Abstract
Reteplase is a segment of tissue plasminogen activator used for the removal of thrombi in blood vessels. In the present study the cloned reteplase gene was used for its expression in competent E. coli. The recombinant plasmid, pET15b/reteplase (rpET-BL21), was transformed into competent E. coli strain BL21 (DE3) cells. Overnight culture of the transformed bacteria was induced by the addition of isopropylthio-ß-Dgalactoside (IPTG) to the final concentrations of 0.25, 0.5, 1 and 1.5 mM. Also, the effects of different temperatures(25, 30, 37 and 39°C), shaking speeds (100, 170 and 190 rpm), and various glucose concentrations (0.25, 0.5, 0.75 and 1 mM) on the expression of reteplase were examined. Samples were analyzed by SDS-PAGE. Maximum amount of protein production was obtained by the addition of 1 mM IPTG at 37°C, 100 rpm of shaking speed in the absence of glucose.
Keywords: Reteplase, Optimization, Expression, Temperature, Shaking speed, Glucose
INTRODUCTION
Tissue plasminogen activator (tPA) is an enzyme natively present in humans which upon activation facilitates the destruction of blood clots through the conversion of plasminogen to plasmin. Plasma tPA has long been considered to be the product of the endothelial cells that line various parts of the vascular system regardless of vessel size or location. Therefore, this substance and its derivatives can be used as the drug of choice for the treatment of thrombotic disorders especially acute myocardial infarction which is named thrombolytic therapy(1,2).
tPA is mainly produced by recombinant DNA technology(3–6). However, due to the high cost of production and other considerations, the details of its production are not available. One of the key points in the production of this drug is its glycosylation and that is the main reason why this drug is not produced in prokaryotic cells. This problem can be addressed by eliminating the glycosylated parts of the enzyme while preserving its active domain. One of these products is reteplase.
There are some reports regarding the production of the recombinant reteplase in prokaryotic systems(7,8). These reports have used systems suitable for expression in E. coli. However, as mentioned above, some studies regarding the production of retaplase in E. coli have not been published nor the details of certain experimental methods are available. Additionally, the influence of different factors affecting its expression has not been fully investigated. Therefore, in the present study, we examined the effect of different conditions such as isopropylthio-ß-Dgalactoside (IPTG) concentration, temperature, shaking speed and glucose concentration on the expression of reteplase gene in E. coli cells.
MATERIALS AND METHODS
Materials
Nutrient agar, nutrient broth, simmon citrate agar, acrylamide, bis acrylamide. TEMED, ammonium persulfate, formamide, EDTA, bromophenol blue, glacial acetic acid, and Luria-Bertani medium (LB) were obtained from Merck, Germany. Ethidium bromide, and loading buffer were purchased from Cinnagen, Tehran, Iran. Taq polymerase was obtained from BioRon, Poland. DNA ligase, DNA molecular size marker and DNA ladder were from Fermentas, Germany. High pure PCR template preparation kit and QIA quick gel extraction kit were obtained from Roche, Germany and Qiagen, Germany, respectively. All other chemicals were from Merck, Germany.
Methods
Expression of reteplase in E. coli
The recombinant plasmid, pET15b/ reteplase (rpET-BL21), was used for the transformation of competent E. coli strain BL21 (DE3) cells prepared using calcium chloride method(9). Transformed cells were first grown on LB agar plates(6) supplemented with 100 mg/ml ampicillin (Roche, Germany) at 37°C. For the expression of reteplase, a single colony was inoculated into 5 ml LB medium. After overnight growth, 200μl of the culture was transferred to the flasks containing 15 ml fresh LB medium and ampicillin. Additionally, 200μl of the overnight culture of BL21/pET15b without recombinant gene was transferred to the flasks containing 15 ml fresh LB medium containing antibiotic. Cells were shook at 170 rpm to a density of OD600 0.5, then over-expression of recombinant protein was induced by the addition of IPTG to the final concentrations of 0.25, 0.5, 1 and 1.5 mM. The cultures were incubated further at 37°C. The OD600 of these cultures were measured at intervals of 1 h for 3 h and 1 ml of sample was taken and each sample was centrifuged. Pellets were resuspended in 100 μl of PBS (0.75 mM Na2HPO4, 0.1 mM NaCl, 1.7 mM KH2PO4, pH=7.2) and 300 μl of SDS sample buffer (0.5 M Tris-HCl, pH=6.8, Glycerol, 10% SDS, 2.9 mM β-mercaptoethanol and 0.5% bromophenol blue) was added and then samples were heated at 100°C for 5 min. 20 μl of each sample was electrophoresed on a 12% SDS-polyacrylamide gel.
