 CG sites in the Hpcal1 5′ flanking region (Fig. 2A). Bisulfite sequencing was unable to detect any significant changes in methylation status of the 5′ end of this CGI with any treatment (data not shown). In contrast, most of the changes occurred in CG sites 34–77 (the promoter), which was the promoter region of all subsequent investigations. In animals treated neonatally with oil (controls), the promoter was moderately methylated (60–70%) at d 10 (Fig. 2B, left panel, gray diamonds) and progressively underwent demethylation with advancing age (∼40% at d 90 and ∼0% at d 200) (Fig. 2, C and D, left panel). Interestingly, the cancer-promoting treatment (T+E2) completely reversed the aging effect, leading to approximately 100% remethylation of the promoter and silencing of Hpcal1 expression in controls treated with oil neonatally (Fig. 2E).
Fig. 2.
Schematic diagram of CpG dinucleotides content (%) in the 5′ flanking region of the Hpcal1 gene. A, A CGI of 805 bp (gray) was identified flanking the transcriptional start site (TSS). The CGI has 77 CG sites (solid double-headed arrow). Our pilot experiments did not show significant changes in methylation status in the 5′ region of the CGI. We therefore only examined a region encompassing CG 34–77 in subsequent studies (red horizontal bracket). Individual CG sites are represented by vertical lines. The methylation status of this region was determined by bisulfite genomic sequencing and MSPCR (broken double-headed arrow; see Fig. 3 for results). B–E, left panel, DNA isolated from the dorsal prostate of animals treated with HEB (blue circles), LEB (red squares), BPA (yellow triangles), or oil (gray diamonds) at postnatal (PN) d 10, 90, or 200 (±T+E2) was subjected to bisulfite genomic sequencing analysis. Methylation status of CG sites 34–77 of the Hpcal1 promoter is indicated as average percentage methylation. B–E, right panel, RNA was isolated from the dorsal prostate of animals treated with HEB (blue bar), LEB (red bar), BPA (yellow bar), or oil (gray bar) at PN d 10, 90, or 200 (±T+E2). Relative level of Hpcal1 transcript was determined by real-time PCR and normalized to Rpl19 gene expression. The relative level of Hpcal1 transcript in PN d 10 oil-treated samples was arbitrary assigned a value of 100; values from all treatment groups at various life stages were normalized to the mean value of this group, and values from all treatment groups at various life stages were normalized to the mean value of this group. Statistical significant differences are indicated (*, P < 0.05). RER, Relative expression ratio.
Compared with control treatment, neonatal treatment with HEB/LEB/BPA-induced hypermethylation (∼100% methylated) of the Hpcal1 promoter at d 10 and silencing of gene expression (Fig. 2B). For animals treated with LEB, the Hpcal1 CGI became demethylated (∼50–60% methylated) when animals reached maturation and throughout life (Fig. 2, C and D, left panel), reversing the early epigenetic pattern and resulting in reexpression of Hpcal1 comparable with that in oil-treated animals (Fig. 2, C and D, right panel). Still, demethylation of the Hpcal1 promoter never achieved the complete loss in control animals. In contrast, the early epigenetic pattern induced by HEB was most persistent and less altered with aging (Fig. 2, C and D, left panel, blue circles) and correlated with very low expression of Hpcal1 (∼0.5% at d 10, ∼25% at d 90, and ∼1% at d 200) in treated animals as compared with controls (∼100%) (Fig. 2, C and D, left panel). For animals treated neonatally with BPA, the promoter was aberrantly hypermethylated (100% methylated) at d 10, similar to the scenario in the two EB-treatment groups. Some degree of demethylation of the promoter began (70–80% methylated at d 90 and 50–60% methylated at d 200) (Fig. 2D, left panel, yellow triangles) as rats aged, but levels of gene expression remained low (∼10–20% of controls) throughout life (Fig. 2D, right panel).
As mentioned earlier, the cancer-promoting T+E2 treatment resulted in significant hypermethylation of the Hpcal1 promoter in control rats. However, this treatment did not change the degree of promoter methylation in the neonatal LEB-treated cohort, which remained at approximately 60%, but did lower gene expression in that cohort as compared with levels in adults (d 90 and 200). In contrast, the T+E2 treatment resulted in demethylation of the promoter in the neonatal HEB-treated animals as compared with all groups without T+E2 treatment. Gene expression in the prostates of the same animals was reduced to a level similar to that in the LEB group. These results differ from those in the neonatal BPA-treated group, in which the promoter CGI was almost completely remethylated, similar to control animals, and gene expression was almost completely silenced (Fig. 2E). Taken together, these results demonstrate that the early epigenetic pattern introduced by neonatal EB/BPA exposure at the Hpcal1 promoter is affected by later-life events and leads to different outcomes depending on the initial exposure.
Increased expression of Nsbp1 and Hpcal1 by 5-AZA-dC treatment was associated with promoter demethylation in prostate epithelial cell lines
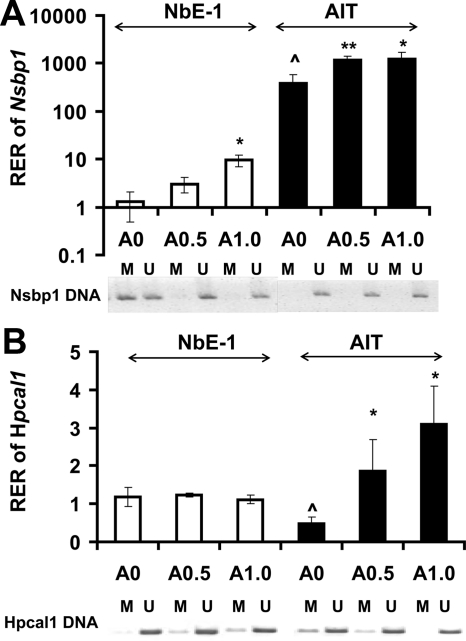
To establish linkage of promoter methylation status to gene transcription for the studied genes, we treated the normal prostatic epithelial cell line NbE-1 (immortalized) and the rat PCa cell line AIT with two different doses of the demethylating agent 5-AZA-dC. The treatments resulted in demethylation of the promoter of Nsbp1 in NbE-1 cells (Fig. 3A) and Hpcal1 in AIT cells (Fig. 3B), as indicated by MSPCR results, and concordant elevation of gene transcript levels. These data suggest a causal relationship between promoter methylation and gene expression for these two genes.
