SUMMARY
Eleven-nineteen Lysine-rich Leukemia (ELL) participates in the Super Elongation Complex (SEC) with the Pol II CTD kinase P-TEFb. SEC is a key regulator in the expression of HOX genes in Mixed Lineage Leukemia (MLL) -based hematological malignancies, in the control of induced gene expression early in development, and in immediate early gene transcription. Here, we identify an SEC-like complex in Drosophila, as well as a distinct ELL-containing complex that lacks P-TEFb and other components of SEC named the “little elongation complex” (LEC). LEC subunits are highly enriched at RNA Polymerase II (Pol II) -transcribed small nuclear RNA (snRNA) genes, and the loss of LEC results in decreased snRNA expression in both flies and mammals. The specialization of the SEC and LEC complexes for mRNA and snRNA-containing genes, respectively, suggests the presence of specific classes of elongation factors for each class of genes transcribed by RNA polymerase II.
INTRODUCTION
Transcription of genes catalyzed by RNA Polymerase II (RNA Pol II) is a multifaceted process requiring the collaborative action of a large number of proteins in order to ensure appropriate initiation, elongation, and termination of transcription. The disparity between the efficiency of transcription by RNA Pol II in cells when compared to its efficiency in the test tube was the basis for the biochemical approaches toward the identification of factors that can stimulate or regulate transcription (Sims et al., 2004; Smith et al., 2011). The product of the Eleven–Nineteen Lysine-rich Leukemia gene (ELL) (Thirman et al., 1994), a common translocation partner of the Mixed-Lineage Leukemia (MLL) gene in acute myeloid leukemia, was biochemically identified and purified from rat liver extracts as a single polypeptide that could increase the catalytic rate of transcription elongation by RNA Pol II in vitro and also function as an elongation factor in vivo (Eissenberg et al., 2002; Gerber et al., 2001; Shilatifard et al., 1996). MLL is found in a number of translocations where its N-terminus is fused to the C-terminal portion of a wide variety of other proteins with very little sequence similarity (Mohan et al., 2010). Recently, ELL was shown to be part of the Super Elongation Complex (SEC) containing other MLL translocation partners, AF9, ENL, AFF1 and AFF4, as well as the Pol II CTD kinase P-TEFb (Lin et al., 2010). SEC has been shown to be one of the active forms of P-TEFb and is required for the misexpression of HOX genes in MLL-AF4 transformed cells (Lin et al., 2010); for HIV transactivation (He et al., 2010; Sobhian et al., 2010); for normal cellular functions such as the induction of HSP70 by heat shock; and for developmental genes’ responses to environmental signals (Lin et al., 2011; Lin et al., 2010).
In a mouse model system, the transformation of hematopoietic cells by MLL-ELL chimeras was shown to require the occludin homology domain at the C-terminus of ELL (DiMartino et al., 2000; Luo et al., 2001; Miller et al., 2000; Shilatifard et al., 1997). In Drosophila, the occludin homology domain was also shown to be sufficient to rescue loss-of-function alleles of the Su(Tpl) gene that encodes ELL (Eissenberg et al., 2002; Gerber et al., 2005). In occludin, this domain interacts with ZO-1 at tight junctions (Furuse et al., 1994). In this study, we sought to identify proteins that associate with this domain of ELL and found that the fly homologs of MLL translocation partners, Lilli (AFF1 and AFF4-related) and Ear (ENL and AF9-related), are found in a SEC-like complex with ELL. However, we also identified a set of relatively uncharacterized proteins in Drosophila that associates with the C-terminus of ELL in a complex that we call LEC, which regulates snRNA transcription from flies to humans.
RESULTS
Identification of proteins that interact with ELL's occludin homology domain
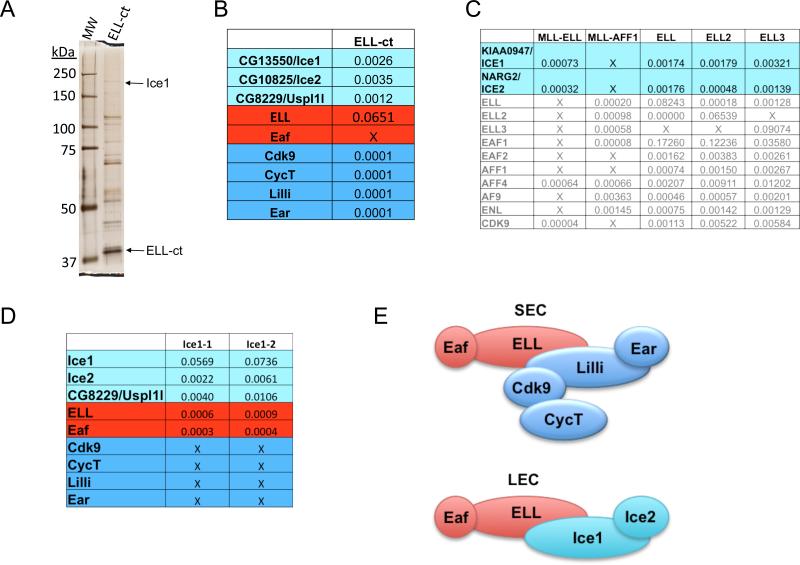
The conserved C-terminus of ELL (ELL-ct), containing the occludin homology domain, was expressed as a Flag-fusion protein in Drosophila S2 cells, isolated from nuclear extracts by Flag-affinity chromatography and processed for Multidimensional Protein Identification Technology (MudPIT). ELL-ct brought down three previously uncharacterized Drosophila proteins, CG13550, CG10825 and CG8229 (Figure 1A-B, Figure S1A). As expected, the Drosophila homologs of SEC components, Cdk9, CycT, Lilli (human AFF1-4 related), and Ear (human ENL and AF9 related), are also pulled down with ELL-ct (Figure 1B).
Figure 1.
Identification of proteins interacting with the C-terminal occludin homology domain of Drosophila ELL. (A) Silver stain gel of a Flag purification of a C-terminal portion of ELL (ELL-ct) from Drosophila S2 cell nuclear extracts. (B) MudPIT analysis of the Flag-affinity purifications shown in (A). CG13550 and CG10825 were enriched in the ELL-ct purifications and are, therefore, referred to as Interacts with the C-terminus of ELL 1 and 2 (Ice1 and Ice2), respectively. Normalized spectral abundance factors (NSAF), which show the relative abundance of each protein in a mixture of proteins (Zhang et al., 2010), are shown. (C) We previously reported the purification of MLL chimeras frequently found in leukemia. NSAF values for proteins identified in the Flag-MLL-ELL and Flag-MLL-AFF1 chimera purifications, and in Flag-ELL, Flag-ELL2, and Flag-ELL3 purifications are shown. Proteins in gray are the previously reported components of SEC (Lin et al. 2010). Two proteins that uniquely co-purified with MLL-ELL, but not the other chimeras tested, are KIAA0947 and NARG2, which based on Blast searches and sequence alignments are the human orthologs of Drosophila Ice1 and Ice2 (please see Figure S1). ICE1 and ICE2 are also found in ELL, ELL2, and ELL3 Flag affinity purifications from human 293T cells. (D) Isolation of Flag-Ice1 (CG13550) from S2 cells and identification of interactors by MudPIT. Ice1-associated proteins include Ice2, Eaf, ELL, and CG8229, a protein sharing limited homology to USPL1 in humans. Two different purifications are indicated as Ice1-1, Ice1-2. (E) Schematic of proposed ELL complexes from Drosophila that are conserved in humans. Top panel, the Drosophila version of the Super Elongation Complex contains ELL, Eaf, Lilli (related to AFF1-4 in humans), Ear (ENL and AF9 related), and the RNA Pol II CTD kinase P-TEFb subunits, Cdk9 and CycT. Bottom panel, the Little Elongation Complex (LEC) contains ELL, Eaf, Ice1 and Ice2.
