Abstract
Background:
The choice of antimicrobial therapy for bloodstream infections is often empirical and based on the knowledge of local antimicrobial activity profiles of the most common bacteria causing such infections.
Aims:
The present study was aimed to investigate frequency of bacterial pathogens causing septicemia and their antimicrobial resistant pattern in hospital admitted patients.
Settings and Design:
It was a prospective study, conducted at Majeedia Hospital, Hamdard University, New Delhi, India.
Material and Methods:
We examined prospectively, 168 bacterial strains isolated from 186 clinically diagnosed septicemia cases admitted at a University Hospital in New Delhi, over a period of six months from July 2009 to December 2009. Antimicrobial susceptibility was performed according to Clinical and Laboratory Standards Institute (CLSI, USA) guidelines.
Results:
The most frequently identified Gram-positive bacteria were coagulase-negative staphylococci 63.5%, Staphylococcus aureus 23.1%, enterococci 5.8% and alpha-haemolytic streptococci 5.8%. The most frequently Gram-negative bacteria identified were Acinetobacter species 31%, Salmonella typhi 24.1%, Escherichia coli 23.3% and Pseudomonas aeruginosa 13.8%. Coagulase-negative staphylococci showed maximum resistance to cefaclor 57.1% and ampicillin 46.9%. Staphylococcus aureus showed maximum resistance to amoxicillin 100% and ampicillin 91.7%. Acinetobacter species showed maximum resistance to amoxicillin 89.7%, amoxiclav 87.1% and ampicillin 85.7%. Salmonella typhi, Escherichia coli, Pseudomonas aeruginosa and Klebsiella pneumoniae showed maximum resistance to ampicillin, 46.4%, 92%, 93.8% and 100%, respectively.
Conclusions:
Gram-negative pathogens predominated in bloodstream infections. Resistance to most of the antimicrobial agents for a number of pathogens implicated in bloodstream infections, especially in Gram-negative bacteria, has reached worrisome levels and continues to increase.
KEY WORDS: Antimicrobial resistance, antimicrobial resistant pattern, antimicrobial susceptibility, bloodstream infection, prospective study, septicemia
The lethality of septicemia in terms of mortality rate has declined in recent two decades. However, the increasing cases of septicemia particularly in developing countries is still a major health problem that creates a biggest challenge for the clinicians in selection of suitable antimicrobial agents, as it is further complicated by the development of resistance in organisms to antimicrobial agents, which is the mainstay of treatment for septicemia. Increasing antimicrobial resistance among bloodstream infections (BSIs) have been reported in many studies conducted in India,[1,2] and other countries worldwide.[3–6] Both Gram-positive and Gram-negative bacteria have been isolated from BSIs and predominance of one type over other varies from place to place and even in the same place over time.[1,2,7] Early diagnosis and rapid administration of antimicrobial therapy to patients with BSIs has been shown to reduce mortality and morbidity.[8,9] The choice of empirical antimicrobial therapy for septicemia must therefore include knowledge of the extent of resistance in common pathogens associated with BSIs. Since the organisms comprise both the Gram-positive and Gram-negative pathogens, knowledge of their resistance patterns is critical in local geographical area.
Therefore, this prospective study was conducted to investigate the frequency of bacterial pathogens causing septicemia in our hospital admitted patients and to provide updated information about antimicrobial resistance to commonly used antimicrobial agents, in order to help clinicians to choose the most suitable therapy.
Materials and Methods
Study design
A prospective study was conducted at a University Hospital (Majeedia Hospital of Hamdard University), New Delhi, India, over a period of six months from July 2009 to December 2009.
Patient population
During the study period, 803 blood samples from patients of clinically suggestive septicemia were evaluated, out of 4162 patients admitted in different units, namely Medicine, Nursery, Pediatrics, Intensive care unit, Orthopedic, Dialysis and Surgery of Majeedia hospital. The research protocol was approved by Jamia Hamdard Institutional Review Board (Approval Letter No. 07/09, JH-IRB dated July 2009). An informed consent was obtained from the patients or patients’ close relative prior to inclusion in the study. All eligible admitted patients were selected based on the following inclusion and exclusion criteria. Patients of either sex, all age group with suspected or proven infection were included in the study. Patients classified as “Do not resuscitate” or “Do not treat” were excluded from the study.