Expression of reteplase at different temperatures
To examine the effects of different temperatures on reteplase expression, the same process as outlined above was utilized. Synthesis was induced by the addition of IPTG to the final concentration of 1 mM. The cultures were incubated for 2 h at 25, 30, 37, and 39°C. At the end of incubation, the OD600 of cultures were measured and cells were harvested by centrifugation. Pellets were then prepared and analyzed by SDS-PAGE.
Expression of reteplase at different shaking speeds
To examine the effects of shaking speeds on reteplase expression, synthesis was induced by the addition of IPTG to the final concentration of 1 mM. The cultures were incubated for 2 h at 37°C at 100, 170 and 190 rpm shaking speeds. At the end of incubation, OD600 of cultures were measured and cells were harvested by centrifugation Pellets were then analyzed by SDS-PAGE.
Expression of reteplase at different glucose concentrations
To examine the effect of glucose concentration on reteplase expression, synthesis was induced by the addition of IPTG to the final concentration of 1 mM. Then, glucose was added to the final concentrations of 0.25, 0.5, 0.75 and 1 mM and the cultures were incubated for 2 h at 37°C at 100 rpm. At the end of incubation, OD600 of cultures were measured and cells were harvested by centrifugation. Pellets were then analyzed by SDS-PAGE.
Western blot
After electrophoresis, in order to make the proteins accessible to antibody detection, the proteins were transferred (400 nA, 1 h) on to a nitrocellulose paper and non-specific binding was blocked by placing the membrane in 2.5% non-fat milk (in PBS) for 1 h. Subsequently, primary mouse anti His antibody (1:1000 dilution in 2.5% non-fat milk) was added and incubated at room temperature for 1 h. After three washes with 2.5% non-fat milk (each for 10 min), secondary antimouse antibody conjugated to horse raddish peroxidase (1:250 dilution) was added and shook at room temperature for 1 h. Then the membrane was washed three times in 2.5% non-fat milk, and based on the manufacturesr's instructions, ECL kit (Roche, Germany) was used for the detection of reteplase.
RESULTS
After obtaining recombinant E. coli cells, the presence of reteplase in these cells was examined by western blotting which revealed that the target protein of 39 kD was expressed (Fig. 1). This size was calculated based on this proteins’ Rf as compared to the molecular weight of the protein marker standards.
Fig. 1.
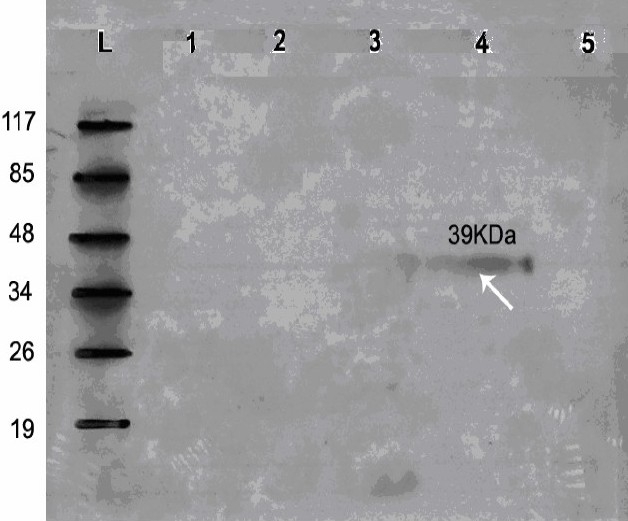
Western blot analysis by ECL method of extracts from E. coli strain DE3 containing recombinant plasmid pET15b/reteplase (rpET-BL21) using mouse anti His antibody. Lane: L) Protein marker, Lane:1 extract from E. coli strain DE3 bacteria containing plasmid pET15b, Lane:2) extract from E. coli strain DE3 bacteria, Lane:3) pET-BL21 induction with 1 mM IPTG, Lane:4) extracts from E. coli strain DE3 bacteria containing recombinant plasmid (rpET15b). A band with molecular mass of approximately 39 kDa was detected. Lane:5) extract from E. coli strain DE3 bacteria
For the optimization of reteplase expression, this protein was induced by the addition of different concentrations of IPTG to the growing culture at various times of incubation.
