Abstract
Adiponectin overexpression in mice increases insulin sensitivity independent of adiposity. Here, we combined stable isotope infusion and in vivo measurements of lipid flux with transcriptomic analysis to characterize fatty acid metabolism in transgenic mice that overexpress adiponectin via the aP2-promoter (ADNTg). Compared with controls, fasted ADNTg mice demonstrated a 31% reduction in plasma free fatty acid concentrations (P = 0.008), a doubling of ketones (P = 0.028), and a 68% increase in free fatty acid turnover in plasma (15.1 ± 1.5 vs. 25.3 ± 6.8 mg/kg · min, P = 0.011). ADNTg mice had 2-fold more brown adipose tissue mass, and triglyceride synthesis and turnover were 5-fold greater in this organ (P = 0.046). Epididymal white adipose tissue was slightly reduced, possibly due to the approximately 1.5-fold increase in the expression of genes involved in oxidation (peroxisome proliferator-activated receptor α, peroxisome proliferator-activated receptor-γ coactivator 1α, and uncoupling protein 3). In ADNTg liver, lipogenic gene expression was reduced, but there was an unexpected increase in the expression of retinoid pathway genes (hepatic retinol binding protein 1 and retinoic acid receptor beta and adipose Cyp26A1) and liver retinyl ester content (64% higher, P < 0.02). Combined, these data support a physiological link between adiponectin signaling and increased efficiency of triglyceride synthesis and hydrolysis, a process that can be controlled by retinoids. Interactions between adiponectin and retinoids may underlie adiponectin's effects on intermediary metabolism.
Achieving glycemic control, managing dyslipidemia, and improving insulin sensitivity are major therapeutic goals in the management of diabetes and metabolic syndrome (1). Controlling both plasma free fatty acid (FFA) and glucose concentrations may be needed to restore insulin sensitivity (2–4) because mixed meal studies and clamp experiments have demonstrated metabolic interactions between glucose, FFA and insulin sensitivity (5–7). For instance, the role of FFA to inhibit the phosphatidylinositol 3-kinase-mediated insulin signaling pathway is balanced by insulin's suppression of lipolysis (2, 4), and mounting evidence demonstrates that FFAs also play a significant role in regulating insulin secretion and hepatic glucose output or gluconeogenesis (8–12). Thus, excess circulating FFA can significantly contribute to metabolic dysregulation by multiple mechanisms (10, 11). Therapeutically, drugs that exert a combined lipid and glucose-lowering effect may be most effective in the management of diabetes [e.g. thiazolidinediones and metformin (14)] (13), but the side effects of these drugs can limit their effectiveness and better agents are needed. Molecules with similar salutary, integrated effects on FFA levels, and glucose homeostasis would offer potential novel therapeutic approaches. Adiponectin, a high molecular weight, secreted adipokine, may be such a molecule (15–17). Plasma concentrations of adiponectin are inversely associated with obesity and type 2 diabetes (18), and the protein can elicit potent antidiabetic effects similar to thiazolidinedione and other peroxisome proliferator-activated receptor γ (PPARγ) agonists (18–20). Adiponectin limits triglyceride (TG) accumulation in liver (21, 22), increases glucose and TG clearance, and suppresses hepatic glucose output (23). These antidiabetic actions of adiponectin may be via its regulation of FFA flux.
Transgenic mice that overexpress adiponectin under the control of the aP2 promoter (ADNTg) provide a model for studying the potential mechanisms through which adiponectin may integrate metabolic responses, particularly FFA flux. In the present study, we investigated how adiponectin regulates TG metabolism and fatty acid turnover in vivo. The present article demonstrates: 1) a significant increase in fatty acid turnover in the adiponectin-overexpressing mouse, 2) an association between adiponectin-induced stimulation of TG synthesis/breakdown and fatty acid disposal, and 3) a pattern of gene expression supporting a possible role for retinoids in the metabolic effects mediated by adiponectin.
Materials and Methods
Mice
Adiponectin-overexpressing (ADNTg) mice and corresponding wild-type (WT) littermate controls on a C57BL/6 background were used for all studies (23). Male animals were used for all the experiments to minimize effects of hormonal fluctuations in female animals. Most animals were between 12 and 16 wk of age, and ADNTg mice were generated as described previously (23). All mice were backcrossed more than 10 times to the C57BL/6 background. For gene expression analysis, mice were maintained on a 12-h light, 12-h dark cycle (0600–1800 h light) and housed in groups of two to four with unlimited access to water and chow (no. 5058; LabDiet, Henderson, CO). To obtain fasting samples, food was removed at 0900 h and the animals killed at 1200 h. The Institutional Animal Care and the Use Committee of University of Texas Southwestern Medical Center approved all animal experiments.
Real-time quantitative PCR
Liver, epididymal white adipose tissue (EWAT), and retroperitoneal subcutaneous fat (SubQ) were harvested, snap frozen, and tissues were stored at −80 C until analysis. RNA extraction and purification were performed using TRIzol reagent (Invitrogen, Carlsbad, CA) and RNeasy Mini kit (QIAGEN, Valencia, CA) as per protocol for tissues. RNA was reverse transcribed to cDNA by the reverse transcriptase reaction using reagents and enzymes from Invitrogen. Gene expression was measured by quantitative PCR using the SYBR green supermix from Bio-Rad (Hercules, CA). The relative expression levels were calculated by the comparative cycle threshold method. Primer sequences can be found in the Supplemental data, published on The Endocrine Society's Journals Online web site at http://endo.endojournals.org.
Microarray experiment and analysis
Previous studies suggested that adipose or liver tissue may be primary target organs of adiponectin (22), and gene transcript profiling was performed on EWAT and liver tissue, with a focus on markers of fatty acid oxidation and mitochondrial activity. These microarray data were generated on the FVB background and hence were used as a reference dataset only. Overall, 1236 and 526 probes (corresponding to 1127 and 459 unique Entrez Gene ID) met the 1.2-fold change and P < 0.05 cutoff in EWAT and liver, when comparing WT and ADNTg animals (data not shown). In addition to informing us on the generalizability of the impact of the transgene on lipid metabolism, genes for retinol metabolism were also inspected. Thus, the array data presented in Table 1 are included for comparison with PCR data and for comparison with observed phenotypes (increased retiny lesters present in tissues). These results are presented (even when the array findings were below a 1.2-fold change or even when not significant) for consideration by the reader within the totality of the data. Animals were euthanized by CO2 asphyxiation and liver, and EWAT were harvested and flash frozen in liquid N2. Total RNA was isolated after homogenizing the frozen tissues in TRIzol reagent (Invitrogen) with a PT10/35 Polytron (Kinematica AG, Clifton, NJ) and processed using RNeasy kits (QIAGEN) according to the manufacturers' instructions. RNA concentrations were quantified from absorbance at 260 nm. Microarray processing was performed as previously described (24). Briefly, labeled cRNA was hybridized for 48 h on Agilent 60-mer two-color spotted microarrays (Agilent Technologies, Santa Clara, CA). Individual samples (including individual WT samples) were hybridized against the WT control pool. Fold-change and P values were generated by averaging replicates (two to three replicates per strain) and using the Rosetta Resolver version 6.0 software (Microsoft, Bellevue, WA). Fold-change values represent the difference in regulation of the ADNTg samples vs. the WT control samples where a positive value signifies up-regulation in the ADNTg compared with WT and vice versa.