Blood collection and organism identification
Blood samples from adults and geriatrics patients were collected by trained laboratory staff at the request of clinician, whereas in case of neonates and pediatrics, the samples were collected by pediatricians. Blood samples were collected after thorough cleaning of the venous site with 70% alcohol and subsequently followed by povidone iodine. Under the aseptic conditions 5 ml of blood was drawn by venipuncture and transferred into two culture bottles each containing 50 ml of 0.5% bile-broth and 50 ml of 0.5% glucose-broth. Both the bottles were incubated aerobically at 37°C for 10 days. Routine subculturing was done on 5% sheep blood agar and McConkey agar after 24 hours, 48 hours, 5th day and 10th day. In between these time points, subculturing was done until there was visible turbidity. The growth of microorganism was considered pathogenic if the same microorganism was isolated from both the broths and contaminated if either the growth was obtained in only one bottle or a mixed growth was obtained. In cases where the microorganisms obtained were coagulase-negative staphylococci, a repeat blood culture was performed. Organisms were identified by cultural characters, morphology and standard biochemical tests.[10]
Susceptibility testing
Susceptibility of the bacterial isolates to different antimicrobials was determined by using the Kirby Bauer disc diffusion method as per Clinical and Laboratory Standards Institute (CLSI) guidelines.[11] On separate plate Muller Hunting Agar medium, control were tested with same antibiotic disc and zone of inhibition were compared. The antibacterial concentration per disc was as follows: ampicillin (10μg), amoxicillin (30μg), amoxiclav (20:10μg), cefuroxime (30μg), cefoxitin (30μg), cefaclor (30μg), cefotaxime (30μg), ceftazidime (30μg), cefoperazone (75μg), gentamicin (10μg), amikacin (30μg), netilmicin (30μg), ciprofloxacin (5μg), ofloxacin (5μg), tetracycline (30μg) and chloramphenicol (30μg) for all organisms. For Gram-positive organisms, penicillin G (2 IU), erythromycin (15μg) and vancomycin (30μg), were added along with antimicrobials mentioned above. No case was investigated for fungi or anaerobes. All the antimicrobial agents were purchased from HiMedia Laboratories, Mumbai, India.
Results
Demographics
Over the period of six month from July 2009 to December 2009, 803 blood samples out of 4162 admissions, from patients of clinically suggestive septicemia were evaluated. The causative organism could be identified in 165 patients, yielding 168 bacterial strains (20.9% of 803 blood samples) as some had polymicrobial infections. In 21 patients the organism could not be isolated. However, these 21 patients showed clinical symptoms of septicemia. Hence, a total of 186 patients were confirmed as septicemia. The age distribution of the patients was as follows: neonates 40 (21.5%), 1 month to 14 years 84 (45.2%), 14-59 years 49 (26.3%) and 59 years and above 13 (7%). Male patients (n=122, 65.6%) were predominant throughout the study period as compared to female (n=64, 34.4%).
Frequency of occurrence of microorganisms causing septicemia
The frequency of isolation of Gram-negative bacteria (n=116, 69%) was found to be more than that of Gram-positive bacteria (n=52, 31%). The most frequently identified Gram-positive bacteria were coagulase-negative staphylococci 33 (63.5%), Staphylococcus aureus 12 (23.1%), enterococci, alpha-haemolytic streptococci, 3 (5.8% each) and Streptococcus pneumoniae 1 (1.9%) [Figure 1]. Gram-negative bacteria identified were Acinetobacter species 36 (31%), Salmonella typhi 28 (24.1%), Escherichia coli 27 (23.3%), Pseudomonas aeruginosa 16 (13.8%), Klebsiella pneumoniae 5 (4.3%) and Salmonella paratyphi A 4 (3.5%) [Figure 2].
Figure 1.