Fig. 3.
Real-time PCR analysis of transcript levels and promoter methylation status of (A) Nsbp1 and (B) Hpcal1 in NbE-1 and AIT cells in response to 5-AZA-dC. Cells were treated with 0.5 or 1.0 μm 5-AZA-dC or with DMSO (0.1%) as the control every 2 d for up to 8 d. Upper panel (A and B), RNA was isolated and reverse transcribed, and levels of Nsbp1 or Hpcal1 transcript were measured by real-time PCR (bar plots). Relative expression ratio (RER) of Nsbp1 or Hpcal1 was calculated by normalizing to Rpl19 transcript levels in the same sample. The RER of gene in DMSO-treated control sample was set as 1.0. Data presented are the means with sd of three independent experiments. *, P < 0.05 or **, P < 0.01 were consider statistically significant (compared with control). ^, Gene expression of NbE-1 and AIT cells differed significantly (P < 0.05). Lower panel (A and B), Methylation status of Nsbp1 or Hpcal1 was analyzed by MSPCR. DNA isolated was modified by sodium bisulfite followed by PCR, with primers specific for methylated DNA (M) or unmethylated DNA (U). Representative results from one of the three independent experiments are shown.
siRNA-mediated decreases in gene transcription were associated with changes in cell viability in prostate cell lines
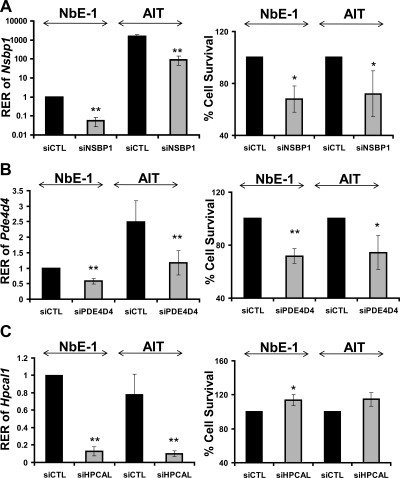
Transient transfection of NbE-1 or AIT cells with siRNA against Nsbp1 effectively lowered the levels of gene transcript in NbE-1 or AIT cells to less than 10% of those in cells transfected with nontargeting siRNA and significantly reduced cell viability by approximately 40% in both cell models (Fig. 4A). RNA interference achieved only a 40–60% reduction in Pde4d4 mRNA levels but a significant approximately 40% reduction in viability in both cell lines (Fig. 4B). Of interest to note, levels of Hpcal1 mRNA were reduced by 90% after transfection of siRNA against the gene, but viability of NbE-1 cells was increased only modestly (15%), suggesting that Hpcal1 may not be a major regulator of cell growth/apoptosis (Fig. 4C).
Fig. 4.
Real-time PCR analysis of transcript levels of (A) Nsbp1, (B) Pde4d4, and (C) Hpcal1 and cell survival of in NbE-1 and AIT cells in response to siRNA knockdown. Cells were treated with 0.2 μm siRNA oligos or with 0.2 μm nontarget control for 72 h. Left panel, RNA was isolated and reverse transcribed. Nsbp1, Pde4d4, or Hpcal1 expression was quantified by real-time PCR and normalized to Rpl19 in the same sample. The 2-ddCt method was used to calculate the relative expression level of transcript [relative expression ratio (RER)] in target-gene knockdown samples. RER of nontarget control was set as 1.0. Right panel, Percentage of viable cells (compared with nontarget control) after siRNA knockdown assay was determined by alamarBlue. No significant toxicity was found in cells treated with Lipofectamine only. Data presented are the mean with sd of four separate experiments. *, P < 0.05 or **, P < 0.01 were considered statistically significant.
The lifelong expression profiles of specific genes encoding DNA methylation-modifying proteins are altered by neonatal exposure to EB or BPA
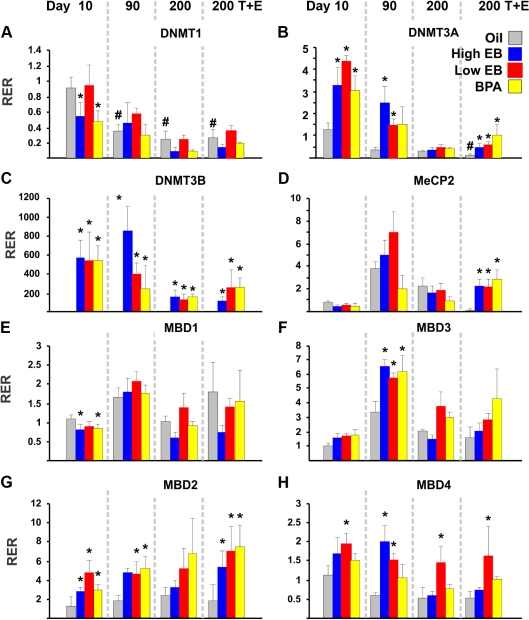
To investigate the mechanism by which neonatal EB/BPA treatment induced epigenetic reprogramming of specific genes through DNA methylation, we analyzed the expression of genes encoding proteins involved in regulating cytosine methylation and chromatin remodeling: DNA methyltransferases (Dnmt1, Dnmt3a, and Dnmt3b) and methyl-CpG binding domain proteins (Mecp2, Mbd1, Mbd2, Mbd3, and Mbd4). We observed three different patterns of expression among these genes: 1) genes not affected or moderately affected by neonatal EB/BPA treatments (Dnmt1, Mecp2, Mbd1, and Mbd3) (Fig. 5A and D–F), 2) genes dramatically induced to overexpress by these treatments (Dnmt3a, Dnmt3b, and Mbd2) (Fig. 5, B, C, and G), and 3) one gene responsive only to EB treatment (Mbd4) (Fig. 5H).
