Abstract
Latent tuberculosis is a subclinical condition caused by Mycobacterium tuberculosis, which affects about one-third of the population across the world. To abridge the chemotherapy of tuberculosis, it is necessary to have active drugs against latent form of M. tuberculosis. Therefore, it is imperative to devise in vitro and models of latent tuberculosis to explore potential drugs. In vitro models such as hypoxia, nutrient starvation, and multiple stresses are based on adverse conditions encountered by bacilli in granuloma. Bacilli experience oxygen depletion condition in hypoxia model, whereas the nutrient starvation model is based on deprivation of total nutrients from a culture medium. In the multiple stress model dormancy is induced by more than one type of stress. In silico mathematical models have also been developed to predict the interactions of bacilli with the host immune system and to propose structures for potential anti tuberculosis compounds. Besides these in vitro and in silico models, there are a number of in vivo animal models like mouse, guinea pig, rabbit, etc. Although they simulate human latent tuberculosis up to a certain extent but do not truly replicate human infection. All these models have their inherent merits and demerits. However, there is no perfect model for latent tuberculosis. Therefore, it is imperative to upgrade and refine existing models or develop a new model. However, battery of models will always be a better alternative to any single model as they will complement each other by overcoming their limitations.
Keywords: In vitro, in vivo, latent tuberculosis, M. tuberculosis, model
INTRODUCTION
Mycobacterium tuberculosis (M. tuberculosis), the causative agent of tuberculosis (TB), has infected about 1.7 billion people worldwide, which accounts for nearly 3 million deaths every year.[1,2] TB remains one of the leading causes of mortality among all infectious diseases.[3] M. tuberculosis is transmitted via respiratory route and infects macrophages in lungs. The host immune system arrests infection by forming a structure called granuloma and stamp down the imminent threat of active infection. However, inside the granuloma M. tuberculosis switches to a dormant or non replicative state under stressful conditions for its survival. This infection persists in humans due to its ability to shift to a clinically latent state without revealing any overt disease symptoms and it is termed as latent tuberculosis infection (LTBI). About one-third of the world population is infected with LTBI and individuals harboring LTBI carry a lifetime risk of reactivation to active disease.[4] If these persons are co infected with HIV, then the risk elevates to 8--10% per year.[5] Granuloma conditions like hypoxia, nutrient starvation, change in pH, free radicals induce mycobacteria to shift its metabolic pathways. Therefore, first-line anti-TB drugs like isoniazid (INH) have limited activity against dormant tubercular bacilli.[6] Latent tuberculosis (LTB) remained a major hindrance in the development of effective chemotherapy against TB. The lengthy drug regime for treatment of TB is due to the presence of latent bacilli which cannot be easily eradicated; therefore, it is necessary to develop effective drugs against latent tubercular bacilli to shorten duration of therapy. It is imperative to develop an ideal model of LTBI to screen potential new chemical entities (NCEs) and existing drugs. Some models have been proposed to check the activity out in vitro, in silico, and in vivo, but they do not simulate ideal conditions. Thus, it is necessory to develop a model that meets the requisites for an ideal model. Models of LTBI have been shown in Figure 1.
Figure 1.
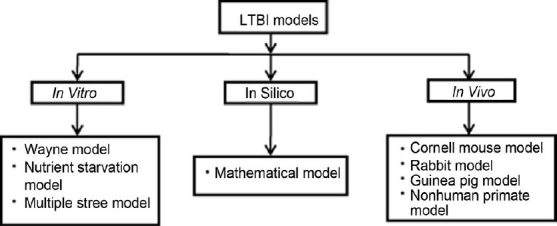
Models of latent tuberculosis infection
In vitro models
Evaluation of NCEs against non--replicating tubercle bacilli requires appropriate in vitro models of latency. Under the given stress conditions, M. tuberculosis may undergo transition to quiescent state which is metabolically silent conditions. Out of these models, the hypoxia and nutrient starvation models of LTBI are well studied and elucidated.