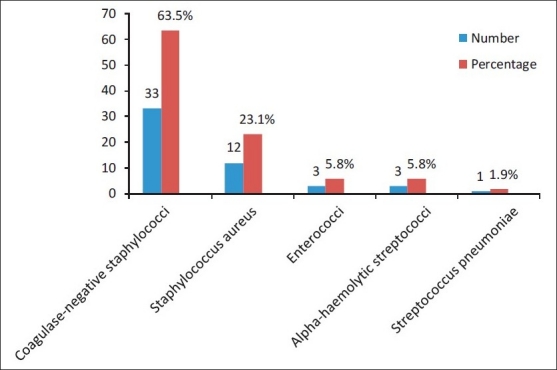
Frequency of occurrence of Gram-positive microbial strains responsible for septicemia during July 2009-December 2009
Figure 2.
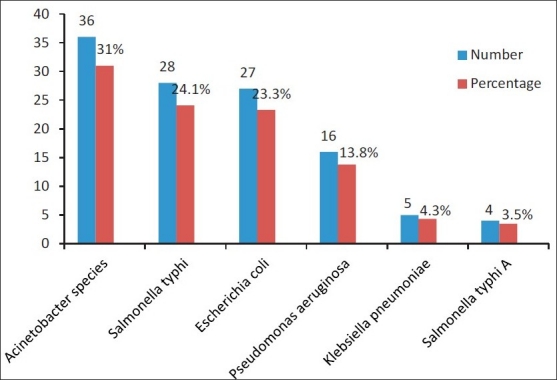
Frequency of occurrence of Gram-negative microbial strains responsible for septicemia during July 2009-December 2009
Resistance of most common Gram-positive microorganisms to antimicrobial agents
Table 1 shows rates of resistance in Gram-positive bacteria to antimicrobial agents.
Table 1.
Resistance pattern in Gram-positive organisms to antimicrobial agents
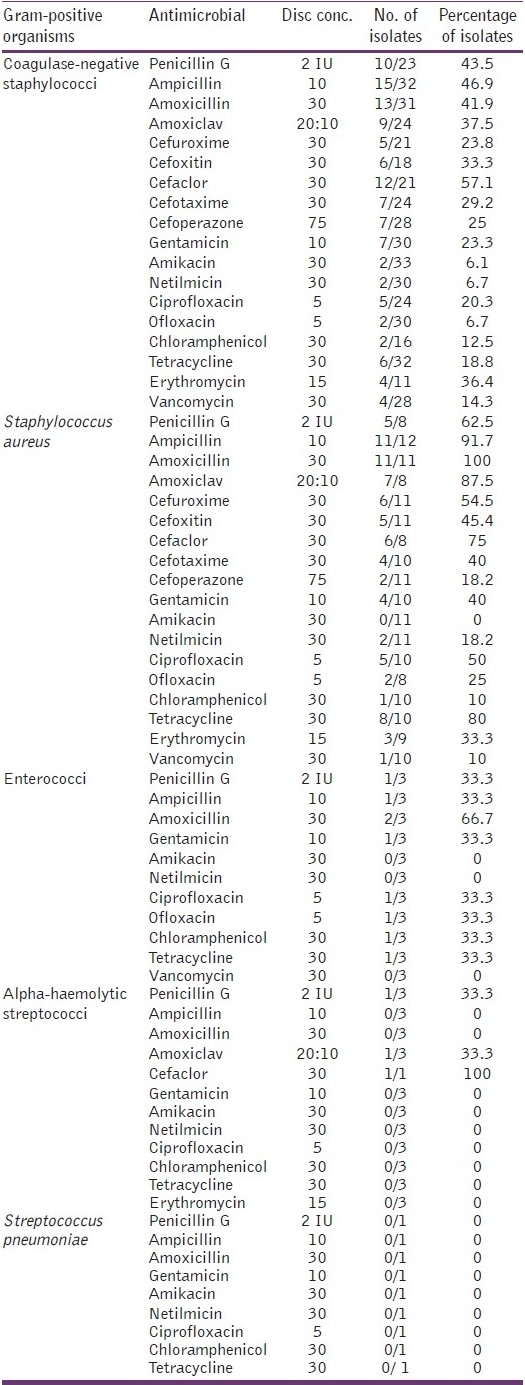
The rates of resistance in coagulase-negative staphylococci to various antimicrobials were as follows: penicillin G (benzyl penicillin) (43.5%), ampicillin (46.9%), amoxicillin (41.9%), amoxiclav (37.5%), cefuroxime (23.8%), cefoxitin (33.3%), cefaclor (57.1%), cefotaxime (29.2%), cefoperazone (25%), gentamicin (23.3%), amikacin (6.1%), netilmicin (6.7%), ciprofloxacin (20.3%), ofloxacin (6.7%), chloramphenicol (12.5%), tetracycline (18.8%), erythromycin (36.4%) and vancomycin (14.3%).