Fig. 5.
Relative gene expression was determined by real-time PCR for (A) Dnmt1, (B) Dnmt3a, (C) Dnmt3b, (D) Mecp2, (E) Mbd1, (F) Mbd3, (G) Mbd2, and (H) Mbd4. RNA was isolated from the dorsal prostate of animals treated with HEB (blue bar), LEB (red bar), BPA (yellow bar), or oil (gray bar) at d 10, 90, or 200 (±T+E2). Relative expression ratio (RER) was calculated by normalizing transcript levels of a gene to that of Rpl19 transcript level in the same sample. The average RER in samples from d 10 oil-treated prostates was arbitrarily assigned a value of 1.0 for each gene analyzed. Levels of transcripts in all other treatment groups at various life stages were normalized to the oil-treated controls. Statistical significant differences are indicated (*, P < 0.05, when comparing values in EB/BPA-treatment groups with oil-treated controls; #, P < 0.05, when values in older control animals were compared with values in d 10 controls).
Dnmt3a and Dnmt3b were generally highly expressed in animals neonatally exposed to EB or BPA in early life (d 10 and 90) but were expressed at very low levels or not at all in the oil-treated controls; the expression of these genes diminished with aging, and some of the overexpression induced in early life was significantly blunted at d 200 in the presence or absence of T+E2 treatments (Fig. 5, B and C). Despite their overlapping function as de novo methylating enzymes, neonatal EB/BPA treatment induced more dramatic overexpression of Dnmt3b (>100- to 800-fold) than that of Dnmt3a (0.5- to 4.5-fold), relative to levels in controls. The marked overexpression of mRNA of these enzymes at d 10 implicates their involvement in early-life reprogramming of DNA methylation patterns in target genes such as Nsbp1 or Hpcal1. The cancer-promoting T+E2 treatment stimulated expression of Dnmt3a and maintained overexpression of Dnmt3b as compared with levels in controls, suggesting a role for these enzymes in prostate tumorigenesis. Similarly, Mbd2 was the other gene exhibiting distinct up-regulation in response to neonatal EB/BPA treatments throughout life. The changes were more noticeable as the rats aged, and protracted T+E2 treatment further enhanced the up-regulation (Fig. 5G). Because these three genes were dramatically affected by neonatal EB/BPA treatments, we checked whether these changes were regulated by DNA methylation of their gene promoter. In silico analysis revealed the absence of CGI in the Dnmt3a promoter region (2-kb flanking region) but its presence in both Dnmt3b and Mbd2. Bisulfite sequencing of the CGI in the 5′ flanking region of each gene revealed no changes in DNA methylation (data not shown) (see Table 1 for primers). We found that the promoter region of both genes remained unmethylated in all treatment groups as well as in the oil-treated controls.
Mbd4 was up-regulated only in animals neonatally exposed to EB (Fig. 5H). The high EB effect was significant only at d 90, but the low EB treatment induced up-regulation throughout life. Interestingly, T+E2 treatment in adult life played a role in silencing Mecp2 expression in oil-treated controls but exerted no effects in groups treated neonatally with EB/BPA.
In summary, our results indicate that neonatal exposure to EB or BPA resulted in significant and persistent overexpression of Dnmt3a and Dnmt3b and Mbd2.
Discussion
Early-life exposure to hormones and hormone-mimicking compounds has been shown to affect the methylation status and expression of key regulatory genes in the prostate and other organs (23, 24, 37–39). We previously demonstrated that the Pde4d4 gene is susceptible to epigenetic reprogramming by neonatal exposure to EB and BPA in a two-hit PCa-induction model (23). Its promoter undergoes progressive hypermethylation in the neonatal oil-treated controls over time, with concordant gene silencing. However, after neonatal exposure to EB/BPA, the Pde4d4 promoter failed to execute this developmental program, resulting in overexpression of Pde4d4 in the experimental groups as animals aged. This deviant phenotype was apparent only during adulthood (not before puberty), persisted throughout life, and was not affected by the T+E treatment (a cancer-promoting regimen during adult life). In the present study, we conducted a careful comparative analysis of the effects of neonatal EB or BPA on promoter methylation status and gene expression of two genes (Nsbp1 and Hpcal1) previously not characterized for their susceptibility to neonatal reprogramming in this model with those of the well-studied Pde4d4. We demonstrated that neonatal EB/BPA-induced epigenetic changes in these genes are unique with respect to time of appearance, persistence during the lifespan, and response to adult-life events (e.g. sexual maturation, aging, and cancer-promoting events). Thus, we could clearly distinguish three patterns of epigenetic changes: 1) a class of genes like Nsbp1/Hmgn5 is a persistent epigenetic mark of early-life exposure, appearing early (at d 10), lasting throughout life, and not altered in adult life; 2) a second class of genes, represented by Pde4d4 (23), which is a concealed mark invisible at early life (d 10) and appears only when the animal reaches maturation at d 90 but persists throughout life; and 3) highly plastic epigenetic marks such as Hpcal1, whose appearance depends on the type of initial exposure (LEB vs. HEB or BPA) and events during adult life, including sexual maturation and hormone exposure. Thus, the fate of these epigenetic marks induced by early-life exposure is dependent on the specific gene, type of environmental exposure, and their responsiveness to modulation by adult-life events. Nevertheless, changes in the methylation status of these three promoters correlated in most cases with changes in gene expression, underscoring the importance of aberrations in promoter methylation driven by early environmental exposure in remodeling of transcriptional programs and development of disease in adult life.