Table 1.
Microarray data from ADNTg mice compared to controls
| Entrez gene name | Fold-change | P value | |
|---|---|---|---|
| EWAT | |||
| PGC1α | Peroxisome proliferator-activated receptor γ, coactivator 1 α | 1.8 | <0.05 |
| PPARβ/δ | Peroxisome proliferator activator receptor δ | 1.2 | <0.05 |
| PPARα | Peroxisome proliferator-activated receptor α | 2.7 | <0.01 |
| ACOX1 | Acyl-coenzyme A oxidase 1, palmitoyl | 1.3 | 0.4600 |
| NCoA1 | Nuclear receptor coactivator 1 | 1.4 | <0.01 |
| HSL | Hormone sensitive lipase | 1.2 | <0.001 |
| PNPLA2 | Patatin-like phospholipase domain containing 2 | 1.7 | <0.01 |
| PRDM16 | PR domain containing 16 | 1.3 | <0.01 |
| CIDEA | Cell death-inducing DFFA-like effector a | 4.2 | <0.001 |
| UCP3 | Uncoupling protein 3 (mitochondrial, proton carrier) | 1.5 | <0.05 |
| STAT5A | Signal transducer and activator of transcription 5A | 1.4 | <0.05 |
| STAT5B | Signal transducer and activator of transcription 5B | 1.3 | <0.001 |
| CYP26B1 | Cytochrome P450, family 26, subfamily B, polypeptide 1 | 1.4 | <0.05 |
| LIPA | Lysosomal acid lipase A (cholesterol ester hydrolase) | −1.6 | <0.01 |
| PEPCK | Phosphoenolpyruvate carboxykinase 1, cytosolic | 2.1 | 0.0810 |
| Liver tissue | |||
| HMGCS1 | 3-Hydroxy-3-methylglutaryl-Coenzyme A synthase 1 | 1.1 | 0.6890 |
| ACAA1b | Ketothiolase, acetyl-Coenzyme A acyltransferase 1B | −1.1 | 0.1730 |
| SREBP1c | Sterol regulatory element binding transcription factor 1 | −1.1 | 0.2240 |
| FAS | Fatty acid synthase | −1.6 | <0.001 |
| ACL | ATP citrate lyase | −1.3 | <0.05 |
| SCD1 | Stearoyl-Coenzyme A desaturase 1 | −2.1 | <0.001 |
| GPAT | Glycerol-3-phosphate acyltransferase, mitochondrial | −1.3 | <0.001 |
| HSL | Hormone sensitive lipase | 1.0 | 0.6740 |
| PNPLA2 | Patatin-like phospholipase domain containing 2 | 1.1 | <0.01 |
| RBP1 | Retinol binding protein 1, cellular | 2.6 | <0.001 |
| RARβ | Retinoic acid receptor, β | 1.4 | <0.001 |
| TRIM24 | Tripartite motif-containing 24 (RXR pathway) | 1.4 | <0.05 |
| DUSP1 | Dual specificity phosphatase 1 (RXR pathway) | −1.7 | <0.001 |
| LIPA | Lysosomal acid lipase A (cholesterol ester hydrolase) | −1.3 | <0.001 |
| PEPCK | Phosphoenolpyruvate carboxykinase 1, cytosolic | −1.5 | 0.1730 |
Differences between ADNTg and WT mice that are significant are listed as <0.05, <0.01, <0.001, and fold changes reflect the level of the ADNTg relative to WT. Microarray data are presented for comparison when transcripts were assessed by PCR (even when microarray data are not significant).
Measurement of tissue retinoids and serum retinol binding protein 4 (RBP4) levels
Retinyl esters were extracted from liver and SubQ tissue (50–100 mg) in a 2:1:1 CHCl3, MeOH, and saline solution using a tissue douncer. After vortexing, samples were centrifuged at 3000 rpm at 4 C for 10 min. The organic layer (bottom) was removed, transferred to a new vial, and tissue was subjected to an additional extraction using 3 ml of hexane. The organic layers were combined, solvents were evaporated under a stream of nitrogen, and extracts were resuspended in 150 μl of ethanol. For SubQ, extracts were resuspended in a 2:1 solution of tetrahydrofuran:ethanol to ensure solubility. Extracts (70 μl) were used immediately for high-performance liquid chromatography. Retinoids were separated using reversed phase high-performance liquid chromatography with a Percosphere-3 C18 83 × 4.6 mm column (PerkinElmer, Inc., Norwalk, CT) and detected with a photodiode array detection system (Waters 996). Mobile phase A consisted of 65% methanol and 35% water, and mobile phase B consisted of 100% methanol. The flow rate for each run was 1 ml/min for the duration of the gradient. The gradient started at 30% A/70% B and linearly increased to 100% B over 7 min, followed by an isocratic gradient of 100% B for 35 min before equilibrating back to 30% A/70% B over 10 min. Standard curves were generated using external standards of all-trans retinol and all-trans retinyl-palmitate to quantify retinoid concentrations in tissues. Serum levels of RBP4 protein were measured using a polyclonal rabbit polyclonal, antimouse RBP4 (Adipogen AG-25A-0036; Adipogen, Incheon, Korea) diluted 1:2000 in Tris-buffered saline with Tween 20, 5% nonfat milk. Serum was diluted 40×, and 10 μl were loaded onto the gel (i.e. 0.25 μl of serum per sample). A goat antirabbit antibody was conjugated with IRDye 800CW (LI-COR Bioscience, Lincoln, NE) in a 1:5000 dilution in PBS with Tween 20. The bands were visualized with an Odyssey instrument (using the 800 channel; LI-COR Bioscience).
Measurement of lipid synthesis, flux and composition, and histology
Rates of fatty acid synthesis were measured in 12-wk-old mice during the early light cycle after a 2-h fast using tritiated water as described (25). For measurement of FFA kinetics, tissue -TG synthesis, and turnover, animals were adapted to a reverse light-dark cycle (2000–0800 h light cycle) for 4 wk, so that studies could be conducted at the end of their acclimated 12-h fast and during the human daylight hours. Jugular catheters were implanted 5 d before the experiment and animals monitored for full recovery. On the day of the experiment, 4-13C-palmitate, complexed with albumin in a molar ratio 2:1, was infused at a rate of 6.50 nmol/min in saline, using a pump rate of 7.5 μl/min from 0900 to 1200 h (final data collected after 16 h with no food). At the end of the infusion, liver, brown adipose tissue (BAT), EWAT, SubQ, and skeletal muscle were dissected, weighed, and frozen in liquid N2 for later isotope analysis. Plasma was isolated from whole blood and tissue-TG, very low-density lipoprotein (VLDL)-TG, and FFA were isolated and prepared for gas chromatography-mass spectrometry (GC/MS) as described previously (26). Briefly, VLDL (density <1.006 g/ml) was isolated from plasma by density gradient ultracentrifugation as described (27). To accomplish this, 300 μl of plasma were adjusted to 1 ml with a NaCL/KBr solution (density 1.006 g/ml), centrifuged at 100,000 rpm for 150 min, and the retrieved top layer (VLDL) was isolated. The lipid was extracted and lipid species separated by thin layer chromatography. Fatty acids in TG and in plasma FFA were derivatized to their methyl esters and analyzed by GC/MS performed on an HP 6890 with a Mass Selective Detector HP 5973 fitted with an ETP electron multiplier (SGE, Inc., Austin, TX) using a HP-1 25 meter column, with a 250-μm inner diameter and 0.33-μm film thickness and helium as the carrier gas. Electron impact was used to selectively monitor ions with mass/charge (m/z) ratios of 270 and 274. Comparable ion peak areas between standards and biological samples were achieved by either adjustment of the volume injected or by dilution or concentration of the sample. Enzyme kits were used to measure plasma FFA (Wako Chemicals, kit nos. 999-34691 and 991-34891) and VLDL-TG (Wako nos. 461-09092 and 461-08992; Wako, Richmond, VA). Plasma ketone concentrations from 6-h fasted mice were measured using the autokit total ketone bodies (Cyclic Enzymatic Method) from Wako Diagnostics (no. 411-73401). To visualize tissue lipids, BAT was collected in Histochoice, embedded in paraffin, and sections were stained with hematoxylin and eosin.