We previously reported the purification of several of the common MLL chimeras associated with acute lymphoid and myeloid leukemia (Lin et al., 2010). These studies led us to identify SEC, a novel P-TEFb-containing complex containing the MLL fusion partners ELL, AFF1 and/or AFF4, and AF9 and/or ENL, as the functional link between these otherwise seemingly disparate MLL translocations (Lin et al., 2010). However, two proteins, KIAA0947 and NARG2, were only identified in the MLL-ELL purifications, but not with MLL-AFF1 or other MLL chimeras (Figure 1C and data not shown). KIAA0947 and NARG2 were also found to co-purify with all three Flag-tagged human ELL paralogs: ELL, ELL2 and ELL3 (Figure 1C). KIAA0947 and NARG2 were also identified from MED26 purifications to be in a complex with ELL and EAF (Takahashi et al., 2011).
Drosophila CG10825 is identified in the Pfam database as being related to human NARG2 (NMDA receptor-regulated gene 2); a gene named for its up-regulation in NMDAR knockout mice, but whose function is unknown (Sugiura et al., 2001). PSI-BLAST searches and sequence alignments identify Drosophila CG13550 and human KIAA0947 as sharing related C-terminal domains. Drosophila CG13550 and human KIAA0947 also have an N-terminal coiled-coil region in common, further indicating that these proteins are orthologs (Figure S1B-C). Given that these proteins were identified in the purifications of the C-terminal portions of ELL from both Drosophila (ELL-ct) and human cells (MLL-ELL), we refer to Drosophila CG13550 and human KIAA0947 as ICE1 and Drosophila CG10825 and human NARG2 as ICE2, with ICE standing for Interacts with the C-terminus of ELL.
Flag-tagged Drosophila Ice1 was also isolated from S2 cells and found to associate with Ice2, ELL and Eaf (Figure 1D). Another protein identified in both the Ice1 and ELL-ct Flag purifications was CG8229. This protein has an N-terminal zinc finger domain related to the N-terminus of USPL1 (Figure S1D), which is of potential interest because USPL1 had been epitope-tagged and purified as part of a large scale proteomics study of ubiquitin-specific protease family members in 293T cells (Sowa et al., 2009). The most significant USPL1 interactors were ELL and ICE1(Sowa et al., 2009). However, USPL1 was not identified in our ELL1-3 or MLL-ELL purifications, so the generality of an USPL1-ICE1-ELL complex remains to be determined. Overall, based on the available biochemical data from Drosophila (present study) and from mammalian cells (Takahashi et al., 2011), ELL is a component of at least two complexes: the SEC and the LEC (Figure 1E).
Ice1 and ELL are positive factors for U11 transcription by Pol II
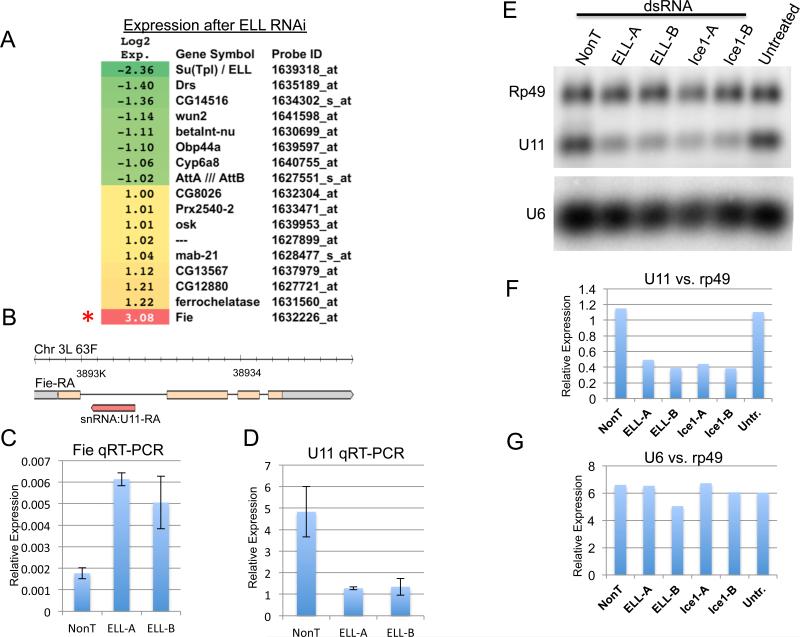
Prior to our biochemical isolation of the ELL complexes, we had performed gene expression analysis in Drosophila S2 cells after depletion of ELL by RNAi knockdown (Figure 2A). Despite ELL's presence at many active sites of transcription, we saw very few genes change 2-fold or more in expression. ELL, the target of the RNAi, was the most down-regulated gene on the array, while Fire exit (Fie) was the most changed gene overall, but was up-regulated. Fie encodes a transmembrane protein expressed in a subset of glial cells (With et al., 2003), but has no obvious connection to known ELL-regulated pathways, such as stress response or Ras signaling (Eissenberg et al., 2002; Lin et al., 2010; Smith et al., 2008). Fie is notable for containing the single copy U11 gene, transcribed antisense to the Fie transcript (Figure 2B). Conceivably, ELL could be a positive regulator of U11, while increased transcription of Fie could be a result of decreased transcriptional interference from antisense U11 transcription (Yuan et al., 2003). RT-PCR of ELL RNAi cells shows that U11 is indeed down-regulated, while confirming the up-regulation of Fie in these cells (Figure 2C-D). We also tested the effect of the knockdown of ELL and Ice1 on U11 snRNA expression by Northern analysis. Two different dsRNA regions were used to reduce the expression of ELL and Ice1 in S2 cells. U11 snRNA levels were reduced by both ELL and Ice1 knockdown when compared to either Rp49 or to the Pol III- transcribed U6 snRNA (Figure 2E-G).
Figure 2.
LEC regulates U11 snRNA gene transcription. (A) Heatmap of gene expression changes after ELL RNAi in S2 cells. Shown are the genes whose expression changed 2- fold. The most changed gene on the array is Fire exit (Fie) which is up-regulated, while Su(Tpl), which encodes ELL, was the second most changed gene, and is down-regulated. (B) Fie is notable for having a snRNA gene in its first intron. The FlyBase annotation for the Fie transcript shows that the U11 snRNA gene is transcribed in the anti-sense direction relative to Fie, which could conceivably antagonize Fie expression. Yellow represents an open reading frame; gray, untranslated regions; thin lines, introns; and the non-coding transcript for U11 is shown in red. (C-D) To determine whether ELL regulates U11 or Fie, we performed quantitative RT-PCR on these two genes after RNAi of ELL using two non-overlapping regions of dsRNA, ELL-A, and ELL-B. While Fie is up-regulated after ELL RNAi (C), U11 is down-regulated (D). Expression is relative to Rp49. Error bars represent the standard deviations. (E-G) Northern blot analysis of ELL and Ice1 RNAi samples shows that U11, but not the Pol III-transcribed U6 snRNA gene, is dependent on LEC. The same Northern blot was probed with Rp49, U11, and U6 32P-labeled riboprobes and analyzed by phosphorimaging and quantitation with Typhoon scanner and ImageQuant software. The scan (E) and quantitations of U11 (F) and U6 (G) relative to Rp49 are shown.