The maximum production of the recombinant protein occurred when 1 mM IPTG was used for 2 h. Protein expression was determined by SDS-PAGE analysis of cell extracts followed by staining with Coomassie Blue, (Figs. 2, and 3).
Fig. 2.
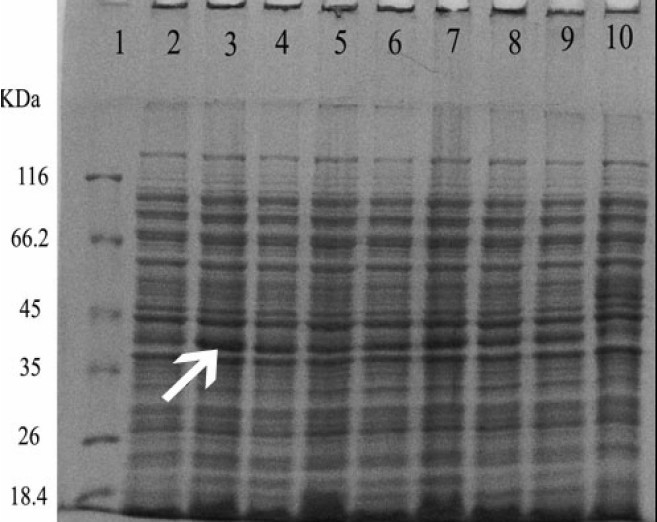
Induction of the expression of reteplase by different concentrations of IPTG in E. coli BL21 cells. Proteins in the whole cell lysates were separated using SDS-PAGE. Line:1) protein marker, Lane:2) rpETBL21 induced with 1.5 mM IPTG, Lane:3) rpET-BL21 induced with 1 mM IPTG . Lane:4) rpET-BL21 induced with 0.5 mM IPTG, Lane:5) rpET-BL21 induced with 0.25 mM IPTG, Lane:6) rpET-BL21 induced with 1.5 mM IPTG, Lane:7) rpET-BL21 induced with 1 mM IPTG. Lane:8) rpET-BL21 induced with 0.5 mM IPTG, Lane:9) rpET-BL21 induced with 0.25 mM IPTG (0.25 mM), Lane:10) BL21 induced with 1 mM IPTG .Arrow shows the 39 kDa reteplase band.
Fig. 3.
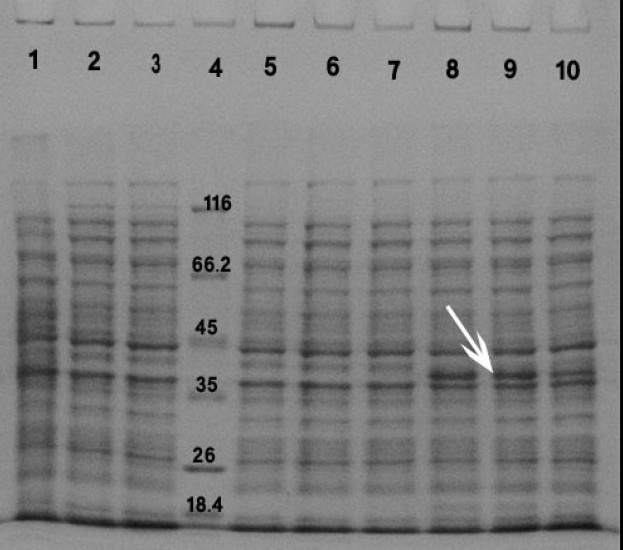
Induction of the expression of reteplase with 1 mM IPTG using different times of incubation. Proteins in the whole cell lysates were separated using SDSPAGE. Lane:1) rpET-BL21 uninduced for 2 h, Lane:2) BL21 uninduced for 1 h, Lane:3) BL21 uninduced for 2 h, Lane:4) protein marker, Lane:5) pET-BL21 induced for 1 h, Lane:6) pET-BL21 induced for 2 h, Lane:7) pET-BL21 induced for 3 h, Lane:8) rpETBL21 induced for 1 h, Lane:9) rpET-BL21 induced for 2 h, Lane:10) rpET-BL21 induced for 3 h. Arrow shows the 39 kDa reteplase band.