Statistical analysis and calculations
Values are presented as mean ± sd with samples sizes noted in the legends. Differences between the mouse strains were tested using StatView (version 5.0.1; SAS Institute, Inc., Berkeley, CA), and P < 0.05 was considered significant. The basis and assumptions for the calculations were as follows. Data presented for fatty acid flux and TG turnover are based on the use of palmitate as the tracer fatty acid. The percentages of TG liver, EWAT, BAT, skeletal muscle, SubQ, and plasma, derived from the plasma FFA palmitate pool, were calculated by dividing the enrichments (%) in TG-palmitate by the enrichments (%) in the plasma FFA pool at 3 h of infusion. The total quantities of TG in liver, VLDL, EWAT, and BAT depots from FFA were derived by multiplying these percentages by the total pool sizes of TG-fatty acids in those tissues. The total pool sizes of skeletal muscle-TG and SubQ-TG were not calculated, because their distributions in the body could not be definitively identified for isolation. The total blood volume in the mice was taken as 7% of body weight (28).
Results
Phenotypes and metabolite concentrations
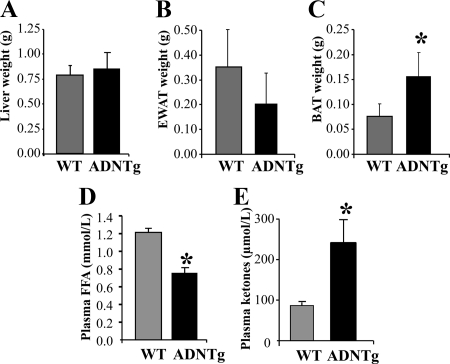
Body weights did not differ between chow-fed WT and ADNTg mice (27.5 ± 2.6 vs. 29.8 ± 4.2 g, respectively; P = 0.298), nor were there differences between the concentrations of fasting plasma VLDL-TG or total liver weights (Fig. 1A) in animals who had been acclimated to a 16-h fast (see Materials and Methods). The tissue weight of EWAT tended to be lower in ADNTg mice (P = 0.058) (Fig. 1B). However, BAT weight was increased more than 2-fold (P = 0.006) (Fig. 1C). Fasting plasma FFA concentrations were 70% that of WT (P = 0.008) (Fig. 1E), whereas ketone concentrations were 2.5-fold higher in the ADNTg mice (P = 0.028) (Fig. 1C).
Fig. 1.
Tissue weights and metabolite concentrations. Liver (A), EWAT (B), and BAT (C) tissue weights measured in chow-fed WT and ADNTg mice, 12 wk of age, along with fasting concentrations of FFA (five per group) (D) and plasma ketones (four per group) (E). *, P < 0.05.
Gene expression analysis
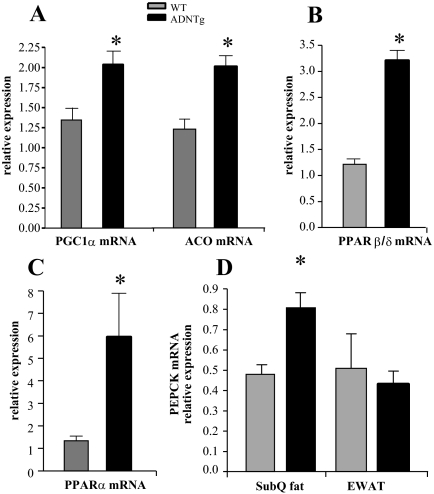
To assess changes in genes associated with fatty acid metabolism in the adiponectin overexpressing mice, mRNA levels were measured in adipose tissue using RT-PCR. In ADNTg animals, EWAT mRNA levels of transcriptional regulators of fatty acid oxidation, PPARγ coactivator 1α (PGC1)α, PPARβ/δ, and PPARα, were all significantly induced (Fig. 2, A–C). A downstream target of these transcription factors, acyl-coenzyme A oxidase (ACO), an enzyme involved in fatty acid oxidation, was also significantly up-regulated (Fig. 2D). Large-scale gene transcript profiling was performed on EWAT and liver tissue, with a focus on markers of fatty acid oxidation and mitochondrial activity. These data were generated on the FVB background and, as described in Materials and Methods, were used as a reference dataset. Analysis of gene expression agreed with the PCR findings (for PGC1α, PPARβ/δ, and PPARα but not for ACOX1) and mRNA levels of nuclear receptor coactivator (NCoA)1/steroid receptor coactivator (SRC)-1, and the lipases hormone sensitive lipase and patatin-like phospholipase domain containing 2 were also significantly induced in EWAT depots of ADNTg animals (Table 1). Thus, when the adipose dataset was analyzed using ingenuity pathway analysis software, oxidative phosphorylation and mitochondrial markers were among the major pathways significantly induced in EWAT depots of ADNTg mice. Interestingly, the specific brown fat markers PRD1-BF1-RIZ1 homologous domain containing 16 (PRDM16) and cell death-inducing DNA fragmentation factor-α-like effector (CIDEA) were induced in the arrays of EWAT derived from ADNTg animals. Higher levels of PRDM16 and Cidea were not associated with induction of uncoupling protein (UCP)1 expression, although a statistically significant 1.5-fold induction of UCP3 was found (Table 1). UCP3 induces fatty acid oxidation in skeletal muscle (29), but less is known about its role in EWAT. Adipose signal transducer and activator of transcription (STAT)5 activation turns on ACOX1 (30), the first enzyme in the peroxisomal β-oxidation pathway. Significant increases were also observed in the expression of the retinoid-responsive, P450 gene family Cyp26A1 (31) and lysosomal acid lipase A, in EWAT of ADNTg mice (Table 1). Retinoids or retinoic acid receptor ligands also up-regulate glyceroneogenesis in adipose tissue (32) via induction of phosphoenolpyruvate carboxykinase (PEPCK), a retinoid-responsive gene (33, 34) and a key regulator of the predominant source of TG-glycerol in adipose tissue (35). Interestingly, expression of the PEPCK mRNA was found to be significantly higher in SubQ but not in the EWAT of ADNTg mice compared with controls (Fig. 2D and in agreement with microarray in Table 1). This is consistent with an enhanced TG accumulation seen in SubQ adipose tissue at the expense of epididymal depots, similar to the effects induced by PPARγ agonists. Overall, the large-scale microarray analysis confirms a “browning” of WAT, i.e. white adipocytes that assume a transcriptional program leaning more toward the transcriptional program more characteristic of brown adipocytes, a pattern that we have previously observed (22, 23).