Genome-wide analyses demonstrate a general requirement for LEC in snRNA transcription by Pol II
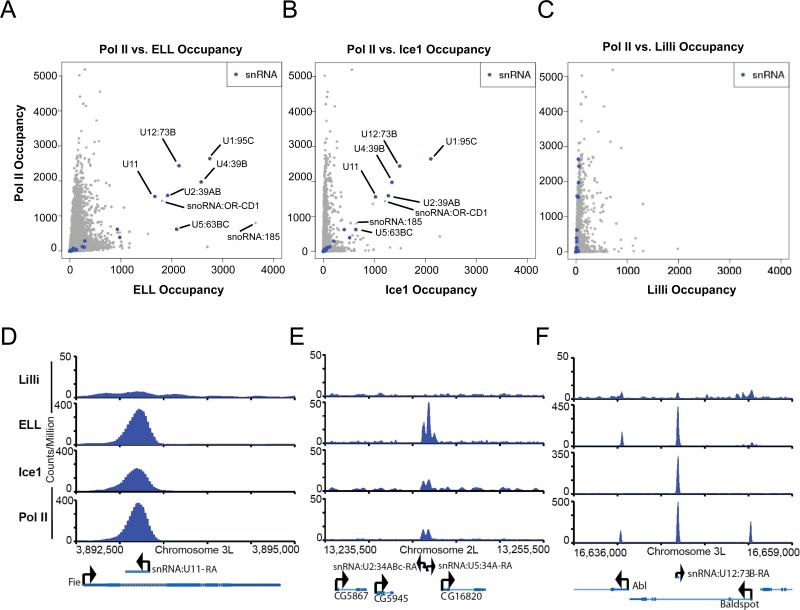
In order to take an unbiased approach to LEC function, we performed ChIP-seq analyses with antibodies to the LEC components ELL and Ice1, and with an antibody to the SEC component Lilli for comparison in Drosophila cells. By plotting Pol II occupancy vs. the occupancy of each of these factors at known transcription start sites, the snRNA genes are identified as sites of strong enrichment of LEC and RNA Pol II (Figure 3A-B). Lilli, a component of SEC, is not strongly enriched at the snRNA genes, not even at those with very high levels of RNA Pol II (Figure 3C). Examples of ELL, Ice1, and Pol II colocalization at snRNA genes are shown in Figure 3D-F. We performed ChIP followed by qPCR with antibodies to ELL and ELL-associated proteins in Drosophila to confirm the occupancy of ELL and Ice, but not Lilli, at snRNA genes (Figure S2A). We also performed ChIP-seq with a different ELL antibody and observed a similar level of enrichment of ELL at the snRNA genes (Figure S2B). Gene ontology analysis of LEC-bound genes shows enrichment for categories linked to mRNA splicing due to the occupancy of the snRNA genes (Figure S2C).
Figure 3.
Drosophila LEC is enriched at Pol II-transcribed snRNA genes. (A) Scatterplot depicting the genome-wide occupancy levels of Pol II and ELL. The highest occupied co-bound genes encode snRNAs. Each point is a transcript from Ensembl62 for fruit fly. The occupancy levels are the sum of extended read fragments per million within +/- 5bp of the transcription start site position. (B) Scatterplot depicting the genome-wide occupancy levels of Pol II and Ice1, showing that the highest co-bound genes encode snRNAs. (C) Scatterplot depicting the genome-wide occupancy levels of Pol II and Lilli, showing that snRNA genes, while bound at varying and high degrees of Pol II, are not bound at high levels by Lilli. (D-F) Examples of snRNA and protein-coding genes showing particularly high enrichment for LEC components, ELL and Ice1, but not the SEC component Lilli at the snRNA genes.
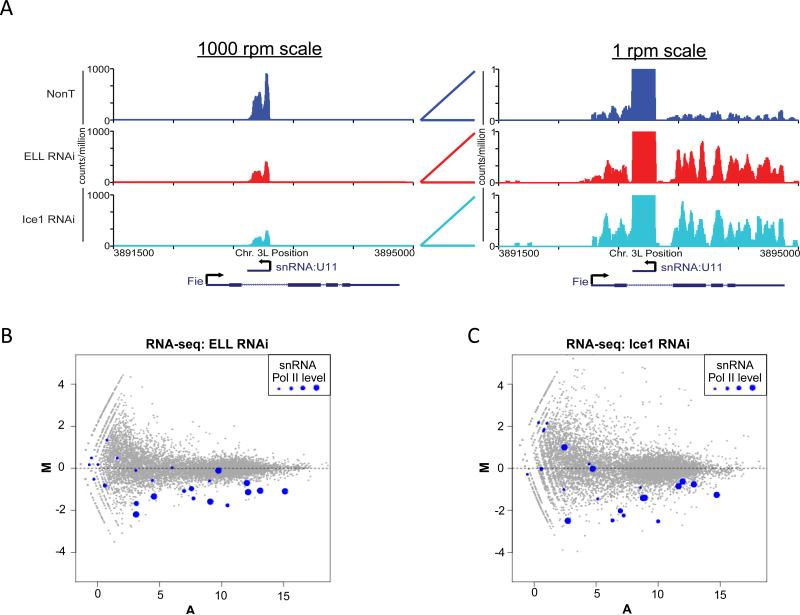
To determine whether LEC is required for snRNA transcription, we used RNAi to knock down ELL and Ice1 levels in S2 cells. In order to allow sequencing of snRNAs, which are not polyadenylated, while avoiding the sequencing of mostly rRNA, total RNA from the RNAi-treated cells was depleted of rRNA using the Ribo-Zero kit from Epicentre Technologies. Ribo-Zero was designed to deplete rRNA from human cells using biotinylated oligos complementary to 5S, 18S, and 28S rRNAs. Bioanalyzer analysis of the Drosophila RNA subjected to the Ribo-Zero procedure indicates that the 18S rRNA is mostly depleted, while the 28S band is reduced by 50% (Figure S3). The rRNA-depleted samples were subjected to RNA-seq. As an initial test of the validity of the RNA-seq results, we looked at the Fie gene and found it to be up-regulated in ELL and Ice1 RNAi, while U11 expression was reduced, thus demonstrating our ability to assay both protein coding and non-polyadenylated genes in Drosophila by RNA-seq (Figure 4A).
Figure 4.