We provided further evidence of a direct association between DNA methylation and gene silencing by employing a normal (NbE-1) and a malignant (AIT) prostate epithelial cell model. Promoter demethylation of Nsbp1 or Hpcal1 induced by 5-AZA-dC in these cell models was associated with increases in levels of gene transcript in these cell models. These data, together with similar findings reported previously for Pde4d4 (23), strongly suggest an important role for DNA methylation in the regulation of these three genes. Moreover, although the scope of the current in vitro studies is limited, we clearly showed that silencing of Nsbp1 and Pde4d4 mediated by RNA interference inhibited cell growth in both models, whereas silencing of Hpcal1 elicited a small increase in cell growth in NbE-1. In a preliminary study, we demonstrated that Hpcal1 also up-regulates cellular cAMP in NbE-1 cells (Tang, W.-y., unpublished data). Collectively, these data suggest that transcriptional changes in these genes may impact growth and other cellular functions of the rat prostate epithelium and likely prostate carcinogenesis or its risk.
The action of estrogen has long been known to involve intracellular cAMP that activates several signaling pathways, which all play critical roles in regulating cell proliferation/death and carcinogenesis (40, 41). Pde4d4 and Hpcal1 collaborate to maintain intracellular cAMP levels but in an opposing manner: although Pde4d4 is involved in degradation of cAMP, Hpcal1 stimulates its production (25, 26). We previously reported in this two-hit PCa-induction model that Pde4d4 is primarily hypomethylated and overexpressed after neonatal EB/BPA exposure (23). Here, we noted that Hpcal1 expression is generally diminished by high EB/BPA exposure. These opposite trends in promoter methylation and gene expression may lead to increased degradation (higher Pde4d4 expression) and reduced production (lower Hpcal1 expression) of cAMP, particularly during adult life. Although we have not yet investigated the consequences of such combined disruption in cAMP regulation, we have reported that neonatal exposure to EB/BPA increases predisposition to prostate carcinogenesis in the exposed rats (23). Our results are in agreement with reports demonstrating tumor-suppressor activities of Hpcal1 for squamous cell carcinoma of the skin and esophagus in rats and for human nonsmall cell lung carcinomas (26–28), although its relationship in PCa remains unknown. In contrast, Pde4d is a probable tumor promoter; it is overexpressed in human PCa specimens and has been shown to promote PCa growth in vitro and in vivo (42). Pde4 has also been implicated in other prostate diseases, including benign prostatic hyperplasia, lower urinary tract symptomatology, and bladder outlet obstruction (43). Last, E2 has been shown to alter cAMP in vivo in prostate tissues (44) and likely to engage in cross talk with androgen-receptor signaling in the prostate (45).
Human NSBP1 (HMGN5) belongs to a family of HMGN proteins, which interact directly with the nucleosome core particles in the euchromatin assembly (46). They can modify the architecture of active chromatin by reducing chromatin compaction, decreasing the repressive activity of histones, and facilitating transcription (47–50). HMGN proteins are tissue and development specific (51) and function as regulators of differentiation and transcription (46, 47). Apropos to NSBP1/HMGN5, its C terminus binds to linker histones (H1 or H5) and reduces their chromatin residence time, leading to chromatin decompaction and activation of transcription (52, 53). NSBP1/HMGN5 may have oncogenic activities, as demonstrated in glioma (29) and bladder (30). Two new studies showed overexpression of human Nsbp1 in PCa and that it promoted cell growth and metastasis and inhibited apoptosis in PCa cell lines (31, 32), a finding in agreement with our finding that RNA interference-mediated silencing of Nsbp1 inhibited rat prostatic epithelial cell growth.
In a previous study in mice (24), we found that exposure of neonates to DES/GEN induced reprogramming of Nsbp1 in the uterus, resulting in aberrant demethylation of the gene promoter and gene overexpression in adulthood (24). Coincidentally, the exposed rats also have a higher propensity to the development of uterine adenocarcinoma (54, 55). Of notable interest is our finding that aberrant Nsbp1 promoter methylation/gene expression and the development of uterine cancer occur only in intact, not ovariectomized, animals (24). Thus, it appears that the epigenetic memories laid down during early life only surface upon activation by an adult event (e.g. sexual maturation). This delay in the appearance of an early-life memory and its dependency on ovarian factors in the mouse uteri is in stark contrast to the epigenetic reprogramming of Nsbp1 by neonatal EB/BPA in the rat prostate, which appears early and persists throughout life, unaffected by adult-life events. The divergence in response of Nsbp1 to neonatal estrogen-reprogramming in the two animal models may be due to tissue-specific outcomes, a species-dependent effect, exposure to distinctly different reprogramming molecules, and/or a gender difference. Taken together, these results underscore the complexity of epigenetic reprogramming by early-life factors and strongly suggest the susceptibility of epigenetic reprogramming to modifications imparted by later-life influences.