Fig. 2.
Effect of adiponectin overexpression on markers of fatty acid metabolism in EWAT. Expression of PGC1α (A), PPARβ/δ (B), PPARα (C), and ACO (ACOX/AOX) (D) from four WT and four ADNTg animals that were 6 h fasted in the light cycle. Fed sc and gonadal adipose expression of PEPCK mRNA (D) was quantified by RT-PCR in tissue from four WT and four ADNTg animals. The levels were determined using β-actin as the internal control. *, P < 0.05.
Impact of adiponectin overexpression on hepatic gene expression, retinyl ester concentrations, and expression of retinoid-associated genes
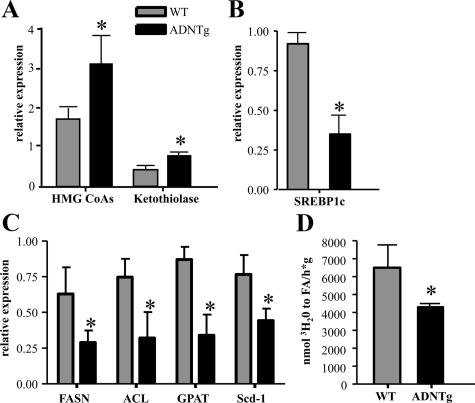
As shown above, low fasting FFA and increased ketone concentrations were observed in ADNTg mice. When liver gene expression was investigated, significant up-regulation of liver mitochondrial 3-hydroxy-3-methylglutaryl (HMG)-CoAs and peroxisomal ketothiolase was found (Fig. 3A). Expression of sterol regulatory element-binding protein (SREBP)-1c, a key transcriptional activator of the lipogenesis pathway (36), was significantly lower in ADNTg liver (Fig. 3B), as were the SREBP-1c target genes, ATP citrate lyase, fatty acid synthase, stearoyl-CoA desaturase-1, and glycerol-3-phosphate acyltransferase (Fig. 3C). Overall, the microarray data agreed with these later findings (Table 1). Consistent with the changes in lipogenic gene expression, hepatic de novo lipogenesis as measured by the incorporation of tritium into newly formed fatty acids was also significantly reduced in the ADNTg mice compared with control (P = 0.034) (Fig. 3D).
Fig. 3.
Expression of key liver lipid metabolism genes and rates of fatty acid synthesis. Expression of HMG-CoA synthase and ketothiolase (A), SREBP-1c (B), and SREBP-1c responsive genes (C) were measured via RT-PCR in four WT and four ADNTg mice that were 6 h fasted in the light cycle using β-actin as the internal control. The rates of hepatic fatty acid synthesis (D) were determined in the livers of five WT and five ADNTg animals using 3H2O ip injection. Measurements were all made in fed state, 2 h into the dark cycle, in 13-wk-old animals. *, P < 0.05.
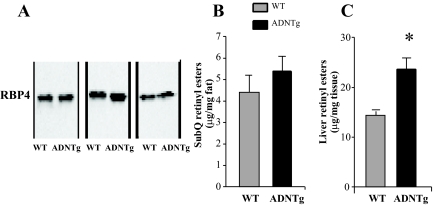
In the liver of ADNTg mice, expression of RBP1 and retinoic acid receptor beta, major components in retinoid metabolism and signaling, were also significantly increased (Table 1). Other genes associated with the retinoic acid receptor pathway, including Trim24, Dusp1, and lysosomal acid lipase A, were also up-regulated (Table 1). When the PEPCK data were inspected, it was not found not to be affected, which may have been due to the probe used. To further investigate a potential new relationship between adiponectin and retinoid metabolism, we measured the RBP4 protein levels in serum, which were similar between the strains (Fig. 4A). We also measured the concentrations of retinyl esters in liver and adipose tissues in ADNTg and WT mice. The retinyl ester concentrations in BAT and EWAT were below the detection limit of the assay, and the ester concentration was not higher in ADNTg SubQ (Fig. 4A). However, the retinyl ester concentrations in livers of ADNTg mice was 64% higher than controls (P = 0.019) (Fig. 4B). This increase in hepatic retinoid content could account for the enhanced expression of genes in retinoid metabolism and could contribute to the phenotype of the ADNTg mice. Retinoid metabolites in the liver may also account for the antisteatotic effect of adiponectin (37). These data provide evidence for an antilipogenic effect of adiponectin that may be associated with the increased retinoid content in the liver.
Fig. 4.
Retinyl esters in sc adipose tissue and liver, adipose PEPCK expression, and serum RBP4 levels. Western blot analysis of plasma RBP4 in three WT and three ADNTg mice (A). Retinyl ester concentrations were measured in SubQ adipose tissue (B) and in liver (C) in three WT and three ADNTg animals that were 6 h fasted in the light cycle. *, P < 0.05.
Kinetics of fatty acid turnover and TG synthesis
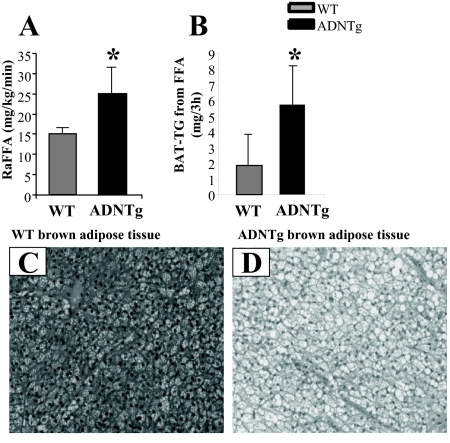
To determine whether the FFA lowering effect of adiponectin resulted from an altered lipolytic rate, we investigated in vivo fatty acid turnover using a 3-h, stable isotope infusion in 16-h fasted animals. The rate of appearance of FFA was increased 68% (P < 0.04) in ADNTg mice compared with WT (Fig. 5A). During the course of the 3-h FFA isotope infusion, significant synthesis of TG can occur (38). Thus, we investigated the labeling patterns of various TG pools to determine the disposal of FFA into TG synthesis. As shown in Table 2, the proportion of VLDL-TG synthesized from plasma FFA was significantly lower in ADNTg compared with controls (32.6 ± 6.6 vs. 43.2 ± 7.3%, respectively; P = 0.018) (Table 2). The same was found for the intrahepatic-TG pool (35.3 ± 5.5 vs. 42.9 ± 2.1%, respectively; P = 0.009), suggesting that plasma FFA entering the liver were trafficked away from TG storage and secretion (VLDL), likely into oxidation pathways. Synthesis of TG from plasma FFA in skeletal muscle and SubQ adipose were not different between the groups. The tendency for EWAT depots to weigh less in the ADNTg (P = 0.084) was associated with a lower absolute amount of TG synthesized from FFA (0.50 ± 0.27 vs. 0.54 ± 0.51), although this did not reach statistical significance (P = 0.099). By contrast, the larger BAT depot in the ADNTg was associated with a greater than 3-fold increase in TG synthesis and turnover (P = 0.046) (Table 2 and Fig. 5B). Histological staining of BAT demonstrated more lipid droplets in the ADNTg mice compared with WT (Fig. 5, C vs. D).