Hypoxia induced model
Dormancy is assumed to be associated with hypoxic conditions inside the granuloma of infected lungs. Wayne carried out a series of experiments in the field of LTB. He cultured M. tuberculosis in tightly closed containers having media and subjected the culture to slow oxygen depletion conditions. Under hypoxic conditions bacilli are believed to enter into non-replicating state. Gradual depletion of the oxygen in media causes microaerophilic levels (oxygen concentration reaches about 1%) which is also termed as non-replicating state I (NRP I) and then it is followed by anaerobic levels which is termed as NRP II state. Wayne also enlightened that when these latent bacilli are again exposed to normal oxygen conditions, there is rebound growth of acid fastness bacilli.[7,8]
In the Wayne model, M. tuberculosis is grown with constant shaking for a defined period of time (usually 24 days) which leads to hypoxic conditions. Initially, tubercle bacilli experience a microaerophilic state (at about 70 hour) and in the end anaerobic conditions are attained (after 200 hour) which can be confirmed by decolorization of methylene blue (1.5 μg/ml), an indicator of anaerobic conditions.[9] Hypoxia-induced latent bacilli are treated with test drugs and growth of mycobacteria is determined by the plating (conventional) method. M. tuberculosis in the NRP state does not divide and/or synthesize DNA and these latent bacilli become resistant to standard antitubercular drugs such as isoniazid, but they are sensitive to metronidazole.[10] Nonetheless, a new nitroimidazopyran compound, PA-824, has shown activity against both aerobic and anaerobic M. tuberculosis in vitro.[11]
it is not a true simulation of the human granuloma conditions, this model partly mimics the hypoxic conditions. It induces only one type of condition for the bacilli, and hence, we cannot rely on one model but there is need to explore other models of latency.
Previously conventional plating was employed to determine the growth which bears some shortcomings like contamination, labor intensive, time consuming, etc. Recently a new method (BACTEC 460 TB system) was used as an alternative to the conventional plating method to overcome the limitations of the plating method. It is less laborious and fast, and chances of contamination are minimized.[12] A new method for confirming latency has been developed which is based on the rate of nitrate reduction in hypoxia induced dormant bacilli.[13]
Nutrient starvation induced model
M. tuberculosis is deprived of nutrients in granuloma; therefore, bacilli shut some of the metabolic pathway down to economize the energy in cell. They set off rescue pathways in starvation conditions like upregulation of those genes which are responsible for synthesis of vital enzymes during starvation. It is assumed that during nutrient starvation conditions; mycobacteria utilize lipid as a sole source of energy.[14] In general, there is a global metabolic shift when bacilli undergo starvation conditions.[15,16]
In a nutrient starvation model, normal-growing mycobacteria are centrifuged and pellets are washed twice with a phosphate buffer saline (PBS). Then these pellets are resuspended in PBS and incubated at 370°C for 6 weeks. During these period, the mycobacteria shift to the latent state. After confirming latency, these starved bacilli were treated with test drugs for 7 days. Then the growth of mycobacteria is determined to know the activity of the drugs. Latency can be confirmed by different methods like CFU enumeration, respiration test (methylene blue test), Ziehl Neelsen staining, and drug resistance test.[15,17] It has been observed that bacteria shut down their respiration within 9-12 days of nutrient starvation. Isoniazid, rifampin, and metronidazole are not effective in the nutrient starvation model whereas activity of pyrazinamide (PRZ) is augmented during nutrient starvation conditions.[15,18] Econazole and clotrimazole are active against the latent form of bacilli in this model.[17]
Like hypoxia model, the nutrient starvation model cannot truly mimic granulomatous environment; though it remains a vital model in screening of potential drugs against latent bacilli. It is a widely accepted model to study dormancy. As this model employs the conventional plating method, it also carries downsides and these limitations can be overcome by employing the BACTEC 460 TB system. Both the models are widely used for studying dormancy as these are complimentary to each other.
Multiple stress model
Single-stress conditions like hypoxia and nutrient starvation may be sufficient to switch mycobacteria to latent form, but this cannot fully simulate the conditions of granuloma. It is always a better alternate to study latent bacilli under multiple stress conditions which may provide better simulation of granuloma conditions like hypoxia, nutrient starvation, changes in pH, etc.[19] A multiple stress model may prove to be an ideal in vitro model of latency because it may give improved simulation. It is observed that lipid accumulation and buoyant density decrease in bacilli under multiple stress conditions.[19]
In a multiple stress model, M. tuberculosis is grown in appropriate nutrient-rich media till its growth reaches to OD of 0.2 (600 nm). The bacilli are pelleted and resuspended into low nutrient media having acidic pH (pH 5) in specialized bottle with standard jolt neck. It is sealed by rubber septa to ensure air tightness. Gas mixture (5% O2+10% CO2+85% N2) is then injected in to the bottle. Oxygen consumption is constantly estimated and the gas mixture is injected at alternate days to maintain oxygen concentration.[19] It seems an appropriate model for latency as it mimics closely to true granuloma conditions. But this model has not been widely used for screening of drugs.