RNA-seq analysis of Drosophila S2 cells after ELL and Ice1 RNAi. (A) RNA-seq analysis confirms the original finding by RT-PCR and Northern analysis, that U11 snRNA is down-regulated by ELL and Ice1 RNAi and Fie is up-regulated. The left panel shows a chromosomal region containing Fie at the 1000 reads per million (rpm) scale. Only U11 transcripts can be seen at this scale and its levels are visibly reduced in the ELL and Ice1 RNAi conditions. The right panel shows the same chromosomal region at a scale of 1 rpm where U11 transcripts are off-scale. Within the Fie exons, transcript levels are visibly higher in the ELL and Ice1 RNAi conditions. (B-C) MA plots of genome-wide expression changes as determined by RNA-seq for ELL RNAi and Ice1 RNAi, respectively. Log2 fold-changes (M) of normalized read abundances of shRNA/wt for each gene are plotted on the y-axis. The log2 average normalized read abundance (A), as reported by DESeq (Anders and Huber, 2010) is plotted on the x-axis. Each dot represents a gene from Ensembl62. Dots highlighted in blue represent snRNA genes. The four sizes of blue dots correspond to quartiles of Pol II occupancy, with the smallest dots representing the lowest levels of Pol II binding (the bottom 25%), and the largest dots representing the highest levels of Pol II (the top 25%) at the snRNA genes. Pol II quartiles were determined from Pol II bound genes.
Our genome-wide RNA-seq analyses from the ELL and Ice1 RNAi knockdown cells show that snRNA genes are mostly down-regulated after reduction of ELL and Ice1 (Figure 4B-C). Gene ontology (GO) analysis of down-regulated genes shows that categories associated with snRNAs and RNA splicing are significantly enriched terms in LEC knockdown cells (Figure S3B). Sorting gene expression by biotype, we observe that snRNA genes are the most changed class of genes in LEC knockdown cells (Figure S3C-D). Together, these data indicate that LEC is generally required for snRNA transcription by RNA Pol II in Drosophila.
Conservation of LEC function in mammals
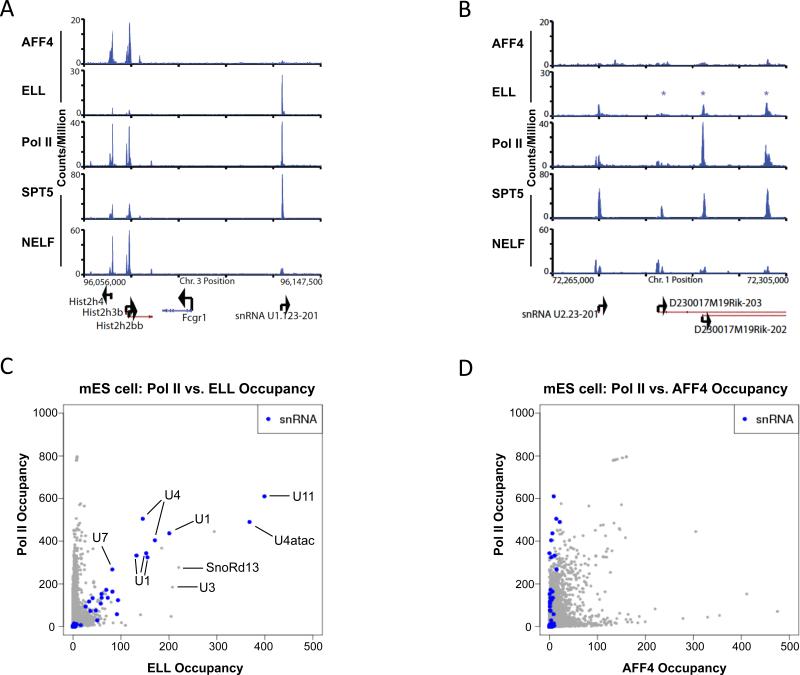
To determine if LEC plays a similar role in snRNA transcription in mammals, we analyzed the global localization patterns of ELL and AFF4 by ChIP-seq in mouse embryonic stem cells. We find that ELL is strongly enriched at snRNA genes (Figure 5A-C). In contrast, AFF4, which was shown to be required for SEC complex stability in HeLa cells (Lin et al., 2010), is present at relatively low levels on snRNA genes when compared to RNA Pol II (Figure 5A-D). This distinction can be clearly visualized at a region of chromosome 3 that has a cluster of histone genes in the vicinity of a copy of a U1 snRNA gene (Figure 5A). While AFF4 and Pol II are strongly enriched at the histone loci, ELL and Pol II are strongly enriched at the U1 snRNA gene. The presence of high levels of ELL and RNA Pol II without high levels of other SEC components could potentially be used to identify novel snRNA genes. For example, on chromosome 1, an annotated copy of the U2 snRNA gene, U2.23-201, is bound by high levels of ELL, Pol II and Spt5, but relatively low levels of AFF4 (Figure 5B). This gene resides within a 30 kb region that includes three other unannotated (as of Ensembl 63) copies of U2 snRNA genes, two of which have high levels of ELL, Pol II and Spt5, but relatively low AFF4 (Figure 5B). Taking a genome-wide view, we compared Pol II and ELL co-occupancy by plotting the reads per million of Pol II by the reads per million of ELL at the TSS of genes, and highlighting the snRNA genes in blue (Figure 5C). This figure shows that many of the highest ELL and Pol II co-occupied genes are snRNA genes. In contrast, plotting Pol II vs. AFF4 occupancy shows that even the snRNA genes with the highest levels of Pol II are not strongly enriched for AFF4 (Figure 5D).
Figure 5.
ELL is enriched at snRNA genes in mouse ES cells. (A-B) Binding profiles in mouse ES cells: the occupancy of the factors indicated on the right show ELL, but not AFF4, associating with Pol II at snRNA genes. (A) AFF4 is strongly co-enriched with ELL and Pol II at a cluster of histone genes on chromosome 3. However, a neighboring U1 snRNA gene only shows strong enrichment for ELL and Pol II. AFF4 and Pol II data from Lin et al. (Lin et al., 2011). (B) A segment of chromosome 1 has a U2 snRNA gene (U2.23-201). To the right, are three other U2 snRNA genes identifiable by BLAST (indicated with asterisks) that are within long intergenic noncoding RNAs (lincRNA). Three of these four U2 snRNA genes are strongly enriched for ELL and Pol II, but not AFF4. SPT5 and NELF data from Rahl et al. (Rahl et al., 2010) are shown for comparison. SPT5 associates with both NELF and elongating Pol II (Yamaguchi et al., 1999). NELF regulates Pol II at the transcription start sites of many genes, but is also required for 3' end processing of some snRNA genes (Egloff et al., 2009). (C) Scatterplot showing genome-wide occupancy levels of Pol II and ELL in mouse ES cells (mES). The highest occupied co-bound genes encode snRNAs. Each point is a transcript from Ensembl62 for mouse. The occupancy levels are the sum of extended read fragments per million within +/- 5bp of the transcription start site position. Since only uniquely mappable reads are used, some multi-copy snRNA genes are underrepresented on the plot. (D) Scatterplot of the genome-wide occupancy levels of Pol II and AFF4 in mES cells, showing that snRNA genes, while bound at varying and high degrees of Pol II, are not bound at high levels by AFF4.