The rates of resistance in Staphylococcus aureus to various antimicrobials were as follows: penicillin G (62.5%), ampicillin (91.7%), amoxicillin (100%), amoxiclav (87.5%), cefuroxime (54.5%), cefoxitin (45.4%), cefaclor (75%), cefotaxime (40%), cefoperazone (18.2%), gentamicin (40%), netilmicin (18.2%), ciprofloxacin (50%), ofloxacin (25%), chloramphenicol (10%), tetracycline (80%), erythromycin (33.3%) and vancomycin (10%). Staphylococcus aureus did not show resistance to amikacin.
The rates of resistance in enterococci are given below: penicillin G (33.3%), ampicillin (33.3%), amoxicillin (66.7%), amoxiclav (33.3%), ciprofloxacin (33.3%), ofloxacin (33.3%), chloramphenicol (33.3%), tetracycline (33.3%). Enterococci did not show resistance to amikacin, netilmicin and vancomycin.
Alpha-haemolytic streptococci showed resistance to penicillin G, amoxiclav (3.3% each) and cefaclor (100%).
Streptococcus pneumoniae did not show resistance to antimicrobials tested.
Resistance of most common Gram-negative microorganisms to antimicrobial agents
Table 2 shows rates of resistance in Gram-negative bacteria to antimicrobial agents.
Table 2.
Resistance pattern in Gram-negative organisms to antimicrobial agents
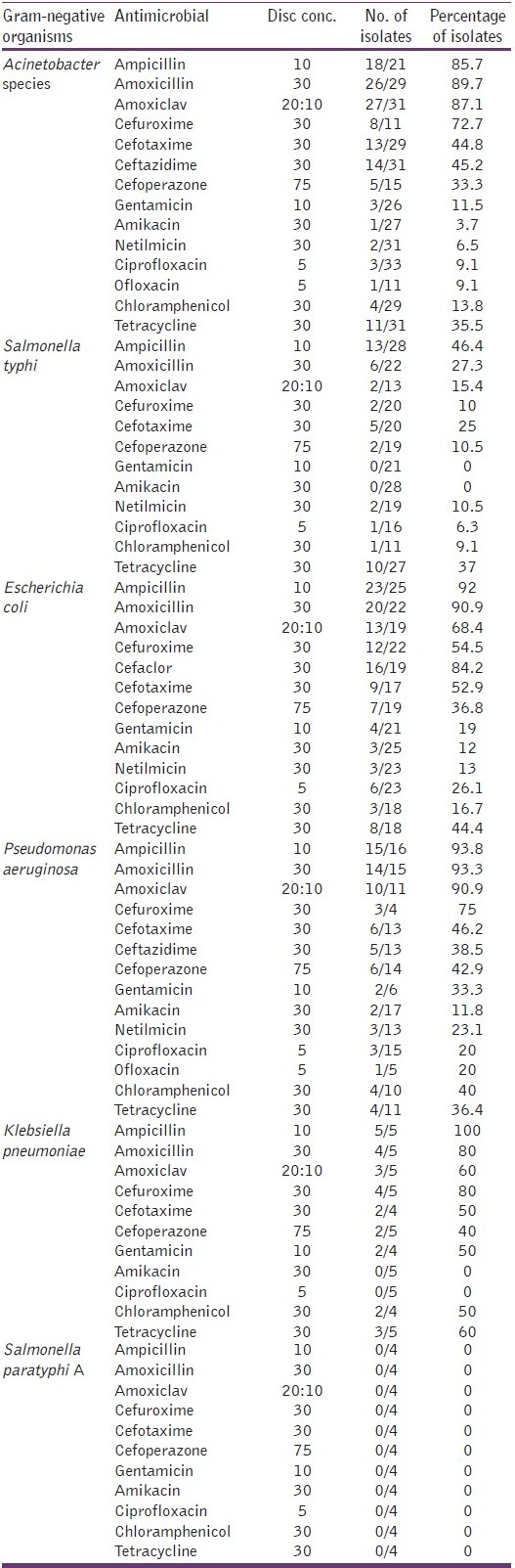
The rates of resistance in Acinetobacter species to various antimicrobials are given below: ampicillin (85.7 %), amoxicillin (89.7%), amoxiclav (87.1%), cefuroxime (72.7%), cefotaxime (44.8%), ceftazidime (45.2%), cefoperazone (33.3%), gentamicin (11.5%), amikacin (3.7%), netilmicin (6.5%), ciprofloxacin (9.1%), ofloxacin (9.1%), chloramphenicol (13.8%) and tetracycline (35.5%).