Fig. 5.
Plasma FFA appearance rate, BAT-TG synthesis, and morphologic assessment of BAT. Plasma FFA turnover was measured in five WT and five ADNTg 16-h fasted animals (A) and the quantity of BAT-TG synthesized (B) during the 3-h isotope infusion assessed by GC/MS. Representative sections are shown for WT (C) and ADNTg (D) BAT. For BAT histology in 6-h fasted animals, sections were stained with hematoxylin and eosin and imaged at a ×20 magnification. *, P < 0.05.
Table 2.
Tissue- and plasma-TG pool sizes and contributions of plasma FFA to TG synthesis
| WT | ADNTg | P value | |
|---|---|---|---|
| VLDL-TG (mg/dl) | 7.5 ± 5.4 | 12.4 ± 12.7 | 0.210 |
| % TG synthesized | 43.2 ± 7.3 | 32.6 ± 6.6 | 0.018 |
| TG pool size (mg) | 0.14 ± 0.09 | 0.23 ± 0.21 | 0.196 |
| Absolute synthesized (μg) | 68 ± 58 | 73 ± 75 | 0.457 |
| Liver tissue weight (g) | 0.786 ± 0.101 | 0.867 ± 0.165 | 0.199 |
| % TG synthesized | 42.9 ± 2.1 | 35.3 ± 5.5 | 0.009 |
| TG pool size (mg) | 11.5 ± 2.2 | 17.4 ± 6.7 | 0.108 |
| Absolute synthesized (mg) | 4.9 ± 0.7 | 6.2 ± 2.6 | 0.152 |
| SubQ % TG synthesized | 0.2 ± 0.1 | 0.3 ± 0.7 | 0.390 |
| Skeletal muscle % TG synthesized | 1.3 ± 1.0 | 1.1 ± 1.1 | 0.422 |
| EWAT tissue weight (g) | 0.352 ± 0.152 | 0.206 ± 0.120 | 0.058 |
| % TG synthesized | 0.2 ± 0.2 | 0.3 ± 0.5 | 0.375 |
| TG pool size (mg) | 268 ± 82 | 163 ± 125 | 0.084 |
| Absolute synthesized (mg) | 0.54 ± 0.51 | 0.50 ± 0.27 | 0.099 |
| BAT tissue weight (g) | 0.076 ± 0.026 | 0.157 ± 0.047 | 0.006 |
| % TG synthesized | 3.1 ± 2.3 | 4.1 ± 2.1 | 0.224 |
| TG pool size (mg) | 51 ± 22 | 124 ± 63 | 0.025 |
| Absolute synthesized (mg) | 1.87 ± 1.96 | 5.71 ± 3.98 | 0.046 |
Data are mean ± sd, and values are derived from five WT and six ADNTg 12-h fasted mice, who had been acclimated for 4 wk to fasting 12 h, then feed being made available for 12 h in the dark. For the fraction of TG derived from the FFA pool, these data were determined after a 3-h infusion of FFA stable isotope in the fasting state. The total pool sizes of skeletal muscle-TG and SubQ-TG were not calculated, because their distributions in the body could not be definitively identified for a single isolated sample.
Discussion
Adiponectin improves insulin sensitivity by exerting integrated effects on whole animal metabolism (15). Beneficial metabolic properties of adiponectin have been demonstrated in clinical conditions, such as type 2 diabetes (39), cardiovascular disease (40), and liver disease (41, 42), and the nature of metabolic improvements related to higher concentrations of adiponectin are thought to be associated with improvements in fat metabolism. To better understand how adiponectin may modulate TG metabolism and fatty acid handling, the present study employed basic molecular studies together with kinetic measurements to test the hypothesis that adiponectin promotes TG synthesis and hydrolysis and fatty acid disposal. Collectively, our data demonstrate that ADNTg mice have higher rates of fatty acid release from adipose and elevated fatty acid disposal in the fasting state, which in liver is converted to ketones, whereas in BAT is cycled into and out of TG. This is consistent with enhanced “metabolic flexibility” that we have recently reported (22), where we demonstrated significantly enhanced sympathetic sensitivity in the ADNTg mice during the fasting state and is in line with recent evidence that higher rates of TG turnover are a hallmark of metabolic flexibility (43–45).
To extend studies implicating EWAT remodeling in the ADNTg mouse, we investigated adipose and liver gene signatures that could explain potential alterations in lipid flux. In adipose, elevated expression of fatty acid oxidation markers, such as PPARα and ACOX, could implicate peroxisomal oxidation pathways, and increased expression of PGC1α and PPARβ/δ indicated potential up-regulation of mitochondrial oxidation; this latter observation is consistent with previous reports of increases in mitochondrial content in the ADNTg mouse (22, 46–48) and in the ability of retinoic acid to shift toward activation of PPARβ/δ during adipocyte differentiation (49). A significant elevation in transcript levels of A and B isoforms of STAT5 were also found in the microarrays of gonadal adipose tissue in ADNTg mice. STAT5 activators are known to regulate ACOX expression (30). Surprisingly, up-regulation of Cidea and PRDM16 was associated with induction of UCP3, rather than UCP1, and induction of NCoA1(SRC-1) further supported the concept of mitochondria enrichment in EWAT (50). Formation of SRC-1-PGC1α promotes transactivation of PGC1α and leads to mitochondrial biogenesis, and the relative ratio of NCoA1/SRC-1 (increases mitochondrial biogenesis) to NCoA2/transcriptional intermediary factor 2 (decreases mitochondrial biogenesis) regulates mitochondrial content in EWAT via its interaction with PGC1α (50). Lastly, in SubQ adipose, PEPCK, a key regulator of the source of TG-glycerol, was significantly induced. Previous studies have shown that flux through PEPCK is both retinol responsive and a target for PPARγ (32, 35).