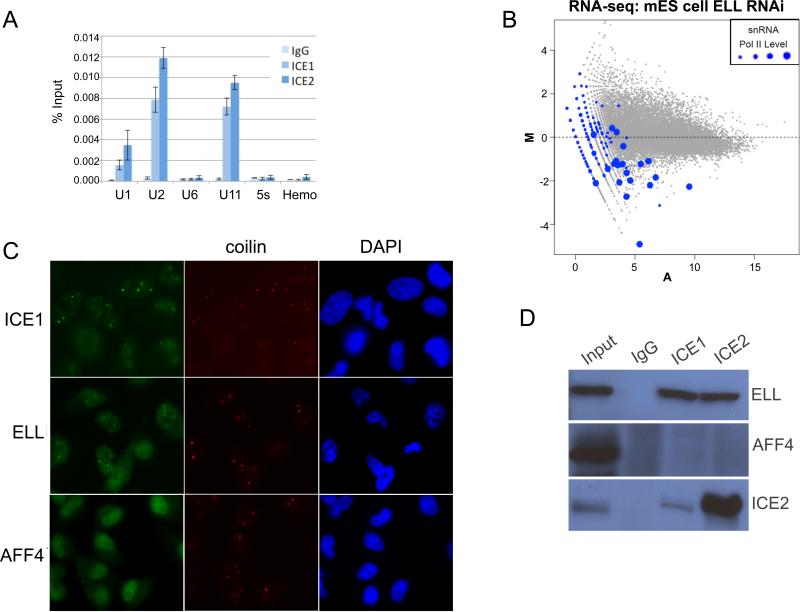
ChIP with human ICE1 and ICE2 antibodies was used to confirm that it is the LEC version of ELL that can be recruited to Pol II transcribed snRNA genes (Figure 6A). Furthermore, RNA-seq analysis after shRNA-mediated knock down of ELL in mouse ES cells results in a general reduction of levels of snRNAs (Figure 6B), demonstrating a conserved function of LEC in snRNA transcription from Drosophila to mammals. Previous studies showed that ELL and Eaf are found to co-localize with coilin at Cajal bodies in HeLa cells (Polak et al., 2003), although the functional significance of this was not clear as Cajal bodies are heterogeneous in composition and may carry out several different functions (Cioce and Lamond, 2005). However, Cajal bodies have been found at sites of snRNA transcription (Frey and Matera, 2001). We, therefore, asked whether ICE1 of LEC and AFF4 of SEC colocalize with coilin in these cells (Figure 6 C). We find that LEC subunits ICE1, but not the SEC component AFF4, colocalize at many sites with coilin, a cytological demonstration that LEC is a distinct complex from SEC. Endogenous immunoprecipitations with 293T cell extracts using ICE1 and ICE2 antibodies further confirm that ELL, but not AFF4, is a component of LEC (Figure 6D).
Figure 6.
LEC regulates snRNA transcription in mammals. (A) ChIP with human ICE1 and ICE2 antibodies from 293T cells confirms that it is the LEC form of ELL that is present at Pol II-transcribed snRNA genes. The Pol III transcribed U6 snRNA and 5s RNA genes are not occupied by ICE1 or ICE2. A hemoglobin gene (Hemo) serves as a non-transcribed control gene for these cells and normal Rabbit IgG serves as a non-specific antibody control. Error bars indicate the standard deviations. (B) shRNA-mediated knockdown of ELL in mouse ES (mES) cells shows that ELL is responsible for the expression of many Pol II-transcribed snRNA genes. Ribosomal RNA was depleted from total RNA by Ribo-Zero treatment and subjected to RNA-seq. The MA plot highlights snRNA genes with different levels of Pol II occupancy as in Figure 4. (C) ICE1 and ELL, but not AFF4 (shown in green), are found in Cajal bodies of HeLa cells. Coilin (shown in red) serves as a marker for Cajal bodies. Nuclei are stained with DAPI. (D) Endogenous co-immunoprecipitation from 293T cells with antibodies to human ICE1 and ICE2 pulls down ELL, but not AFF4.
DISCUSSION
Transcription of snRNA genes has been a valuable model system for understanding transcription regulation by Pol II and Pol III (Hernandez, 2001; Jawdekar and Henry, 2008). The sequences of the promoters of Pol II and Pol III-transcribed genes are very similar and they both recruit and require the same SNAP complex (SNAPc). Relatively minor sequence changes, such as the addition of a TATA box to a Pol II transcribed snRNA promoter, can result in the gene being transcribed by RNA Pol III (Hernandez, 2001). Addition of a mRNA promoter to a snRNA can result in the transcript being processed by the mRNA 3' cleavage and polyadenylation factor (Hernandez, 2001). These studies provided important clues to the interconnectedness of initiation, termination and processing of mRNAs.
In metazoans, Pol II-transcribed snRNA genes also recruit the Integrator complex, a large multiprotein complex that interacts with the Pol II CTD (Baillat et al., 2005). The Integrator complex possesses an endonuclease activity required for 3' end processing of U1 and U2 snRNAs. Integrator is well characterized for its role in 3' end formation, but as a large 12-protein complex, could perform other roles in the transcription regulation of snRNA genes. Integrator interacts with a Ser7 phosphorylated form of the RNA Pol II CTD, a site of phosphorylation shown to be required for U1 and U2 snRNA transcription (Egloff et al., 2007). snRNA transcription is relatively insensitive to P-TEFb inhibitors (Medlin et al., 2005), which indicates distinct regulatory requirements for this class of Pol II-transcribed genes.
How does LEC specifically associate with Pol II transcribed snRNA genes?
Genome-wide ChIP-seq and RNA-seq experiments in both flies and mammals demonstrate a requirement for LEC, but not SEC for snRNA transcription by Pol II. An important question is how LEC specifically associates with Pol II at this class of genes. Although ELL can directly interact with Pol II, and snRNA genes are among the highest in Pol II levels, this interaction would not explain the differential recruitment of LEC vs. SEC. Similarly, Med26, a metazoan-specific component of Mediator, can interact with Eaf (Takahashi et al., 2011), but Eaf is also a common component of both LEC and SEC. Additional candidates for recruiting LEC to snRNA genes include the Integrator complex (Baillat et al., 2005; Egloff et al., 2007) and the unique modification states of Pol II at these genes (Egloff et al., 2010; Sims et al., 2011). A combination of some of the above interactions could contribute to LEC's localization to Pol II-transcribed snRNA genes.
Why would short snRNA genes require an elongation complex for their transcription?
The ELL-ICE complex is called LEC for its proposed role in transcriptional regulation of the “little” snRNA genes. It is not immediately evident why an elongation factor such as ELL is required for Pol II to transcribe these short genes. ELL was originally identified as a factor that stimulated the elongation rate of Pol II in vitro by decreasing transient pausing by Pol II along the transcribed regions by keeping the 3' OH of the nascent transcript properly aligned with the catalytic site of Pol II (Shilatifard et al., 2003; Shilatifard et al., 1996). Perhaps the short, yet highly transcribed, snRNA genes require the same activity of ELL as first observed in the “stressed” conditions of the in vitro assays. Highly transcribed genes are more susceptible to DNA damage, which can result in stalled polymerases (Datta and Jinks-Robertson, 1995; Svejstrup, 2003). Stalling of a polymerase on a short gene would need to be quickly resolved if high rates of transcription were to be maintained. Short, highly transcribed genes may also present unique topological challenges to the transcription machinery, since transcription induces supercoils before and after the polymerase and these could interfere with the coupling of termination with reinitiation on shorter genes.