DNA methylation, an integral part of epigenetic reprogramming, involves at least two classes of proteins, DNMT and MBD proteins (56). Here, we studied the expression of eight genes encoding these proteins after neonatal exposure to EB/BPA with and without T+E2 treatment during adulthood. Of the eight genes analyzed, levels of expression of Dnmt3a, Dnmt3b, Mbd2, and Mbd4 were those most affected by neonatal exposure to BPA and/or EB. Neonatal exposure resulted in the up-regulation of all four genes, with the greatest increase in expression in Dnmt3b. DNMT1 functions as a maintenance DNA methyltransferase, whereas DNMT3A and DNMT3B are involved in de novo methylation (57). Because Dnmt1 expression was unchanged by hormone treatments, altered DNA methylation in the affected genes after neonatal EB/BPA treatments must have relied on de novo methylation via Dnmt3a and Dnmt3b. Waves of genome-wide demethylation and de novo remethylation take place during germ-cell development and early embryogenesis to establish specific DNA methylation patterns in various tissues (58). But after birth, the expression of these enzymes is significantly diminished. The continued overexpression of Dnmt3a and Dnm3b in neonatal EB/BPA-treated groups, as compared with controls, may be the culprit in the aberrant methylation of susceptible genes involved in early-life reprogramming in this rat model. Furthermore, the fact that Dnmt3a and Dnmt3b were also overexpressed in response to the cancer-promoting T+E2 treatment implies that they also play a role during prostate carcinogenesis.
Although all members of the MBD family of proteins share a highly conserved methylated DNA binding domain and function to establish a locally compact chromatin and transcriptional repression (59), each member has been shown to play a distinct role in epigenetic regulation (60). Specifically, MBD2 and MBD4 appear to possess demethylating activities. Although debates continue, MBD2 has been shown to catalyze demethylation of methylated DNA and to mediate gene activation (61, 62), whereas MBD4 has a glycosylase domain that, upon activation of protein kinase C, promotes the incision of methylated DNA and base-excision repair, resulting in demethylation of MBD4-bound promoters (63). In this study, neonatal EB/BPA elicited significant overexpression of Mbd2 and Mbd4 throughout life, suggesting that they may promote aberrant promoter demethylation in target genes.
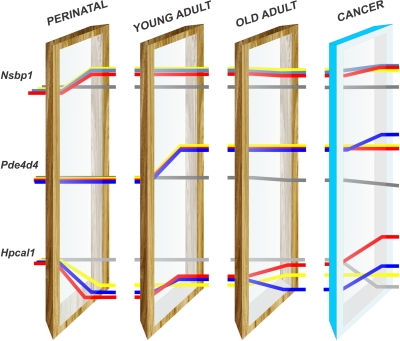
To summarize, we have identified three distinct patterns of epigenetic changes in genes responsive to neonatal exposure to EB/BPA (Fig. 6). In Fig. 6, we have represented the genetic makeup as light that passes through developmental windows. It is possible that, at each window, the genetic information is read and interpreted with epigenetic editing as if it is bent to follow a different path. In our rat model, methylation of the Nsbp1 promoter is a permanent epigenetic mark of early EB/BPA exposure and once edited in early life remains unchanged for life. On the other hand, methylation of the Pde4d4 promoter is a concealed epigenetic mark that surfaces only at sexual maturation. Most interestingly, methylation of the Hpcal1 promoter is a highly nimble mark, with its expression reflecting the interplay of early- and adult-life experiences. Its susceptibility to modifications by adult-life events makes it suitable for development into a biomarker for monitoring lifestyle changes in adulthood that can reverse early-life reprogramming, a key premise of disease prevention. Last, early-life modifications of key genes involved in DNA methylation and demethylation, such as Dnmt3a/b and Mbd2/4, may be responsible for early-life reprogramming as well as permitting a more dynamic regulation of the epigenome throughout life. The key question for the future is whether the epigenetics-induced changes in transcriptional programs impart a more adaptive or chaotic phenotype in the prostate of the adult rats. It is apparent from our findings that the complex interplay of the type of estrogen, timing of exposure, reproductive status, and aging significantly adds up to estrogen reprogramming of the prostate gland in early life and its later-life entropy, highlighting previously unrecognized mechanisms in prostate carcinogenesis.
Fig. 6.
Persistent, concealed, and nimble epigenetic marks were laid down in the prostates of rats exposed neonatally to EB/BPA. The genetic makeup is represented as white light encountering different developmental/life-stage windows and then diffracted through epigenetic modification to form a spectrum of responses. Some genes pass through with minimal diffraction, others are maximally bent. Nsbp1 is altered at the very first developmental window and represents an epigenetic mark that persists throughout life unaffected by later events (puberty, aging, or a cancer promoting regimen). At this life stage, the expression of Nsbp1 is increased in all the cohorts examined (EB/BPA). The increase in gene expression is illustrated by the position of the lines relative to where they started (at birth). Pde4d4 was unaffected by the neonatal window and was overexpressed after passing through the second panel (young adult; d 90); apparently, the light was diffracted only until it encountered the second window (puberty). At this point, adult hormones interact with the early epigenetic modifications and affect the phenotype (gene expression). The changes in Pde4d4 expression are not further affected by later-life events, such as aging or a cancer-promoting regimen (T+E2 treatment). Hpcal1 is highly nimble mark; the path of the light is altered both by the early event and by later-life events (puberty, aging, and T+E2 treatment), which is reflected in the phenotype (changes in gene expression). This illustrates the reversibility of early epigenetic reprogramming and the difference between EB and BPA exposure. Only the LEB (red line) could be reversed by later-life events. Furthermore, the response to cancer-promoting treatment clearly illustrates the differences between EB and BPA exposure. Only the EB cohorts showed increased expression of Hpcal1 (red and blue lines).
Acknowledgments
We thank Nadia Khan and Hong Xiao for technical support in sample preparation, Dr. Marian Miller for assistance in graphic design, Dr. Linda Levin for helpful discussion on the statistics, and Dr. Sonia Godoy-Tundidor for assistance in the preparation of the manuscript. We also thank Nancy Voynow for her professional editing of the manuscript.