Lower expression of key lipogenic genes and reduced rates of fatty acid synthesis were found in livers of ADNTg mice. Further, increased expression of mitochondrial HMG-CoA synthetase and ketothiolase supported the observations of lower FFA and higher plasma ketone concentrations in the ADNTg. The presence of elevated levels of adiponectin seems to impose, at least at the level of the hepatic lipid metabolism, a metabolic fingerprint reminiscent of the fasting state. However, the improvements in systemic metabolism are critically dependent upon changes in both liver and adipose tissues. Analysis of the microarray data also revealed surprising changes in a group of genes related to retinoid metabolism in both liver and adipose tissue. Indeed, the molecular signatures observed in the ADNTg animals, namely increased retinoic acid receptor beta in liver, appearance of BAT markers in the WAT, and up-regulation of PEPCK (for glyceroneogenesis), demonstrated similarities to phenotypes associated with retinoid agonism (32, 51–54) and would serve to improve fatty acid trafficking in this animal. Other data are converging to implicate connections between adiponectin and retinoid pathways, and two examples are the potential for both antifibrotic and antiapoptotic roles of adiponectin. Specifically, adiponectin has recently been shown to prevent activation of stellate cells, although the mechanism is unclear. Stellate cells from adiponectin-overexpressing animals are less susceptible to thioacetamide-mediated fibrosis, and the reverse was seen in adiponectin null animals (55). With regard to an antiapoptotic role shown for adiponectin, retinoic acid ligated to the retinoic acid alternate receptor PPARβ/δ also lowers apotosis (56). These associations drive attention to the possible important connections of retinoids and adiponectin, but the directionality of these relationships must be further investigated.
Lastly, using a novel measurement of fatty acid turnover in conscious mice, we found a striking 68% increase in plasma FFA flux in the ADNTg compared with WT controls. These data were also echoed by the up-regulation of lipases HSL and PNPLA2 in EWAT found in the reference microarray database we used. Further, both the rate of FFA release and its clearance from plasma were significantly increased during fasting, concurrent with reductions in plasma FFA and increases in ketone concentrations. We have focused our analysis here on liver and adipose tissue, but we cannot rule out effects on metabolic processes in the heart and important target organ for adiponectin as well. The present results provide the first in vivo kinetic evidence for phenomena described in past studies of adiponectin overexpression, including lower fasting and post-TG gavage plasma FFA concentrations (22), reductions in visceral fat mass (21, 23), and morphological changes in adipose appearance (23). After infusion of the isotope to perform the kinetic studies, significant quantities of label were detected in tissue-TG stores. For VLDL-TG and liver-TG, the lower percentages of TG synthesized from plasma FFA flux suggest that FFA entering the liver were rerouted away from esterification, potentially to ketone synthesis. Additionally, we have recently demonstrated that adiponectin potently stimulates a ceramidase activity associated with its receptors adiponectin receptor 1 and 2 (57). The diversion of FFA into the ceramide pathway and the subsequent conversion to sphingosine-1-phosphate and its derivatives could account for differences observed in the FFA flux seen in the ADNTg livers. ADNTg mice have less gonadal WAT (22), and the metabolic basis for that observation is lower TG synthesis shown here. By contrast, the significantly greater quantity of BAT found in the present study, and in past studies (23) results, at least in part, from a 3-fold increase in TG synthesis rate. These data, when combined with past observations of faster FFA suppression after a glucose load in ADNTg mice (22), and greater fat oxidation at the onset of a long fast in adiponectin transgenic ob/ob mice (21), provide in vivo support for the concept that fatty acids are being exported from adipose at a higher rate in the fasted state, and within BAT, the TG synthesis and reesterification rates are elevated. Evidence for adiponectin to increase BAT lipid metabolism is novel and suggests that this role be studied further given the recent emphasis on BAT physiology in human body weight regulation (58).
In summary, we demonstrate that the adiponectin-overexpressing mouse exhibits improved fatty acid handling in vivo. Importantly, we have focused our studies on mice exposed to a standard chow diet in the fasted state. The relevant observation here is that even in this unchallenged state, the effects of adiponectin are quite notable, consistent with euglycemic clamp studies performed on unchallenged adiponectin knockout mice that also highlighted significant effects on insulin sensitivity (59). The present results are likely to become even more striking when the mice are challenged with a high-fat diet. The sum of these data supports a physiologic link between adiponectin-induced stimulation fatty acid clearance and TG synthesis/breakdown. These effects may be also partially mediated through retinoid-controlled pathways. Due to significant similarities in the effects of adiponectin and retinoid agonism, potential interactions between adiponectin and retinoids could be the common denominator of several observed characteristics induced by adiponectin. The specifics of these interactions will be key areas for future investigation.
Supplementary Material
Acknowledgments
We thank Norma Anderson, Ingrid Wernstedt, Nishanth Sunny, and Vidya Vaidyanathan for assistance with data collection; Jie Song for mouse genotyping; and Steven Kliewer for insightful discussions of the microarray data.
This work was supported by National Institutes of Health Grants 5RL1DK081187 (to E.J.P.); R01-DK55758, RC1 DK086629, and P01DK088761 (to P.E.S.); 5PL1DK081182 (David Russell; University of Texas Southwestern Medical Center); and 5UL1DE019584 (to J.D.H.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ACO
- Acyl-coenzyme A oxidase
- BAT
- brown adipose tissue
- CIDEA
- Cell death-inducing DNA fragmentation factor-α-like effector
- EWAT
- epididymal white adipose tissue
- FFA
- free fatty acid
- GC/MS
- gas chromatography-mass spectrometry
- HMG
- 3-hydroxy-3-methylglutaryl
- NCOA
- nuclear receptor coactivator
- PEPCK
- phosphoenolpyruvate carboxykinase
- PGC1α
- PPARγ coactivator 1α
- PPARγ
- peroxisome proliferator-activated receptor γ
- PRDM16
- PRD1-BF1-RIZ1 homologous domain containing 16
- RBP4
- retinol binding protein 4
- SRC
- steroid receptor coactivator
- SREBP
- sterol regulatory element-binding protein
- STAT
- signal transducer and activator of transcription
- SubQ
- subcutaneous fat
- TG
- triglyceride
- UCP
- uncoupling protein
- VLDL
- very low-density lipoprotein
- WT
- wild type.