The rates of resistance in Salmonella typhi to various antimicrobials are given below: ampicillin (46.4%), amoxicillin (27.3%), amoxiclav (15.4%), cefuroxime (10%), cefotaxime (25%), cefoperazone (10.5%), netilmicin (10.5%), ciprofloxacin (6.3%), chloramphenicol (9.1%) and tetracycline (37%). Salmonella typhi did not show resistance to gentamicin and amikacin.
The rates of resistance in Escherichia coli to various antimicrobials are given below: ampicillin (92%), amoxicillin (90.9%), amoxiclav (68.4%), cefuroxime (54.5%), cefaclor (84.2%), cefotaxime (52.9%), cefoperazone (36.8%), gentamicin (19%), amikacin (12%), netilmicin (13%), ciprofloxacin (26.1%), chloramphenicol (16.7%) and tetracycline (44.4%).
The rates of resistance in Pseudomonas aeruginosa to various antimicrobials are given below: ampicillin (93.8%), amoxicillin (93.3%), amoxiclav (90.9%), cefuroxime (75%), cefotaxime (46.2%), cefazidime (38.5%), cefoperazone (42.9%), gentamicin (33.3%), amikacin (11.8%), netilmicin (23.1%), ciprofloxacin (20%), ofloxacin (20%), chloramphenicol (40%) and tetracycline (36.4 %).
The rates of resistance in Klebsiella pneumoniae to various antimicrobials are given below: ampicillin (100%), amoxicillin (80%), amoxiclav (60%), cefuroxime (80%), cefotaxime (50%), cefoperazone (40%), gentamicin (50%), chloramphenicol (50%) and tetracycline (60%). Klebsiella pneumoniae did not show resistance to amikacin and ciprofloxacin.
Salmonella paratyphi A did not show resistance to antimicrobials tested.
Discussion
Bacterial resistance to antimicrobial agents is a continuing serious problem in the treatment of BSIs. BSIs caused by bacterial pathogens are often due to strains that are resistant to a broader range of antimicrobial agents. The present study examines the antimicrobial resistance patterns of 168 bacterial strains isolated from blood of septicemia patients. The data highlights the prevalence of antimicrobial resistance among bacterial pathogens isolated from BSIs and confirms that Delhi is no exception to progressive antimicrobial resistance against major bacterial pathogens.
Septicemia is significantly more frequent in males than in females in a wide range of age group patients. Gram-negative bacterial strains have been the most prevalent causative pathogens of septicemia in the present findings. This result is in contrast to the microbiology of BSIs, where Gram-positive microorganisms were the predominant pathogens.[6] Gram-negative microorganisms were also the commonest cause of BSIs in other studies.[1,2,7,12] The changing etiological agents of septicemia may reflect the changing demography of septicemia in developing countries, which might be related to the geographical variations.