Present address for W.-y.T.: Division of Molecular and Translational Toxicology, Department of Environmental Health Sciences, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland 21205.
Present address for L.M.M.: Department of Biology, Canisius College, Buffalo, New York 14208.
This work is in part supported by National Institutes of Health Grants CA136023 (to G.S.P.), DK40890 (to G.S.P.), ES015584 (to G.S.P. and S.M.H.), ES018758 (to G.S.P. and S.M.H.), CA112532 (to S.M.H.), ES018789 (to S.M.H.), ES006096 (to S.M.H.), ES019480 (to S.M.H.), ES020988 (to S.M.H.), and ES016817 (to W.-y.T.) and by the Department of Veterans Affairs Award BX000675 (to S.M.H.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- 5-AZA-dC
- 5-Aza-deoxycytidine
- BPA
- bisphenol A
- BW
- body weight
- CGI
- CpG island
- Ct
- threshold cycle
- DES
- diethylstilbestrol
- DMSO
- dimethylsulfoxide
- Dnmt3a
- DNA methyltransferase-3a
- DP
- dorsal lobe
- E2
- 17β-estradiol
- EB
- 17β-estradiol-3-benzoate
- GEN
- genistein
- HEB
- high-dose EB
- Hmgn5
- high-mobility group nucleosome binding domain 5
- Hpcal1
- hippocalcin-like 1
- LEB
- low-dose EB
- Mbd2
- methyl-CpG binding domain protein 2
- Mecp2
- methyl CpG binding protein 2
- MSPCR
- methylation-specific PCR
- Nsbp1
- nucleosome binding protein-1
- PCa
- prostate cancer
- Pde4d4
- phosphodiesterase 4D variant 4
- Rpl19
- ribosomal protein L19
- siRNA
- small interfering RNA
- T
- testosterone.
References
- 1. Bostwick DG, Burke HB, Djakiew D, Euling S, Ho SM, Landolph J, Morrison H, Sonawane B, Shifflett T, Waters DJ, Timms B. 2004. Human prostate cancer risk factors. Cancer 101:2371–2490 [DOI] [PubMed] [Google Scholar]
- 2. Brawley OW, Jani AB, Master V. 2007. Prostate cancer and race. Curr Probl Cancer 31:211–225 [DOI] [PubMed] [Google Scholar]
- 3. Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, Rafnar T, Gudbjartsson D, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Blondal T, Stacey SN, Helgason A, Gunnarsdottir S, Olafsdottir A, Kristinsson KT, Birgisdottir B, Ghosh S, Thorlacius S, Magnusdottir D, Stefansdottir G, Kristjansson K, et al. 2007 Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet 39:977–983 [DOI] [PubMed] [Google Scholar]
- 4. Crawford ED. 2003. Epidemiology of prostate cancer. Urology 62:3–12 [DOI] [PubMed] [Google Scholar]
- 5. Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. 2000. Environmental and heritable factors in the causation of cancer-analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343:78–85 [DOI] [PubMed] [Google Scholar]
- 6. Verkasalo PK, Kaprio J, Koskenvuo M, Pukkala E. 1999. Genetic predisposition, environment and cancer incidence: a nationwide twin study in Finland, 1976–1995. Int J Cancer 83:743–749 [DOI] [PubMed] [Google Scholar]
- 7. Tang WY, Ho SM. 2007. Epigenetic reprogramming and imprinting in origins of disease. Rev Endocr Metab Disord 8:173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Prins GS. 2008. Endocrine disruptors and prostate cancer risk. Endocr Relat Cancer 15:649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang X, Ho SM. 2011. Epigenetics meets endocrinology. J Mol Endocrinol 46:R11–R32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Price D. 1963. Comparative aspects of development and structure in the prostate. Natl Cancer Inst Monogr 12:1–27 [PubMed] [Google Scholar]
- 11. Prins GS. 1989. Differential regulation of androgen receptors in the separate rat prostate lobes: androgen independent expression in the lateral lobe. J Steroid Biochem 33:319–326 [DOI] [PubMed] [Google Scholar]
- 12. Bateson P, Barker D, Clutton-Brock T, Deb D, D'Udine B, Foley RA, Gluckman P, Godfrey K, Kirkwood T, Lahr MM, McNamara J, Metcalfe NB, Monaghan P, Spencer HG, Sultan SE. 2004. Developmental plasticity and human health. Nature 430:419–421 [DOI] [PubMed] [Google Scholar]
- 13. Cnattingius S, Lundberg F, Sandin S, Grönberg H, Iliadou A. 2009. Birth characteristics and risk of prostate cancer: the contribution of genetic factors. Cancer Epidemiol Biomarkers Prev 18:2422–2426 [DOI] [PubMed] [Google Scholar]
- 14. Ekbom A, Wuu J, Adami HO, Lu CM, Lagiou P, Trichopoulos D, Hsieh C. 2000. Duration of gestation and prostate cancer risk in offspring. Cancer Epidemiol Biomarkers Prev 9:221–223 [PubMed] [Google Scholar]
- 15. Ross RK, Henderson BE. 1994. Do diet and androgens alter prostate cancer risk via a common etiologic pathway? J Natl Cancer Inst 86:252–254 [DOI] [PubMed] [Google Scholar]
- 16. Ekbom A. 1998. Growing evidence that several human cancers may originate in utero. Semin Cancer Biol 8:237–244 [DOI] [PubMed] [Google Scholar]
- 17. Sequoia JS, Wright ME, McCarron P, Pietinen P, Taylor PR, Virtamo J, Albanes D. 2006. A prospective investigation of height and prostate cancer risk. Cancer Epidemiol Biomarkers Prev 15:2174–2178 [DOI] [PubMed] [Google Scholar]