The effect of different temperatures on reteplase expression was examined by induction with 1 mM IPTG for 2 h at different incubation temperatures. The most intense band on SDS-PAGE was detected at 37°C (Fig. 4).
Fig. 4.
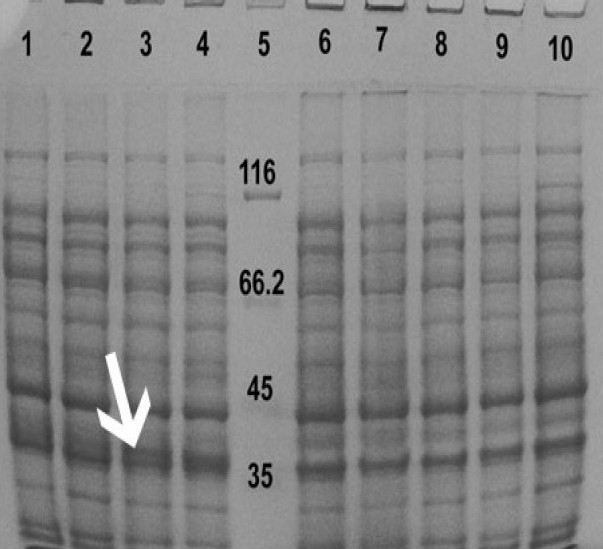
Induction of the expression of reteplase with 1 mM IPTG for 2 h using different incubation temeratures. Proteins in the whole cell lysates were separated using SDS-PAGE. Lane:1) rpET-BL21 induced at 25°C, Lane:2) rpET-BL21 induced at 30°C, Lane:3) rpET-BL21 inducaed at 37°C, Lane:4) rpETBL21 induced at 39°C, Lane:5) protein marker. Lane:6) pET-BL21 induced at 25°C, Lane:7) pETBL21 induced at 30°C, Lane:8) pET-BL21 induced at 37°C, Lane:9) pET-BL21 inducted at 39°C and Lane:10) Ecoli BL21 cells grown at 37°C. Arrow shows the 39 kDa reteplase band.
The effect of shaking speed on reteplase expression was also examined. Maximum production of reteplase was detected at 100 rpm (Fig. 5).
Fig. 5.
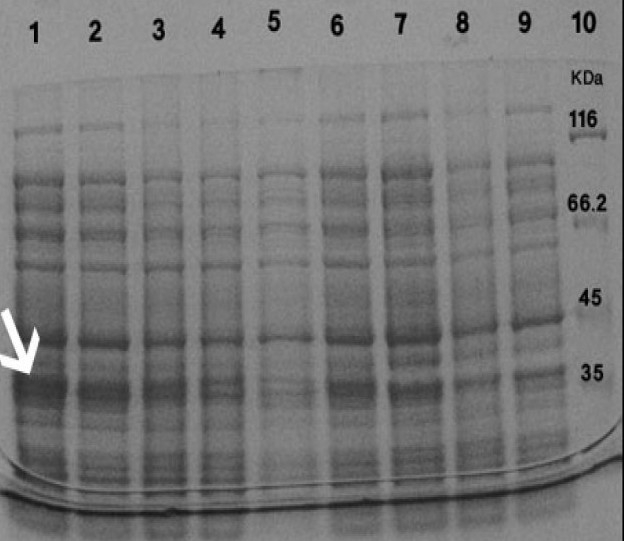
Induction of the expression of reteplase with 1 mM IPTG for 2 h using different shaking speeds. Proteins in the whole cell lysates were separated using SDS-PAGE Lanes:1 and 2): rpET-BL21 induced at 100 rpm, Lanes:3 and 4): rpET-BL21 induced at 170 rpm, Lanes:5 and 6): rpET-BL21 induced at 190 rpm. Lane:7) pET-BL21 induced at 100 rpm, Lane:8) pETBL21 induced at 170 rpm, Lane:9) pET-BL21 induced at 190 rpm and Lane:10) protein marker. Arrow shows the 39 kDa reteplase band.