Within the SEC, ELL is involved in the regulation of the expression of genes during development in a process that involves the phosphorylation of the Pol II CTD by P-TEFb (Lin et al., 2011). In metazoans, snRNA genes can also be considered to be developmentally regulated genes. For example, two forms of U1 snRNA are differentially regulated during development in both Drosophila and mammals (Caceres et al., 1992; Lo and Mount, 1990). Disruption of Integrator subunits in a variety of organisms leads to developmental defects (Ezzeddine et al., 2011). In humans, disruption of proteins needed for snRNP biogenesis leads to spinal muscular atrophy (Khoo and Krainer, 2009). RB, P53 and BRCA1 have all been linked to the regulation of snRNA transcription, suggesting that the control of snRNA transcription is tightly linked with the control of cell proliferation during development (Jawdekar and Henry, 2008). As part of a highly regulated control of snRNA transcription in metazoans, a control step at the elongation stage could be built into snRNA transcriptional regulation. In this scenario, part of the essential function of ELL in development could be through its association with LEC at snRNA genes.
Other possible roles of LEC at Pol II-transcribed snRNA genes
LEC could also function at snRNA genes at a step distinct from transcription elongation. The ICE2 protein has distant homology to the translation factor Eif3d and to two different classes of proteins that bind to the 5' ends of RNAs (Margelevicius et al., 2010), so LEC could interact with snRNAs to facilitate snRNP biogenesis. The ICE1 protein sequence does not yield any clues to its function at snRNA genes and may form a scaffold for the rest of the complex and/or participate in targeting ELL to these genes. Most factors that have been found to affect snRNA expression are found to primarily affect 3' end termination (Baillat et al., 2005; Egloff et al., 2007; Egloff et al., 2008; Ezzeddine et al., 2011; Medlin et al., 2005). However, our Northern blotting and RNA-seq experiments after ELL and Ice1 RNAi and have not revealed any larger “read-through” snRNA species characteristic of termination defects.
Gene specific classes of elongation factors for RNA polymerase II
The last several years have seen many important developments in the unique regulatory mechanisms of snRNA transcription. The Integrator complex was found to be a 3' processing factor for Pol II transcribed snRNAs (Baillat et al., 2005). A new site of phosphorylation of the Pol II CTD repeat, Serine 7, was discovered and shown to help recruit Integrator (Chapman et al., 2007; Egloff et al., 2007). Another novel modification within the CTD, a single conserved arginine, was shown to be methylated by CARM1, an enzyme with a widespread role in mRNA splicing regulation (Sims et al., 2011). Methylation of the CTD by CARM1 appears to be generally repressive of snRNA and snoRNAs, as these genes were mostly up-regulated when the only active Pol II in cells had a point mutant of arginine to alanine (Sims et al., 2011). Together, these findings support the earlier studies that indicated the existence of a tight integration of initiation and termination machineries for transcriptional control of snRNA expression (Hernandez, 2001). Our finding that ELL is part of an evolutionarily conserved complex required for Pol II-mediated snRNA transcription, suggests that transcriptional elongation could be yet another step in the regulation of snRNA expression. For protein-coding genes, multiple links between transcription elongation, RNA processing and transcription termination have been well established (Moore and Proudfoot, 2009). Therefore, for any class of genes that has unique initiation, termination or processing machineries, there may also be a unique elongation factor to help integrate these events. Since ELL can function with P-TEFb at developmental genes containing paused RNA Pol II as part of SEC (Lin et al., 2011), or as part of LEC at snRNA genes (demonstrated in this study), the context in which these elongation factors are recruited to genes clearly matters.
EXPERIMENTAL PROCEDURES
Flag-Purifications and Proteomics
ELL-ct-Flag (759-1059) was made as a Flag C-terminal fusion. Flag-Ice1 was tagged with a Flag-HA tag at the N-termini as a full-length protein. Vectors were a gift from the Workman Laboratory. Proteins were expressed under the control of the Metallothionein promoter. Dmel2 (Invitrogen) S2 cells were grown in SFX media in spinner flasks at room temperature. Cells were pelleted and washed in 20 mM HEPES pH 7.5, and 140 mM NaCl. A 3-4 ml pellet was resuspended in a 25 ml lysis buffer (20 mM HEPES pH 7.5, 3 mM MgCl2, 0.1% Triton X-100, 1 mM DTT, and protease inhibitors from Sigma and Roche). Cells were swollen on ice for 5 minutes, Dounce homogenized with 15 strokes of the tight pestle of a Wheaton 40 ml Dounce homogenizer. Nuclei were pelleted 2000 × g for 10 minutes. Nuclei were resuspended with 5 times the swollen nuclear volume with salt extraction buffer (20 mM HEPES pH 7.5, 0.42 M NaCl, 10% Glycerol, 1 mM MgCl2, 0.2 mM DTT, and protease inhibitors). The nuclear suspension was rocked for 1-2 hours at 4°C. Nuclear extracts were cleared by centrifugation at 40,000 rpm for 30 minutes in a Ti50 rotor (Beckman). Extracts were mixed with 100 μl packed M2 Flag agarose beads (Sigma) overnight at 4 degrees with rocking. Beads were washed 5 times for 5 minutes with wash buffer (20 mM HEPES, 350 mM NaCl, 10% Glycerol, 1 mM MgCl2, 0.1% Triton X-100 and protease inhibitors). Proteins were eluted with 200μg/ml Flag peptide (Sigma). Proteins were separated by SDS-PAGE for silver-staining or Trichloroacetic acid precipitated for MudPIT analysis. TCA-precipitated protein mixtures from purifications were digested with endoproteinase Lys-C and trypsin (Roche) and subjected to MudPIT analysis as previously described (Wu et al., 2008a, b). Endogenous immunoprecipitations from 293T cells were performed with RIPA buffer whole cell lysates. Extracts were incubated with affinity-purified antibodies overnight and captured on protein A agarose, washed with RIPA buffer, and eluted in SDS sample buffer.
Antibodies
Antibodies recognizing Drosophila proteins were raised against bacterially expressed, His-tagged recombinant proteins. Rabbit anti-ELL-fl (aa 1-1079) used in Fig. S2B was previously described (Gerber et al., 2001). Rabbit anti-ELL-MR (aa 301-584) was used in Fig. 3 and Fig. S2A. Guinea Pig (Fig. S1) and Rabbit (Fig. 3 and S2A) antibodies were raised against Ice1 (CG13550) aa 1-192. Guinea Pig anti-Ice2 (CG10825) was raised against aa 1-261. Guinea Pig anti-Lilli-CT (aa 1427-1673) was previously described (Lin et al., 2010). Rabbit anti-Drosophila Rpb1 antibody was previously described (Lin et al., 2010). Rabbit anti-human ICE1 (KIAA0947) was raised against aa 1872-2266. Rabbit anti-human ICE2 (NARG2) was raised against aa 666-982. Mouse anti-coilin is from Abcam. Antibodies recognizing human AFF4, ELL and Rpb1 proteins were previously described (Lin et al., 2010).
Immunostaining
Human ICE1 and coilin immunofluorescence in HeLa cells was performed with 3.7% formaldehyde fixation of cells for 10 minutes, 0.5% Triton X-100 permeabilization, and blocking with 3% BSA for 30 minutes before antibody incubations.