- 18. Rohrmann S, Sutcliffe CG, Bienstock JL, Monsegue D, Akereyeni F, Bradwin G, Rifai N, Pollak MN, Agurs-Collins T, Platz EA. 2009. Racial variation in sex steroid hormones and the insulin-like growth factor axis in umbilical cord blood of male neonates. Cancer Epidemiol Biomarkers Prev 18:1484–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bonasio R, Tu S, Reinberg D. 2010. Molecular signals of epigenetic states. Science 330:612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berger SL. 2009. Epigenetic inheritance and replicative cellular aging. In: Saddone-Corsi P. ed. Epigenetic control and neuronal function 1–5 Washington, DC: Society for Neuroscience [Google Scholar]
- 21. Jablonka E, Lamb MJ. 2002. The changing concept of epigenetics. Ann NY Acad Sci 981:82–96 [DOI] [PubMed] [Google Scholar]
- 22. Anway MD, Cupp AS, Uzumcu M, Skinner MK. 2005. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308:1466–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ho SM, Tang WY, Belmonte de Frausto J, Prins GS. 2006. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res 66:5624–5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tang WY, Newbold R, Mardilovich K, Jefferson W, Cheng RY, Medvedovic M, Ho SM. 2008. Persistent hypomethylation in the promoter of nucleosomal binding protein 1 (Nsbp1) correlates with overexpression of Nsbp1 in mouse uteri neonatally exposed to diethylstilbestrol or genistein. Endocrinology 149:5922–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Braunewell KH. 2005. The darker side of Ca2+ signaling by neuronal Ca2+-sensor proteins: from Alzheimer's disease to cancer. Trends Pharmacol Sci 26:345–351 [DOI] [PubMed] [Google Scholar]
- 26. Gonzalez Guerrico AM, Jaffer ZM, Page RE, Braunewell KH, Chernoff J, Klein-Szanto AJ. 2005. Visinin-like protein-1 is a potent inhibitor of cell adhesion and migration in squamous carcinoma cells. Oncogene 24:2307–2316 [DOI] [PubMed] [Google Scholar]
- 27. Wickborn C, Klein-Szanto AJ, Schlag PM, Braunewell KH. 2006. Correlation of visinin-like-protein-1 expression with clinicopathological features in squamous cell carcinoma of the esophagus. Mol Carcinog 45:572–581 [DOI] [PubMed] [Google Scholar]
- 28. Fu J, Fong K, Bellacosa A, Ross E, Apostolou S, Bassi DE, Jin F, Zhang J, Cairns P, Ibañez de Caceres I, Braunewell KH, Klein-Szanto AJ. 2008. VILIP-1 downregulation in non-small cell lung carcinomas: mechanisms and prediction of survival. PLoS One 3:e1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qu J, Yan R, Chen J, Xu T, Zhou J, Wang M, Chen C, Yan Y, Lu Y. 2011. HMGN5: a potential oncogene in gliomas. J Neurooncol 104:729–736 [DOI] [PubMed] [Google Scholar]
- 30. Wahafu W, He ZS, Zhang XY, Zhang CJ, Yao K, Hao H, Song G, He Q, Li XS, Zhou LQ. 2011. The nucleosome binding protein NSBP1 is highly expressed in human bladder cancer and promotes the proliferation and invasion of bladder cancer cells. Tumour Biol 32:931–939 [DOI] [PubMed] [Google Scholar]
- 31. Jiang N, Zhou LQ, Zhang XY. 2010. Downregulation of the nucleosome-binding protein 1 (NSBP1) gene can inhibit the in vitro and in vivo proliferation of prostate cancer cells. Asian J Androl 12:709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Song G, Zhou LQ, Weng M, He Q, He ZS, Hao JR, Pan BN, Na YQ. Zhonghua Yi Xue Za Zhi 2006 Expression of nucleosomal binding protein 1 in normal prostate, benign prostate hyperplasia, and prostate cancer and significance thereof. Zhonghua Yi Xue Za Zhi 86:1962–1965 [PubMed] [Google Scholar]
- 33. Prins GS, Ye SH, Birch L, Ho SM, Kannan K. 2011. Serum bisphenol A pharmacokinetics and prostate neoplastic responses following oral subcutaneous exposures in neonatal Sprague-Dawley rats. Reprod Toxicol. 31:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee C, Prins GS, Henneberry MO, Grayhack JT. 1981. Effect of estradiol on the rat prostate in the presence and absence of testosterone and pituitary. J Androl 2:293–299 [Google Scholar]
- 35. Chang SM, Chung LW. 1989. Interaction between prostatic fibroblast and epithelial cells in culture: role of androgen. Endocrinology 125:2719–2727 [DOI] [PubMed] [Google Scholar]
- 36. Ho SM, Leav I, Damassa D, Kwan PW, Merk FB, Seto HS. 1988. Testosterone-mediated increase in 5α-dihydrotestosterone content, nuclear androgen receptor levels, and cell division in an androgen-independent prostate carcinoma of Noble rats. Cancer Res 48:609–614 [PubMed] [Google Scholar]
- 37. Bromer JG, Zhou Y, Taylor MB, Doherty L, Taylor HS. 2010. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. FASEB J 24:2273–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li S, Washburn KA, Moore R, Uno T, Teng C, Newbold RR, McLachlan JA, Negishi M. 1997. Developmental exposure to diethylstilbestrol elicits demethylation of estrogen-responsive lactoferrin gene in mouse uterus. Cancer Res 57:4356–4359 [PubMed] [Google Scholar]
- 39. Li S, Hansman R, Newbold R, Davis B, McLachlan JA, Barrett JC. 2003. Neonatal diethylstilbestrol exposure induces persistent elevation of c-fos expression and hypomethylation in its exon-4 in mouse uterus. Mol Carcinog 38:78–84 [DOI] [PubMed] [Google Scholar]