References
- 1. Committee E. 2010. Executive summary: standards of medical care in diabetes. Diabetes Care 33:S4–S10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boden G. 2008. Obesity and free fatty acids. Endocrinol Metab Clin North Am 37:635–646, viii-ix [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Florentin M, Elisaf MS, Mikhailidis DP, Liberopoulos EN. 2009. Drug combinations for dyslipidemia and obesity treatment in metabolic syndrome. Curr Pharm Des 15:3446–3462 [DOI] [PubMed] [Google Scholar]
- 4. Delarue J, Magnan C. 2007. Free fatty acids and insulin resistance. Curr Opin Clin Nutr Metab Care 10:142–148 [DOI] [PubMed] [Google Scholar]
- 5. Rickels MR, Naji A, Teff KL. 2006. Insulin sensitivity, glucose effectiveness, and free fatty acid dynamics after human islet transplantation for type 1 diabetes. J Clin Endocrinol Metab 91:2138–2144 [DOI] [PubMed] [Google Scholar]
- 6. Soriguer F, García-Serrano S, García-Almeida JM, Garrido-Sánchez L, García-Arnés J, Tinahones FJ, Cardona I, Rivas-Marín J, Gallego-Perales JL, García-Fuentes E. 2009. Changes in the serum composition of free-fatty acids during an intravenous glucose tolerance test. Obesity 17:10–15 [DOI] [PubMed] [Google Scholar]
- 7. Hennes MM, Dua A, Kissebah AH. 1997. Effects of free fatty acids and glucose on splanchnic insulin dynamics. Diabetes 46:57–62 [DOI] [PubMed] [Google Scholar]
- 8. Collins QF, Xiong Y, Lupo EG, Jr, Liu HY, Cao W. 2006. p38 Mitogen-activated protein kinase mediates free fatty acid-induced gluconeogenesis in hepatocytes. J Biol Chem 281:24336–24344 [DOI] [PubMed] [Google Scholar]
- 9. Kehlenbrink S, Tonelli J, Koppaka S, Chandramouli V, Hawkins M, Kishore P. 2009. Inhibiting gluconeogenesis prevents fatty acid-induced increases in endogenous glucose production. Am J Physiol 297:E165–E173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boden G. 1998. Free fatty acids (FFA), a link between obesity and insulin resistance. Front Biosci 3:d169–d175 [DOI] [PubMed] [Google Scholar]
- 11. Lam TK, Carpentier A, Lewis GF, van de Werve G, Fantus IG, Giacca A. 2003. Mechanisms of the free fatty acid-induced increase in hepatic glucose production. Am J Physiol 284:E863–E875 [DOI] [PubMed] [Google Scholar]
- 12. Dobbins RL, Chester MW, Daniels MB, McGarry JD, Stein DT. 1998. Circulating fatty acids are essential for efficient glucose-stimulated insulin secretion after prolonged fasting in humans. Diabetes 47:1613–1618 [DOI] [PubMed] [Google Scholar]
- 13. Lebovitz HE, Banerji MA. 2001. Insulin resistance and its treatment by thiazolidinediones. Recent Prog Horm Res 56:265–294 [DOI] [PubMed] [Google Scholar]
- 14. DeFronzo RA, Abdul-Ghani M. 2011. Type 2 diabetes can be prevented with early pharmacological intervention. Diabetes Care 34(Suppl 2):S202–S209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pajvani UB, Scherer PE. 2003. Adiponectin: systemic contributor to insulin sensitivity. Curr Diab Rep 3:207–213 [DOI] [PubMed] [Google Scholar]
- 16. Leth H, Andersen KK, Frystyk J, Tarnow L, Rossing P, Parving HH, Flyvbjerg A. 2008. Elevated levels of high-molecular-weight adiponectin in type 1 diabetes. J Clin Endocrinol Metab 93:3186–3191 [DOI] [PubMed] [Google Scholar]
- 17. Semple RK, Soos MA, Luan J, Mitchell CS, Wilson JC, Gurnell M, Cochran EK, Gorden P, Chatterjee VK, Wareham NJ, O'Rahilly S. 2006. Elevated plasma adiponectin in humans with genetically defective insulin receptors. J Clin Endocrinol Metab 91:3219–3223 [DOI] [PubMed] [Google Scholar]
- 18. Shetty S, Kusminski CM, Scherer PE. 2009. Adiponectin in health and disease: evaluation of adiponectin-targeted drug development. Trends Pharmacol Sci 30:234–239 [DOI] [PubMed] [Google Scholar]
- 19. Yamauchi T, Kamon J, Waki H, Murakami K, Motojima K, Komeda K, Ide T, Kubota N, Terauchi Y, Tobe K, Miki H, Tsuchida A, Akanuma Y, Nagai R, Kimura S, Kadowaki T. 2001. The mechanisms by which both heterozygous peroxisome proliferator-activated receptor γ (PPARγ) deficiency and PPARγ agonist improve insulin resistance. J Biol Chem 276:41245–41254 [DOI] [PubMed] [Google Scholar]
- 20. Combs TP, Berg AH, Obici S, Scherer PE, Rossetti L. 2001. Endogenous glucose production is inhibited by the adipose-derived protein Acrp30. J Clin Invest 108:1875–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim JY, van de Wall E, Laplante M, Azzara A, Trujillo ME, Hofmann SM, Schraw T, Durand JL, Li H, Li G, Jelicks LA, Mehler MF, Hui DY, Deshaies Y, Shulman GI, Schwartz GJ, Scherer PE. 2007. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J Clin Invest 117:2621–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Asterholm IW, Scherer PE. 2010. Enhanced metabolic flexibility associated with elevated adiponectin levels. Am J Pathol 176:1364–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Combs TP, Pajvani UB, Berg AH, Lin Y, Jelicks LA, Laplante M, Nawrocki AR, Rajala MW, Parlow AF, Cheeseboro L, Ding YY, Russell RG, Lindemann D, Hartley A, Baker GR, Obici S, Deshaies Y, Ludgate M, Rossetti L, Scherer PE. 2004. A transgenic mouse with a deletion in the collagenous domain of adiponectin displays elevated circulating adiponectin and improved insulin sensitivity. Endocrinology 145:367–283 [DOI] [PubMed] [Google Scholar]
- 24. Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, Kobayashi S, Davis C, Dai H, He YD, Stephaniants SB, Cavet G, Walker WL, West A, Coffey E, Shoemaker DD, Stoughton R, Blanchard AP, Friend SH, Linsley PS. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol 19:342–347 [DOI] [PubMed] [Google Scholar]
- 25. Shimano H, Horton JD, Hammer RE, Shimomura I, Brown MS, Goldstein JL. 1996. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J Clin Invest 98:1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Donnelly KL, Margosian MR, Sheth SS, Lusis AJ, Parks EJ. 2004. Increased lipogenesis and fatty acid reesterification contribute to hepatic triacylglycerol stores in hyperlipidemic Txnip−/− mice. J Nutr 134:1475–1480 [DOI] [PubMed] [Google Scholar]
- 27. Grefhorst A, Parks EJ. 2009. Reduced insulin-mediated inhibition of VLDL secretion upon pharmacological activation of the liver X receptor in mice. J Lipid Res 50:1374–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hoff J. 2000. Methods of blood collection in the mouse. Lab Animal 29:47–53 [Google Scholar]
- 29. Bézaire V, Seifert EL, Harper ME. 2007. Uncoupling protein-3: clues in an ongoing mitochondrial mystery. FASEB J 21:312–324 [DOI] [PubMed] [Google Scholar]
- 30. Coulter AA, Stephens JM. 2006. STAT5 activators modulate acyl CoA oxidase (AOX) expression in adipocytes and STAT5A binds to the AOX promoter in vitro. Biochem Biophys Res Comm 344:1342–1345 [DOI] [PubMed] [Google Scholar]
- 31. Haque M, Anreola F. 1998. The cloning and characterization of a novel cytochrome P450 family, CYP26, with specificity toward retinoic acid. Nutr Rev 56:84–85 [DOI] [PubMed] [Google Scholar]