Development of in vitro models
Though the models described herein are in use since long time but there is a scope to further refine these models to have better simulation of granulomatous environment. In the present models, we can ameliorate to make them efficient and reproducible. Some of the researchers have employed the BACTEC 460 TB system but the use of radioactive materials limits the use of this system.[12,20] Moreover, due to hazards of radioactive materials its use ha s been forbidden in developed countries. In the near future its use is going to be discontinued in developing countries also. Therefore, it is provident to employ some other advanced techniques like MGIT 960, BACTEC 9000 MB, and MB/BacT systems.[21] The MGIT 960 system employs a mycobacterial growth indicator tube based on the principle of fluorescence. It have slew of merits over the BACTEC 460 system. It is a fully automated system having facility of taking reading at hourly intervals which circumvents manual task of BACTEC 460. MGIT 960 has already replaced the BACTEC 460 TB system in the developed countries and very soon it will replace it in the developing countries also.
In silico model
In recent times, the role of computational prediction in biological research has increased considerably to streamline and expedite the drug discovery process. Several mathematical models are available to study different areas of tuberculosis. They illustrate the interaction of macrophages with mycobacteria, based on the available knowledge of the host and pathogen system.[22] The available data are depicted mathematically with a variety of environmental factors to make an in silico model that can simulate the real conditions. In this model, the effect of change of one parameter like bacterial factor or more than one factor can be predicted. The final outcome can be confirmed by performing a series of experiments. The strength of this model depends on various programmed factors like size of input data, complex interactions. Mathematical models are fast and can easily mimic the real ones based on the data. Despite these advantages, the accuracy of in silico models is still debatable.[23]
In vivo model
It is imperative to have an ideal in vivo model of LTB that can replicate the actual human latency to study the host--parasite interaction, the activity of drugs, and mechanism of latency. It is important to develop and establish a new animal model and/or improve an existing model of LTBI for evaluating anti-tuberculosis activity of novel and existing drugs. There are some common animal models of LTBI such as mouse, guinea pig, rabbit, nonhuman primate (NHP), and zebra fish.[24] Though there is not a single ideal model, still these models are used for studying in vivo activity of potential drugs. In the present section, we have discussed commonly available in vivo animal models.
Mouse model
Mouse is the most popular and widely used animal model of M. tuberculosis infection for long time. The relatively resistant nature of mice to develop TB has been exploited for easy establishment of latent infection. The Cornell mouse model, one of the most accepted mouse model for studying dormancy, was developed by McCune and colleagues at Cornell University in the 1950s. In this model, mice are infected with a high dose of M. tuberculosis (3.95 × 106 CFU per mouse) and then treated with anti-TB drugs like isoniazid and pyrazinamide to kill replicating bacilli in organs. Culturable tubercle bacilli from organs homogenates are determined to confirm the state of dormancy. However, 3 months after cessation of the chemotherapy, active infection is relapsed in one-third of the mice and, when immunosuppressant therapy is given, relapse takes place in almost all the mice.[25] The bacilli recovered from relapsed mice are acid fast and fully susceptible to INH or PZA. This implies that dormant bacilli do not develop stable genetic drug resistance, but they are phenotypically resistant to drugs in latency.[24] To screen the activity of drugs against LTBI, test drugs are given to the mice having LTBI. Immunosuppressant treatment is given to the drug-treated mice to augment the response of reactivation to detect latent bacilli. In this model, metronidazole is inactive in preventing reactivation, whereas econazole and clotrimazole has shown mycobactericidal effect in developing and developed LTBI.[17,26] Some researchers have modified the Cornell mouse model by optimizing various parameters namely M. tuberculosis inoculums, duration of drugs therapy, drug dosages, and interval between cessation of drugs.[27]
A mouse model can detect very low numbers of bacilli and also replicates many facets of the human immune responses. Nevertheless, mice do not develop a well-formed granuloma and lack the caseous necrotic centers as observed in human infection.[28] Moreover, this model lacks stability and consistency, and does not simulate host immunity-induced LTBI. Despite all the limitations, investigations by using the Cornell mouse model have made significant contributions in understanding latency and reactivation.