Also, supplementing media with glucose showed a negative effect on the protein expression having the most intense protein band in the absence of glucose (Fig. 6).
Fig. 6.
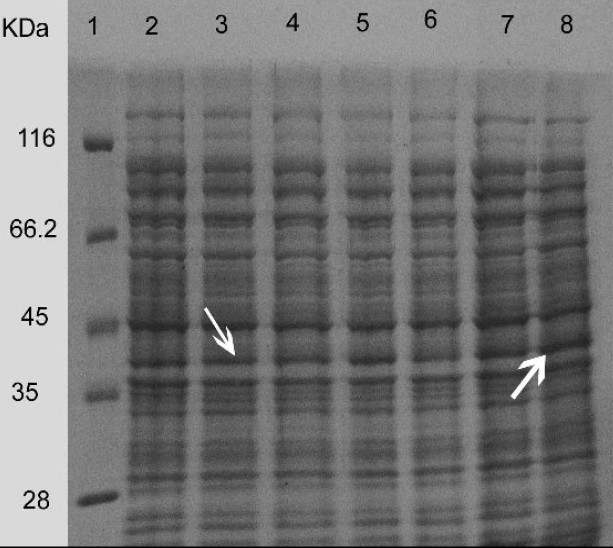
Induction of the expression of reteplase with 1 mM IPTG for 2 h at 37°C using different concentrations of glucose. Proteins in the whole cell lysates were separated using SDS-PAGE. Lane:1) protein marker, Lane:2) rpET-BL21 induced in the presence of.25 mM glucose, Lane:3) rpET-BL21 induced in the presence of 0.5 mM glucose, Lane:4) rpET-BL21 induced in the presence of 0.75 mM glucose, Lane:5) rpET-BL21 induced in the presence of 1 mM glucose, Lane:6) rpET-BL21 induced in the presence of 1.5 mM glucose, Lane:7) rpET-BL21 induced in the presence of 2 mM glucose and Lane:8) rpET-BL21 induced in the absence of glucose. Arrows show the 39 kDa reteplase band.
DISCUSSION
The purpose of this investigation was to examine the effect of different experimental conditions on the expression of reteplase in E. coli. pET system has several advantages that persuaded us to use this system for the expression of our gene(1,9). pET is one of the most powerful systems developed for the expression of recombinant proteins in E. coli. Strain BL21 (DE3) is also the most widely used host for target gene expression. Cell mass and target protein yields are often increased several-fold using IPTG induction(10). By adjusting the concentration of IPTG, expression of proteins can be regulated at different levels, lower level expression can increase the solubility and activity of some target proteins(9). The concentration of IPTG used to induce lac repressor-regulated promoters can dramatically influence expression. With some proteins, it is important to slowly induce transcription of the expression plasmid (with lower IPTG concentrations) while with others, production of high amounts of protein is desired(11). Therefore in the current study the effect of different concentrations of IPTG on reteplase expression was examined and 1 mM concentration was selected as being optimum.
In our experiments, the optimal temperature for protein expression was 37°C. Schein and coworkers reported that growth at 37°C caused some proteins to accumulate as inclusion bodies, while incubation at 30°C lead to soluble and active protein. Prolonged (e.g., overnight) induction at low temperatures (15-20°C) may prove optimal for the yield of soluble protein(12). Regarding shaking speeds, in our experiments low speeds resulted in better protein expression. Also, the addition of glucose is not recommended during reteplase expression.