- 32. Cadoudal T, Glorian M, Massias A, Fouque F, Forest C, Benelli C. 2008. Retinoids upregulate phosphoenolpyruvate carboxykinase and glyceroneogenesis in human and rodent adipocytes. J Nutr 138:1004–1009 [DOI] [PubMed] [Google Scholar]
- 33. Rosenberg IH, Russell RM, Wood RJ. 1992. Retinoic acid, bound to its nuclear receptor, enhances the expression of the gene for phosphoenolpyruvate carboxykinase. Nutr Rev 50:50–52 [DOI] [PubMed] [Google Scholar]
- 34. Lucas PC, O'Brien RM, Mitchell JA, Davis CM, Imai E, Forman BM, Samuels HH, Granner DK. 1991. A retinoic acid response element is part of a pleiotropic domain in the phosphoenolpyruvate carboxykinase gene. Proc Natl Acad Sci USA 88:2184–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nye CK, Hanson RW, Kalhan SC. 2008. Glyceroneogenesis is the dominant pathway for triglyceride glycerol synthesis in vivo in the rat. J Biol Chem 283:27565–27574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Horton JD, Goldstein JL, Brown MS. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109:1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Amengual J, Ribot J, Bonet ML, Palou A. 2010. Retinoic acid treatment enhances lipid oxidation and inhibits lipid biosynthesis capacities in the liver of mice. Cell Physiol Biochem 25:657–666 [DOI] [PubMed] [Google Scholar]
- 38. Baar RA, Dingfelder CS, Smith LA, Bernlohr DA, Wu C, Lange AJ, Parks EJ. 2004. Investigation of in vivo fatty acid metabolism in AFABP/aP2−/− mice. Am J Physiol 288:E187–E193 [DOI] [PubMed] [Google Scholar]
- 39. Vasseur F, Helbecque N, Lobbens S, Vasseur-Delannoy V, Dina C, Clément K, Boutin P, Kadowaki T, Scherer PE, Froguel P. 2005. Hypoadiponectinaemia and high risk of type 2 diabetes are associated with adiponectin-encoding (ACDC) gene promoter variants in morbid obesity: evidence for a role of ACDC in diabesity. Diabetologia 48:892–899 [DOI] [PubMed] [Google Scholar]
- 40. Wang ZV, Scherer PE. 2008. Adiponectin, cardiovascular function, and hypertension. Hypertension 51:8–14 [DOI] [PubMed] [Google Scholar]
- 41. Yoneda M, Iwasaki T, Fujita K, Kirikoshi K, Inamori M, Nozaki Y, Maeyama S, Wada K, Saito S, Terauchi S, Nakajima A. 2007. Hypoadiponectinemia plays a crucial role in the development of nonalcoholic fatty liver disease in patients with type 2 diabetes mellitus independent of visceral adipose tissue. Alcohol Clin Exp Res 31(1 Suppl):S15–S21 [DOI] [PubMed] [Google Scholar]
- 42. Rogers CQ, Ajmo JM, You M. 2008. Adiponectin and alcoholic fatty liver disease. IUBMB Life 60:790–797 [DOI] [PubMed] [Google Scholar]
- 43. Bergman BC, Perreault L, Hunerdosse DM, Koehler MC, Samek AM, Eckel RH. 2010. Increased intramuscular lipid synthesis and low saturation relate to insulin sensitivity in endurance-trained athletes. J Appl Physiol 108:1134–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oakes ND, Thalén PG, Jacinto SM, Ljung B. 2001. Thiazolidinediones increase plasma-adipose tissue FFA exchange capacity and enhance insulin-mediated control of systemic FFA availability. Diabetes 50:1158–1165 [DOI] [PubMed] [Google Scholar]
- 45. Storlien L, Oakes ND, Kelley DE. 2004. Metabolic flexibility. Proc Nutr Soc 63:363–368 [DOI] [PubMed] [Google Scholar]
- 46. Wang YX, Lee CH, Tiep S, Yu RT, Ham J, Kang H, Evans RM. 2003. Peroxisome-proliferator-activated receptor δ activates fat metabolism to prevent obesity. Cell 113:159–170 [DOI] [PubMed] [Google Scholar]
- 47. Akter MH, Razzaque MA, Yang L, Fumoto T, Motojima K, Yamaguchi T, Hirose F, Osumi T. 2006. Identification of a gene sharing a promoter and peroxisome proliferator-response elements with acyl-CoA oxidase gene. PPAR Res 71916:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fan CY, Pan J, Chu R, Lee D, Kluckman KD, Usuda N, Singh I, Yeldandi AV, Rao MS, Maeda N, Reddy JK. 1996. Hepatocellular and hepatic peroxisomal alterations in mice with a disrupted peroxisomal fatty acyl-coenzyme A oxidase gene. J Biol Chem 271:24698–24710 [DOI] [PubMed] [Google Scholar]
- 49. Berry DC, Noy N. 2009. All-trans-retinoic acid represses obesity and insulin resistance by activating both peroxisome proliferation-activated receptor β/δ and retinoic acid receptor. Mol Cell Biol 29:3286–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Picard F, Géhin M, Annicotte J, Rocchi S, Champy MF, O'Malley BW, Chambon P, Auwerx J. 2002. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell 111:931–941 [DOI] [PubMed] [Google Scholar]
- 51. Mercader J, Madsen L, Felipe F, Palou A, Kristiansen K, Bonet ML. 2007. All-trans retinoic acid increases oxidative metabolism in mature adipocytes. Cell Physiol Biochem 20:1061–1072 [DOI] [PubMed] [Google Scholar]
- 52. Mercader J, Ribot J, Murano I, Felipe F, Cinti S, Bonet ML, Palou A. 2006. Remodeling of white adipose tissue after retinoic acid administration in mice. Endocrinology 147:5325–5332 [DOI] [PubMed] [Google Scholar]
- 53. Okuno M, Kojima S, Akita K, Matsushima-Nishiwaki R, Adachi S, Sano T, Takano Y, Takai K, Obora A, Yasuda I, Shiratori Y, Okano Y, Shimada J, Suzuki Y, Muto Y, Moriwaki Y. 2002. Retinoids in liver fibrosis and cancer. Front Biosci 7:d204–d218 [DOI] [PubMed] [Google Scholar]
- 54. Villarroya F. 1998. Differential effects of retinoic acid on white and brown adipose tissues: an unexpected role for vitamin A derivatives on energy balance. Ann NY Acad Sci 839:190–195 [DOI] [PubMed] [Google Scholar]
- 55. Shafiei MS, Shetty S, Scherer PE, Rockey DC. 2011. Adiponectin regulation of stellate cell activation via PPARγ-dependent and -independent mechanisms. Am J Pathol 178:2690–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wolf G. 2008. Retinoic acid as cause of cell proliferation or cell growth inhibition depending on activation of one of two different nuclear receptors. Nutr Rev 66:55–59 [DOI] [PubMed] [Google Scholar]
- 57. Holland WL, Miller RA, Wang ZV, Sun K, Barth BM, Bui HH, Davis KE, Bikman BT, Halberg N, Rutkowski JM, Wade MR, Tenorio VM, Kuo MS, Brozinick JT, Zhang BB, Birnbaum MJ, Summers SA, Scherer PE. 2011. Receptor-mediated activation of ceramidase activity initiates the pleiotropic actions of adiponectin. Nat Med 17:55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ravussin E, Galgani JE. 2011. The implication of brown adipose tissue for humans. Ann Rev Nutr 31:33–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nawrocki AR, Rajala MW, Tomas E, Pajvani UB, Saha AK, Trumbauer ME, Pang Z, Chen AS, Ruderman NB, Chen H, Rossetti L, Scherer PE. 2006. Mice lacking adiponectin show decreased hepatic insulin sensitivity and reduced responsiveness to peroxisome proliferator-activated receptor γ agonists. J Biol Chem 281:2654–2660 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.