Analysis of frequency of occurrence for Gram-positive pathogens revealed that coagulase-negative staphylococci and Staphylococcus aureus were the major causative organism responsible for septicemia. A study reported coagulase-negative staphylococci among Gram-positive organisms as the commonest cause of septicemia.[6] While, study from Chandigarh, North India reported Staphylococcus aureus among Gram-positive microorganisms as the commonest pathogen implicated in septicemia.[1] Similarly frequency analysis of Gram-negative pathogens showed that Acinetobacter species, Salmonella typhi and Escherichia coli were the most frequent causative organisms implicated in septicemia.
The rates of resistance in coagulase-negative staphylococci to beta-lactam antibiotics were higher than the rates of resistance to other classes of antimicrobials, except erythromycin which showed resistance rate of 36.4%. Staphylococcus aureus showed significantly higher rates of resistance to most of the tested antimicrobials with 100% resistance to ampicillin. Enterococci did not show resistance to vancomycin, is in contrast to the report by Sadar et al, who have reported (2.4%) resistance in enterococci to vancomycin.[3]
The rate of resistance was higher in Gram-negative microorganisms than that in Gram-positive microorganisms to the most of the tested antimicrobial agents. Acinetobacter species showed very high rates of resistance to beta-lactams in the range of 33.3-89.7%. However, it showed lower rates of resistance to aminoglycoside in the range between 3.7-11.5%. Also, low level resistance rates were observed in this organism to quinolones. Similarly, Escherichia coli showed markedly higher rates of resistance to beta-lactam antibiotics than other classes of antimicrobials in the range between 36.8-92%. The organism showed maximum resistance to ampicillin and minimum to amikacin. A study reported similar findings from Iran.[13] Pseudomonas aeruginosa, showed approximately same pattern of resistance as that of Escherichia coli. Klebsiella pneumoniae showed 100% resistant to ampicillin and no resistance to amikacin and ciprofloxacin.
Wide prevalence of antimicrobial resistance rates were noted in our study, particularly in Gram-negative bacteria, is in accordance with other study reports.[14,15] The high prevalence of antimicrobial resistance rates in Delhi region might be due to indiscriminate and over use of drugs in our country because of their easy over the counter availability.[16] The other reason might be prevalence of extended spectrum beta lactamase production among Gram-negative isolates from neonatal septicemia patients.[17] Furthermore, other reasons could be the changing patterns of antibiotic use and changes in lifestyle. In the light of above findings there remains a growing need for novel agents, although a recent study reported polymyxin, chloramphenicol and cotrimoxazole are being revisited as possible choices for the treatment.[18] Antimicrobial resistance and their genetic relatedness was not performed, which might be one of the limitations of the present study.
Conclusion
Early initiation of suitable antimicrobial therapy for BSIs is critical in decreasing morbidity and mortality among patients of BSIs due to bacteria. Therefore, accurate microbiological diagnosis and their antimicrobial sensitivity pattern become relevant to the prompt initiation of adequate therapy for BSIs.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
References
- 1.Mehta M, Dutta P, Gupta V. Antimicrobial susceptibility pattern of blood isolates from a teaching hospital in north India. Jpn J Infect Dis. 2005;58:174–6. [PubMed] [Google Scholar]
- 2.Kaistha N, Mehta M, Singla N, Garg R, Chander J. Neonatal septicemia isolates and resistance patterns in a tertiary care hospital of North India. J Infect Dev Ctries. 2009;4:55–7. doi: 10.3855/jidc.625. [DOI] [PubMed] [Google Scholar]