Zhao and coworkers have reported the cloning and expression of reteplase in E. coli using pET22b vector(13). As compared to our study, they used a fixed experimental setting (1 mM IPTG, 37°C, shook for 14 h at 150 rpm) and did not investigate the influence of different factors on protein expression. On the other hand, a group of Chinese researchers seem to have studied some of the parameters measured in our investigation. However, since their work was not written in English language, it was difficult for us to properly compare the results of their study with ours. The main difference between the above mentioned studies and ours is the used expression vector. In those studies, pET22b was utilized which adds pelB leader peptide to the N-terminal section of reteplase favoring secretion of the soluble recombinant protein in the periplasmic space. However, in the current investigation, pET15b was used which does not contain this sequence and thus the produced recombinant protein would precipitate as inclusion bodies inside the cell. Therefore the strategies for purification and refolding of protein could completely differ in these cases. The main advantage of inclusion bodies is that they are mostly composed of the recombinant protein and can be easily isolated from the cell debris(11).
CONCLUSION
We investigated various conditions including IPTG concentrations (0.25, 0.5, 1 and 1.5 mM) various temperatures (25, 30, 37 and 39°C), shaking speeds (100, 170 and 190 rpm) and glucose concentrations (0.25, 0.5, 0.75 and 1 mM) on the expression of reteplase. Maximum amount of protein production was obtained by the addition of 1 mM IPTG at 37°C, and 100 rpm of shaking speed in the absence of glucose.
The influence of the above mentioned variables on the the expression of reteplase in complete E. coli cells would allow us to study the parameters affecting the function of this protein in future experiments.
ACKNOWLEDGMENT
This work was financially supported by the research council of Isfahan University of Medical Sciences (grant No. 188048).
REFERENCES
- 1.Lievadot J, Giugliano RP, Antman EM. Bolus fibrin-olytic therapy in acute myocardial infraction. Clin Cardiol. 2001;286:442–449. doi: 10.1001/jama.286.4.442. [DOI] [PubMed] [Google Scholar]
- 2.Qiu J, Swartz RJ, Georgiou G. Expression of active human tissue-type plasminogen activator in Escherichia coli. Appl Environ Microbiol. 1998;64:4891–4896. doi: 10.1128/aem.64.12.4891-4896.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glick B, Pasternak J. Mulecular Biotechnology: principles and application of recombinant DNA. 2nd ed. Washington D. C: ASM press; 1998. [Google Scholar]
- 4.Makrides SC. Sterategis for achiving high-level expression of gene in Escherichia Coli. Microbiol Rev. 1996;60:512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pennica D, Holmes WE, Kohr WI, Harkins RN, Vehar GA, Ward CA, et al. Cloning and expression of human tissue-type plasminogen activator cDNA in E. coli. Nature. 1983;301:214–221. doi: 10.1038/301214a0. [DOI] [PubMed] [Google Scholar]
- 6.Vallejo LF, Rinas U. Strategies for the recovery of active proteins through refolding of bacterial inclusion body proteins. Microb Cell Fact. 2004;3:1–12. doi: 10.1186/1475-2859-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandi N, Sundaram KR, Tandra SK, Bandy-opadhyay S, Padmanabhan S. Asn12 and Asn278: Critical residues for in vitro biological activity of reteplase. Adv Hematol. 2010;301:172–484. doi: 10.1155/2010/172484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Youchun Z, Wang G, Yang K, Changkai Z. Cloning, expression, and renaturation studies of reteplase. J Microbiol Biotechnol. 2003;13:989–992. [Google Scholar]
- 9.Dormiani K, Khazaie K, Mir MohamadSadeghi H, Rabbani M, Moazen F. Cloning and expression of a human tissue plasminogen activator variant: K2S in Escherchia coli. Pakistan J Biol Sci. 2007;10:946–949. doi: 10.3923/pjbs.2007.946.949. [DOI] [PubMed] [Google Scholar]
- 10.Grabski A, Mehler M, Drott D. The overnight express autoinduction system: high-density cell growth and protein expression while you sleep. Nat Meth. 2005;2:233–235. [Google Scholar]
- 11.Sambrook J, Russell DW. Molecular Cloning a Laboratory Manual. 3rd ed. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 12.Schein CH. Production of soluble recombinant proteins in bacteria. Nat Biotechnol. 1989;7:1141–1149. [Google Scholar]
- 13.Zhao Y, Wang G, Yang K, Changkai Z. Cloning, expression, and renaturation studies of reteplase. J Microbiol Biotechnol. 2003;13:989–992. [Google Scholar]