Guinea pig
pigs do not establish latent infection as they are highly susceptible to low-dose inoculum of M. tuberculosis.[29] But a recent study conducted by Lenaerts et al. showed existence of non replicating tubercle bacilli in lung lesions of a guinea pig, which have striking similarities to latent bacilli present in human beings.[30] In this model guinea pigs are inoculated with 107 CFU of M. tuberculosis 18b which is streptomycin-auxotrophic mutant and then streptomycin therapy is given for 3 months. This model is reproducible but the strain is limited to some laboratories only. Sugawara et al. illustrated a new model, in which guinea pigs are infected by a subcutaneous route with 100-1000 CFU of M. tuberculosis H37Rv which expresses a green fluorescent protein (GFP).[31] In the next 10 months, a tuberculin skin test is performed on these mice to make sure their infection status. In this model, higher dose of inoculum (1000 CFU or more) is not apposite for developing the LTBI since these guinea pigs are prone to reactivation after 10 months.[32]
Rabbits
There is close similarity in different stages of TB progression among rabbits and human beings. Unlike the mouse model, it produces a caseous necrosis center in response to mycobacterial infection. Rabbits show intense resistance to M. tuberculosis infection resulting into latent infection. Therefore, rabbits may be a better model to study LTBI. In this model rabbits are infected by aerosol route. Bacilli thrive in lungs for next 5 weeks and their number declines over the period of 36 weeks in contrast to the other models. Initially, culturable bacilli are seen in rabbits but they are cleared off in 4 months and interestingly, some rabbits develop infections in 6 months.[33] Despite host immune response, infection persists due to the confinement of bacteria within the necrotic center. Following infection, the lung CFU count attains zenith and apparently it is cleared off bacilli. This suggests that like human beings, rabbits carry LTBI and reactivation to active TB takes place in a similar fashion.[24] M. bovis has been employed in some of the studies on rabbits and has shown close similarities to human infection with formation of caseous foci and cavities.[33] One more advantage of this model is that it does not have the problem of spontaneous reactivation.
Nonhuman primate model
In the last decade, nonhuman primate (NHP) models are preferred over all of the above-mentioned models of latency due to their close resemblance with human TB infection.[24] When a monkey is infected via bronchoscopic instillation with low dose of inoculum, there is similar pattern of disease progression as in human beings. It has shown that about 60% of infected NHPs develop LTBI; latency is confirmed by a positive tuberculin skin test (TST) and lymphocyte proliferation assay. After 6 months of infection, NHPs can be grouped in either latent or active TB, depending upon reactivation. Necropsy of NHPs with LTBI suggests that there are small numbers of fibrotic granuloma in lungs.[34,35] This model is also having certain limitations like requirement of biosafety level III and trained staff; moreover genotype variability and small sample size impact the reproducibility in this model. Aside from all above downsides, the NHP model simulates the human dormancy to the best extent than any other model.[36]
LTBI models have enormous significance in the studying the action of the molecules/drugs, mechanism of latency, and role of host immune responses. Although many models exist for studying LTBI in vitro and in vivo, attempts are needed in the direction of building an ideal model to elucidate those unresolved questions in a field of latent TB. All these models have their inherent merits and demerits. However, all of these models can be employed in conjunction to minimize their drawbacks. It is critical to evolve an ideal model which can mimic the true human conditions of latency.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- 1.Miyazaki E, Chaisson RE, Bishai WR. Analysis of rifapentine for preventive therapy in the Cornell mouse model of latent tuberculosis. Antimicrob Agents Chemother. 