- 3.Sader HS, Jones RN, Andrade-Baiocchi S, Biedenbach DJ SENTRY Participants Group (Latin America) Four-year evaluation of frequency of occurrence and antimicrobial susceptibility patterns of bacteria from bloodstream infections in Latin American medical centers. Diagn Microbiol Infect Dis. 2002;44:273–80. doi: 10.1016/s0732-8893(02)00469-8. [DOI] [PubMed] [Google Scholar]
- 4.Kato-Maeda M, Bautista-Alavez A, Rolón-Montes-de-Oca AL, Ramos-Hinojosa A, Ponce-de-León A, Bobadilla-del-Valle M, et al. Increasing trend of antimicrobial drug-resistance in organisms causing bacteremia at a tertiary-care hospital: 1995 to 2000. Rev Invest Clin. 2003;55:600–5. [PubMed] [Google Scholar]
- 5.Biedenbach DJ, Moet GJ, Jones RN. Occurrence and antimicrobial resistance pattern comparisons among bloodstream infection isolates from the SENTRY Antimicrobial Surveillance Program (1997-2002) Diagn Microbiol Infect Dis. 2004;50:59–69. doi: 10.1016/j.diagmicrobio.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Falagas ME, Kasiakou SK, Nikita D, Morfou P, Georgoulias G, Rafailidis PI. Secular trends of antimicrobial resistance of blood isolates in a newly founded Greek hospital. BMC Infect Dis. 2006;6:99. doi: 10.1186/1471-2334-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agnihotri N, Kaistha N, Gupta V. Antimicrobial susceptibility of isolates from neonatal septicemia. Jpn J Infect Dis. 2004;57:273–5. [PubMed] [Google Scholar]
- 8.Leibovici L, Konisberger H, Pitlik SD, Samra Z, Drucker M. Bacteremia and fungemia of unknown origin in adults. Clin Infect Dis. 1992;14:436–43. doi: 10.1093/clinids/14.2.436. [DOI] [PubMed] [Google Scholar]
- 9.Diekema DJ, Pfaller MA, Jones RN, Doern GV, Winokur PL, Gales AC, et al. Survey of bloodstream infections due to gram-negative bacilli: Frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, and Latin America for the SENTRY Antimicrobial Surveillance Program, 1997. Clin Infect Dis. 1999;29:595–607. doi: 10.1086/598640. [DOI] [PubMed] [Google Scholar]
- 10.Collee JG, Miles RS, Watt B. Test for identification of bacteria. In: Collee JG, Fraser AG, Marmion BP, Simmons A, editors. Mackie and McCartney Practical Medical Microbiology. 14th ed. New York: Churchill Livingstone; 1996. pp. 131–49. [Google Scholar]
- 11.CLSI document M2-A9. 9th ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2009. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. [Google Scholar]
- 12.Joshi SJ, Ghole VS, Niphadkar KB. Neonatal gram-negative bacteremia. Indian J Pediatr. 2000;67:27–32. doi: 10.1007/BF02802632. [DOI] [PubMed] [Google Scholar]
- 13.Mamishi S, Pourakbari B, Ashtiani MH, Hashemi FB. Frequency of isolation and antimicrobial susceptibility of bacteria isolated from bloodstream infections at Children's Medical Center, Tehran, Iran, 1996-2000. Int J Antimicrob Agents. 2005;26:373–9. doi: 10.1016/j.ijantimicag.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Luzzaro F, Viganò EF, Fossati D, Grossi A, Sala A, Sturla C, et al. Prevalence and drug susceptibility of pathogens causing bloodstream infections in northern Italy: A two-year study in 16 hospitals. Eur J Clin Microbiol Infect Dis. 2002;21:849–55. doi: 10.1007/s10096-002-0837-7. [DOI] [PubMed] [Google Scholar]
- 15.Goosens H. European status of resistance in nosocomial infections. Chemotherapy. 2005;51:177–81. doi: 10.1159/000086919. [DOI] [PubMed] [Google Scholar]
- 16.Roy I, Jain A, Kumar M, Agarwal SK. Bacteriology of neonatal septicemia in a tertiary care hospital of northern India. Indian J Med Microbiol. 2002;20:156–9. [PubMed] [Google Scholar]
- 17.Bhattacharjee A, Sen MR, Prakash P, Gaur A, Anupurba S. Increased prevalence of extended spectrum beta lactamase producer in neonatal septicemic cases at a tertiary referral hospital. Indian J Med Microbiol. 2008;26:356–60. doi: 10.4103/0255-0857.43578. [DOI] [PubMed] [Google Scholar]
- 18.Vergidis PI, Falagas ME. New antibiotic agents for bloodstream infections. Int J Antimicrob Agents. 2008;32(Suppl 1):S60–5. doi: 10.1016/j.ijantimicag.2008.06.003. [DOI] [PubMed] [Google Scholar]