Chromatin Immunoprecipitation
Drosophila chromatin immunoprecipitation was performed with S2 cells grown in Schneider's media with 10% FBS at room temperature. Cells were fixed for 10 minutes in 1% formaldehyde, quenched with the addition of 1/10 volume of 2.5 M Glycine in PBS for 10 minutes, cells were washed twice in PBS, resuspended in hypotonic buffer (Orlando et al., 1998) and nutated for 10 minutes. 3×10e7 cells were pelleted and resuspended in 1.3 ml RIPA buffer with 0.5% Sarkosyl. Cells were sonicated in polyethylene 15 ml tubes for 15 minutes, high output, in a Bioruptor (Diagenode) at 4° C. Debris was pelleted 10 minutes at 20,000 × g 4° C. ChIP was performed with 200 μl of chromatin and 5 μg of antibodies, and scaled up 10- fold for ChIP-seq. Mouse ES cell chromatin immunopreciptiation was performed with KH2 cells cultured under feeder-free medium ESGRO (Millipore). ChIP was performed according to previously described protocol (Wang et al., 2009). Briefly, ES cells were cross-linked by 1% formaldehyde and sonicated. 10 μg of antibodies were used in the ChIP assays. ChIP libraries were prepared with the Illumina DNA Sample Kit (Part# 0801-0303) according to Illumina's instructions and sequenced on the Genome Analyzer IIx following the manufacturer's protocols. ChIP from HeLa cells was performed according to previously described protocols (Wang et al., 2009).
RNAi
Drosophila RNAi was performed by feeding S2 cells with dsRNA prepared with the Megascript T7 dsRNA synthesis kit from Ambion. S2 cells were grown in Schneider's media with 10% FBS at room temperature. Cells at a density of 2e6 cells/ml were serum starved treated with 10 μg of dsRNA. Additional boosts of 10 μg dsRNA were added at 48 and 96 hours. Cells were harvested at 120 hours. Targets for dsRNA were from the ORF as follows: ELL-A (1200-1800), ELL-B (1826-2306), Ice1-A (1436-1992), Ice1-5 (923-1452) and a non-targeting control region corresponding to a section of the B-lactamase gene (nt 1632-1981 of the pUC18 vector). RNAi in mouse ES cells (V6.5) was performed for 3 days by infecting cells with concentrated lentiviral particles with polybrene at a concentration of 4 μg/ml. ES Cells were grown one passage off of feeder cells before harvesting.
RT-PCR
Total RNA for RT-PCR was isolated with RNeasy (Qiagen), DNase-treated, and re-purified with RNeasy. Quantitative RT-PCR was performed with Sybr Green 1-step RT-PCR kit from Qiagen on MyIQ (Bio-rad). Relative expression to housekeeping genes was calculated assuming 2-fold primer efficiencies.
Northern Blotting
Northern blotting was performed as previously described (Gerber et al., 2001) by separating total RNA on a formaldehyde-containing 2% agarose gel, transferring to Hybond N membrane (GE Healthcare). Northern Blots were hybridized at 65 degrees in 50% formamide-containing hybridization buffer, with 32P-labeled riboprobes complementary to U11 (nt 24-156) and U6 (nt 1-82) and a probe to Rp49 (Gerber et al., 2001). Northern blots were probed sequentially with U11, Rp49 and U6 probes. Probe signal was imaged on a phosphor storage screen from 3-90 minutes and scanned on a Typhoon Trio (GE Healthcare) and quantitated with ImageQuant software at exposures without saturated pixels.
Microarrays
Affymetrix Drosophila Genome 2 arrays were analyzed in R, version 2.11.1, using the packages affy (Gautier et al., 2004), version 1.26.1, and limma (Smyth et al., 2005), version 3.4.3. Normalization was done using rma. Annotation information for the probes was taken from Ensembl 62. RNAi was performed with the ELL-A dsRNA. A region of the B-lactamase gene was used as a non-targeting control.
Ribo-Zero and RNA-seq
Two micrograms of DNase-treated total RNA was depleted of Ribosomal RNA with the Ribo-Zero kit from Epicentre and library preparation was made using the Tru-seq mRNA kit from Illumina.
Solexa/Illumina Data Analysis
Sequencing reads were acquired through the primary Solexa image analysis pipeline, where bases were called and reads were filtered for quality, according to default Solexa standards. Filtered reads were then aligned to the fly genome (UCSC dm3) or mouse genome (UCSC mm9) using the Bowtie (Langmead et al., 2009) alignment tool, version 0.12.7. Only those sequences that matched uniquely to the genome with up to two mismatches were retained for subsequent analyses.
Enriched regions of ChIP-seq signal were determined by the ‘MACS’ (Zhang et al., 2008) peak-finding program, version 1.4.0rc2. Sequence reads for each ChIP-seq dataset and their associated whole-cell extract controls were used for the input and control file, respectively. The effective genome size was configured appropriately for the fly datasets, and the p-value cutoff was set to 1.00e-05 and a fold-change greater than five, or FDR < 1%. All other MACS parameters were left default.
RNA-seq analysis was done using TopHat v1.2.0 (Trapnell et al., 2009) and Bowtie v0.12.7. Only uniquely mapping reads were used. Fly transcript annotations were from Ensembl 62, and mouse transcript annotations were from Ensembl 63. Differentially expressed genes after RNAi were called with an adjusted p-value < 0.001 and MA plots were determined by DESeq (Anders and Huber, 2010), v1.4.1. For the Drosophila RNA-seq experiments, two different dsRNA regions for each target were treated as biological replicates. A non-targeting control dsRNA-treated sample and untreated S2 cells were treated as biological replicates for the “control” condition. The MA plot for the ELL knockdown in mouse ES cells was performed from an ELL shRNA and two non-targeting shRNA knockdowns.
Gene Annotation
Genes were called bound for all ChIP-seq samples if an enriched peak region was found within 1kb of the transcription start site for any transcript isoform of the gene. Plots depicting dots, which correspond to Pol II occupancy, were determined by first selecting Pol II-bound genes, and then by determining the quartiles of RPM levels at those genes.
Track Figures
Read coverage information in the track figures was created using R by extending the reads 150 bases toward the interior of the sequenced fragment and then by computing the number of extended reads in 25 bp windows as the count of extended reads per million reads sequenced (RPM; counts/million). The resulting coverage object was exported and visualized using the UCSC genome browser (Kent et al., 2002).
Supplementary Material
HIGHLIGHTS.
ELL's conserved C-terminal domain interacts with ICE1 and ICE2.
ELL and ICE1-2 form a complex, named LEC, distinct from the P-TEFb-containing SEC.
In both Drosophila and mammalian cells, LEC, but not SEC, is enriched at snRNA genes.
LEC is required for Pol II-mediated snRNA transcription in flies and mammals.
ACKNOWLEDGMENTS
We are grateful to Karin Zueckert-Gaudenz and Allison Peak for Ribo-Zero treatment and RNA-seq library preparation, and Ka-Chun Lai and Eliza Hodes for assistance with the generation of antibodies and cell lines. We thank Dr. Dominik Handler and the EpiCentral blog for communicating data on the efficacy of Ribo-Zero in Drosophila. We are also grateful to the Conaways’ group for conversations in regard to the RNA polymerase II elongation complexes. We thank Laura Shilatifard for editorial assistance. This work was done to fulfill, in part, requirements for C.L.'s Ph.D. thesis research as a student registered with the Open University. These studies were supported in part by the National Cancer Institute, 5R01CA150265 and R01CA89455 to A.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Affymetrix microarray, RNA-seq and ChIP-seq data are available at the gene expression omnibus (GEO) with accession number GSE32120.
SUPPLEMENTAL INFORMATION
Supporting online materials include 3 supplemental figures and supplemental references.