- 40. Aronica SM, Kraus WL, Katzenellenbogen BS. 1994. Estrogen action via the cAMP signaling pathway: stimulation of adenylate cyclase and cAMP-regulated gene transcription. Proc Natl Acad Sci USA 91:8517–8521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ciullo I, Diez-Roux G, Di Domenico M, Migliaccio A, Avvedimento EV. 2001. cAMP signaling selectively influences Ras effectors pathways. Oncogene 20:1186–1192 [DOI] [PubMed] [Google Scholar]
- 42. Rahrmann EP, Collier LS, Knutson TP, Doyal ME, Kuslak SL, Green LE, Malinowski RL, Roethe L, Akagi K, Waknitz M, Huang W, Largaespada DA, Marker PC. 2009. Identification of PDE4D as a proliferation promoting factor in prostate cancer using a sleeping beauty transposon-based somatic mutagenesis screen. Cancer Res 69:4388–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Uckert S, Oelke M, Stief CG, Andersson KE, Jonas U, Hedlund P. 2006. Immunohistochemical distribution of cAMP- and cGMP-phosphodiesterase (PDE) isoenzymes in the human prostate. Eur Urol 49:740–745 [DOI] [PubMed] [Google Scholar]
- 44. Nakhla AM, Khan MS, Romas NP, Rosner W. 1994. Estradiol causes the rapid accumulation of cAMP in human prostate. Proc Natl Acad Sci USA 91:5402–5405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Merkle D, Hoffmann R. 2011. Roles of cAMP and cAMP-dependent protein kinase in the progression of prostate cancer: cross-talk with the androgen receptor. Cell Signal 23:507–515 [DOI] [PubMed] [Google Scholar]
- 46. Bustin M. 2001. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem Sci 26:431–437 [DOI] [PubMed] [Google Scholar]
- 47. Bustin M, Trieschmann L, Postnikov YV. 1995. The HMG-14/-17 chromosomal protein family: architectural elements that enhance transcription from chromatin templates. Semin Cell Biol 6:247–255 [DOI] [PubMed] [Google Scholar]
- 48. Gerlitz G. 2010. HMGNs, DNA repair and cancer. Biochim Biophys Acta 1799:80–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pogna EA, Clayton AL, Mahadevan LC. 2010. Signalling to chromatin through post-translational modifications of HMGN. Biochim Biophys Acta 1799:93–100 [DOI] [PubMed] [Google Scholar]
- 50. Rochman M, Malicet C, Bustin M. 2010. HMGN5/NSBP1: a new member of the HMGN protein family that affects chromatin structure and function. Biochim Biophys Acta 1799:86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shirakawa H, Landsman D, Postnikov YV, Bustin M. 2000. NBP-45, a novel nucleosomal binding protein with a tissue-specific and developmentally regulated expression. J Biol Chem 275:6368–6374 [DOI] [PubMed] [Google Scholar]
- 52. Rochman M, Postnikov Y, Correll S, Malicet C, Wincovitch S, Karpova TS, McNally JG, Wu X, Bubunenko NA, Grigoryev S, Bustin M. 2009. The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol Cell 35:642–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Malicet C, Rochman M, Postnikov Y, Bustin M. 2011. Distinct properties of human HMGN5 reveal a rapidly evolving but functionally conserved nucleosome binding protein. Mol Cell Biol 31:2742–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Newbold RR, Bullock BC, McLachlan JA. 1990. Uterine adenocarcinoma in mice following developmental treatment with estrogens: a model for hormonal carcinogenesis. Cancer Res 50:7677–7681 [PubMed] [Google Scholar]
- 55. Newbold RR, Banks EP, Bullock B, Jefferson WN. 2001. Uterine adenocarcinoma in mice treated neonatally with genistein. Cancer Res 61:4325–4328 [PubMed] [Google Scholar]
- 56. Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. 2005. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol Cell 19:667–678 [DOI] [PubMed] [Google Scholar]
- 57. Nimura K, Ishida C, Koriyama H, Hata K, Yamanaka S, Li E, Ura K, Kaneda Y. 2006. Dnmt3a2 targets endogenous Dnmt3L to ES cell chromatin and induces regional DNA methylation. Genes Cells 11:1225–1237 [DOI] [PubMed] [Google Scholar]
- 58. Reik W, Dean W, Walter J. 2001. Epigenetic reprogramming in mammalian development. Science 293:1089–1093 [DOI] [PubMed] [Google Scholar]
- 59. Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. 1989. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell 58:499–507 [DOI] [PubMed] [Google Scholar]
- 60. Dhasarathy A, Wade PA. 2008. The MBD protein family-reading an epigenetic mark? Mutat Res 647:39–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hamm S, Just G, Lacoste N, Moitessier N, Szyf M, Mamer O. 2008. On the mechanism of demethylation of 5-methylcytosine in DNA. Bioorg Med Chem Lett 18:1046–1049 [DOI] [PubMed] [Google Scholar]
- 62. Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. 1999. A mammalian protein with specific demethylase activity for mCpG DNA. Nature 397:579–583 [DOI] [PubMed] [Google Scholar]
- 63. Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, Matsumoto T, Fujiyama S, Shirode Y, Yamaoka I, Kitagawa H, Takeyama K, Shibuya H, Ohtake F, Kato S. 2009. DNA demethylation in hormone-induced transcriptional derepression. Nature 461:1007–1012 [DOI] [PubMed] [Google Scholar]