1999;43:2126–30. doi: 10.1128/aac.43.9.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karakousis PC, Yoshimatsu T, Lamichhane G, Woolwine SC, Nuermberger EL, Grosset J, et al. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J Exp Med. 2004;200:647–57. doi: 10.1084/jem.20040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Currie CS, Floyd K, Williams BG, Dye C. Cost, affordability and cost-effectiveness of strategies to control tuberculosis in countries with high HIV prevalence. BMC Public Health. 2005;5:130–46. doi: 10.1186/1471-2458-5-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parrish NM, Dick JD, Bishai WR. Mechanisms of latency in Mycobacterium tuberculosis. Trends in Microbiology. 1998;6:107–12. doi: 10.1016/s0966-842x(98)01216-5. [DOI] [PubMed] [Google Scholar]
- 5.Flynn JL, Chan J. Tuberculosis: Latency and Reactivation. Infect Immun. 2001;69:4195–201. doi: 10.1128/IAI.69.7.4195-4201.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karakousis PC, Williams EP, Bishai WR. Altered expression of isoniazid-regulated genes in drug-treated dormant Mycobacterium tuberculosis. J Antimicrob Chemo. 2008;61:323–31. doi: 10.1093/jac/dkm485. [DOI] [PubMed] [Google Scholar]
- 7.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–9. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu YM, Butcher PD, Sole K, Mitchison DA, Coates AR. Protein synthesis is shutdown in dormant Mycobacterium tuberculosis and is reversed by oxygen or heat shock. FEMS Microbiol Lett. 1998;158:139–45. doi: 10.1111/j.1574-6968.1998.tb12813.x. [DOI] [PubMed] [Google Scholar]
- 9.Wayne LG, Sramek HA. Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38:2054–8. doi: 10.1128/aac.38.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wayne LG. Synchronized replication of Mycobacterium tuberculosis. Infect Immun. 1977;17:528–30. doi: 10.1128/iai.17.3.528-530.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lenaerts AJ, Gruppo V, Marietta KS, Johnson CM, Driscoll DK, Tompkins NM, et al. Preclinical testing of the nitroimidazopyran PA-824 for activity against Mycobacterium tuberculosis in a series of in vitro and in vivo models. Antimicrob Agents Chemother. 2005;49:2294–301. doi: 10.1128/AAC.49.6.2294-2301.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jhamb SS, Singh RP, Singh PP. A comparison of conventional and radiometric methods for the assessment of anti-tubercular activity of drugs against Mycobacterium tuberculosis in mice and macrophage models. Indian J Tuberc. 2008;55:70–6. [PubMed] [Google Scholar]
- 13.Wayne LG, Hayes LG. Nitrate reduction as a marker for hypoxic shiftdown of Mycobacterium tuberculosis. Tuberc Lung Dis. 1998;79:127–32. doi: 10.1054/tuld.1998.0015. [DOI] [PubMed] [Google Scholar]
- 14.Archuleta RJ, Yvonne Hoppes P, Primm TP. Mycobacterium avium enters a state of metabolic dormancy in response to starvation. Tuberculosis. 2005;85:147–58. doi: 10.1016/j.tube.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–31. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 16.Murphy DJ, Brown JR. Identification of gene targets against dormant phase Mycobacterium tuberculosis infections. BMC Infect Dis. 2007;7:84–100. doi: 10.1186/1471-2334-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmad Z, Sharma S, Khuller GK. The potential of azole antifungals against latent/persistent tuberculosis. FEMS microbiology letters. 2006;258:200–3. doi: 10.1111/j.1574-6968.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- 18.Huang Q, Chen ZF, Li YY, Zhang Y, Ren Y, Fu Z, et al. Nutrient-starved incubation conditions enhance pyrazinamide activity against Mycobacterium tuberculosis. Chemotherapy. 2007;53:338–43. doi: 10.1159/000107723. [DOI] [PubMed] [Google Scholar]