REFERENCES
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Caceres JF, McKenzie D, Thimmapaya R, Lund E, Dahlberg JE. Control of mouse U1a and U1b snRNA gene expression by differential transcription. Nucleic Acids Res. 1992;20:4247–4254. doi: 10.1093/nar/20.16.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- Cioce M, Lamond AI. Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- Datta A, Jinks-Robertson S. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- DiMartino JF, Miller T, Ayton PM, Landewe T, Hess JL, Cleary ML, Shilatifard A. A carboxy-terminal domain of ELL is required and sufficient for immortalization of myeloid progenitors by MLL-ELL. Blood. 2000;96:3887–3893. [PubMed] [Google Scholar]
- Egloff S, Al-Rawaf H, O'Reilly D, Murphy S. Chromatin structure is implicated in “late” elongation checkpoints on the U2 snRNA and beta-actin genes. Mol Cell Biol. 2009;29:4002–4013. doi: 10.1128/MCB.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Murphy S. Expression of human snRNA genes from beginning to end. Biochem Soc Trans. 2008;36:590–594. doi: 10.1042/BST0360590. [DOI] [PubMed] [Google Scholar]
- Egloff S, Szczepaniak SA, Dienstbier M, Taylor A, Knight S, Murphy S. The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J Biol Chem. 2010;285:20564–20569. doi: 10.1074/jbc.M110.132530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Ma J, Gerber MA, Christensen A, Kennison JA, Shilatifard A. dELL is an essential RNA polymerase II elongation factor with a general role in development. Proc Natl Acad Sci U S A. 2002;99:9894–9899. doi: 10.1073/pnas.152193699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzeddine N, Chen J, Waltenspiel B, Burch B, Albrecht T, Zhuo M, Warren WD, Marzluff WF, Wagner EJ. A subset of Drosophila integrator proteins is essential for efficient U7 snRNA and spliceosomal snRNA 3'-end formation. Mol Cell Biol. 2011;31:328–341. doi: 10.1128/MCB.00943-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Matera AG. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J Cell Biol. 2001;154:499–509. doi: 10.1083/jcb.200105084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Itoh M, Hirase T, Nagafuchi A, Yonemura S, Tsukita S. Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J Cell Biol. 1994;127:1617–1626. doi: 10.1083/jcb.127.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Gerber M, Ma J, Dean K, Eissenberg JC, Shilatifard A. Drosophila ELL is associated with actively elongating RNA polymerase II on transcriptionally active sites in vivo. Embo J. 2001;20:6104–6114. doi: 10.1093/emboj/20.21.6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber MA, Shilatifard A, Eissenberg JC. Mutational analysis of an RNA polymerase II elongation factor in Drosophila melanogaster. Mol Cell Biol. 2005;25:7803–7811. doi: 10.1128/MCB.25.17.7803-7811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell. 2010;38:428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J Biol Chem. 2001;276:26733–26736. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- Jawdekar GW, Henry RW. Transcriptional regulation of human small nuclear RNA genes. Biochim Biophys Acta. 2008;1779:295–305. doi: 10.1016/j.bbagrm.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo B, Krainer AR. Splicing therapeutics in SMN2 and APOB. Curr Opin Mol Ther. 2009;11:108–115. [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev. 2011;25:1486–1498. doi: 10.1101/gad.2059211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo PC, Mount SM. Drosophila melanogaster genes for U1 snRNA variants and their expression during development. Nucleic Acids Res. 1990;18:6971–6979. doi: 10.1093/nar/18.23.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo RT, Lavau C, Du C, Simone F, Polak PE, Kawamata S, Thirman MJ. The elongation domain of ELL is dispensable but its ELL-associated factor 1 interaction domain is essential for MLL-ELL-induced leukemogenesis. Mol Cell Biol. 2001;21:5678–5687. doi: 10.1128/MCB.21.16.5678-5687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margelevicius M, Laganeckas M, Venclovas C. COMA server for protein distant homology search. Bioinformatics. 2010;26:1905–1906. doi: 10.1093/bioinformatics/btq306. [DOI] [PubMed] [Google Scholar]
- Medlin J, Scurry A, Taylor A, Zhang F, Peterlin BM, Murphy S. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. Embo J. 2005;24:4154–4165. doi: 10.1038/sj.emboj.7600876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Williams K, Johnstone RW, Shilatifard A. Identification, cloning, expression, and biochemical characterization of the testis-specific RNA polymerase II elongation factor ELL3. J Biol Chem. 2000;275:32052–32056. doi: 10.1074/jbc.M005175200. [DOI] [PubMed] [Google Scholar]
- Mohan M, Lin C, Guest E, Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer. 2010;10:721–728. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Orlando V, Jane EP, Chinwalla V, Harte PJ, Paro R. Binding of trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. Embo J. 1998;17:5141–5150. doi: 10.1093/emboj/17.17.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak PE, Simone F, Kaberlein JJ, Luo RT, Thirman MJ. ELL and EAF1 are Cajal body components that are disrupted in MLL-ELL leukemia. Mol Biol Cell. 2003;14:1517–1528. doi: 10.1091/mbc.E02-07-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A, Conaway RC, Conaway JW. The RNA polymerase II elongation complex. Annual review of biochemistry. 2003;72:693–715. doi: 10.1146/annurev.biochem.72.121801.161551. [DOI] [PubMed] [Google Scholar]
- Shilatifard A, Duan DR, Haque D, Florence C, Schubach WH, Conaway JW, Conaway RC. ELL2, a new member of an ELL family of RNA polymerase II elongation factors. Proc Natl Acad Sci U S A. 1997;94:3639–3643. doi: 10.1073/pnas.94.8.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. An RNA polymerase II elongation factor encoded by the human ELL gene. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Rojas LA, Beck D, Bonasio R, Schuller R, Drury WJ, 3rd, Eick D, Reinberg D. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332:99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Winter B, Eissenberg JC, Shilatifard A. Regulation of the transcriptional activity of poised RNA polymerase II by the elongation factor ELL. Proc Natl Acad Sci U S A. 2008;105:8575–8579. doi: 10.1073/pnas.0804379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura N, Patel RG, Corriveau RA. N-methyl-D-aspartate receptors regulate a group of transiently expressed genes in the developing brain. J Biol Chem. 2001;276:14257–14263. doi: 10.1074/jbc.M100011200. [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ. Rescue of arrested RNA polymerase II complexes. J Cell Sci. 2003;116:447–451. doi: 10.1242/jcs.00271. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirman MJ, Levitan DA, Kobayashi H, Simon MC, Rowley JD. Cloning of ELL, a gene that fuses to MLL in a t(11;19)(q23;p13.1) in acute myeloid leukemia. Proc Natl Acad Sci U S A. 1994;91:12110–12114. doi: 10.1073/pnas.91.25.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- With S, Rice T, Salinas C, Auld V. Fire exit is a potential four transmembrane protein expressed in developing Drosophila glia. Genesis. 2003;35:143–152. doi: 10.1002/gene.10177. [DOI] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008a;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008b;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Yuan G, Klambt C, Bachellerie JP, Brosius J, Huttenhofer A. RNomics in Drosophila melanogaster: identification of 66 candidates for novel non-messenger RNAs. Nucleic Acids Res. 2003;31:2495–2507. doi: 10.1093/nar/gkg361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.