- 19.Deb C, Lee CM, Dubey V, Daniel J, Abomoelak B, Sirakova T, et al. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. pLoS ONE. 2009;4:e6077. doi: 10.1371/journal.pone.0006077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy M, Srinivasan S, Andersen B, PR G. Rapid assessment of mycobacterial growth inside macrophages and mice, using the radiometric (BACTEC) method. Tuber Lung Dis. 1994;75:127–31. doi: 10.1016/0962-8479(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 21.Alcaide F, Benıtez MA, Escriba JM, Martın R. Evaluation of the BACTEC MGIT 960 and the MB/BacT systems for recovery of mycobacteria from clinical specimens and for species identification by DNA Accu-Probe. J Clin Microbiol. 2000;38:398–401. doi: 10.1128/jcm.38.1.398-401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin Ling P, Kirschner D, Flynn JL. Modeling pathogen and host: in vitro, in vivo and in silico models of latent Mycobacterium tuberculosis infection. Inflamm Infect Dis. 2005;2:149–54. [Google Scholar]
- 23.Kirschner D, Marino S. Mycobacterium tuberculosis as viewed through a computer. Trends Microbiol. 2005;13:206–11. doi: 10.1016/j.tim.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 24.Gupta UD, Katoch VM. Animal models of tuberculosis. Tuberculosis (Edinb) 2005;85:277–93. doi: 10.1016/j.tube.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Miyazaki E, Chaisson R, Bishai W. Analysis of Rifapentine for preventive therapy in the cornell mouse model of latent tuberculosis. Antimicrob Agents Chemother. 1999;43:2126–30. doi: 10.1128/aac.43.9.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhillon J, Allen BW, Hu YM, Coates AR, Mitchison DA. Metronidazole has no antibacterial effect in Cornell model murine tuberculosis. Int J Tuberc Lung Dis. 1998;2:736–42. [PubMed] [Google Scholar]
- 27.Flynn JL, Scanga CA, Tanaka KE, Chan J. Effects of aminoguanidine on latent murine tuberculosis. J Immunol. 1998;160:1796–803. [PubMed] [Google Scholar]
- 28.Phyu S, Mustafa T, Hofstad T, Nilsen R, Fosse R, Bjune G. A mouse model for latent tuberculosis. Scand J Infect Dis. 1998;30:59–68. doi: 10.1080/003655498750002321. [DOI] [PubMed] [Google Scholar]
- 29.Ordway D, Henao-Tamayo M, Shanley C, Smith EE, Palanisamy G, Wang B, et al. Influence of Mycobacterium bovis BCG vaccination on cellular immune response of guinea pigs challenged with Mycobacterium tuberculosis. Clin Vaccine Immunol. 2008;15:1248–58. doi: 10.1128/CVI.00019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenaerts AJ, Hoff D, Aly S, Ehlers S, Andries K, Cantarero L, et al. Location of persisting mycobacteria in a Guinea pig model of tuberculosis revealed by r207910. Antimicrob Agents Chemother. 2007;51:3338–45. doi: 10.1128/AAC.00276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugawara I, Udagawa T, Aoki T, Mizuno S. Establishment of a guinea pig model of latent tuberculosis with GFP-introduced Mycobacterium tuberculosis. Tohoku J Exp Med. 2009;219:257–62. doi: 10.1620/tjem.219.257. [DOI] [PubMed] [Google Scholar]
- 32.Kashino SS, Napolitano DR, Skobe Z, Campos-Neto A. Guinea pig model of Mycobacterium tuberculosis latent/dormant infection. Microbes Infect. 2008;10:1469–76. doi: 10.1016/j.micinf.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manabe YC, Kesavan AK, Lopez-Molina J, Hatem CL, Brooks M, Fujiwara R, et al. The aerosol rabbit model of TB latency, reactivation and immune reconstitution inflammatory syndrome. Tuberculosis. 2008;88:187–96. doi: 10.1016/j.tube.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin PL, Kirschner D, Flynn JL. Modeling pathogen and host: in vitro, in vivo and in silico models of latent Mycobacterium tuberculosis infection. Drug Discov Today: Dis Models. 2005;2:149–54. [Google Scholar]
- 35.Capuano SV, 3rd, Croix DA, Pawar S, Zinovik A, Myers A, Lin PL, et al. Experimental Mycobacterium tuberculosis infection of cynomolgus macaques closely resembles the various manifestations of human M.tuberculosis infection. Infect Immun. 2003;71:5831–44. doi: 10.1128/IAI.71.10.5831-5844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flynn JL, Capuano SV, Croix D, Pawar S, Myers A, Zinovik A, et al. Non-human primates: A model for tuberculosis research. Tuberculosis (Edinb) 2003;83:116–8. doi: 10.1016/s1472-9792(02)00059-8. [DOI] [PubMed] [Google Scholar]


