Abstract
The importance of PTEN (phosphatase and tensin homolog located on chromosome ten) in cancer has surpassed all predictions and expectations from the time it was discovered, and has qualified this gene as one of the most commonly mutated and deleted tumor suppressors in human cancer. PTEN levels are frequently found down-regulated in cancer also in the absence of genetic loss or mutation. PTEN is heavily regulated by transcription factors, microRNAs, ceRNAs (competitive endogenous RNAs, such as the PTEN pseudogene) and methylation, while the tumor suppressive activity of the PTEN protein can be altered at multiple levels through aberrant phosphorylation, ubiquitination and acetylation. These regulatory cues are presumed to play a key role in tumorigenesis through the alteration of the appropriate levels, localization and activity of PTEN. The identification of all these levels of PTEN regulation raises in turn a key corollary question: How low should PTEN level(s) or activity drop in order to confer cancer susceptibility at the organismal level? Our lab and others have approached this question through the genetic manipulation of Pten in the mouse. This work has highlighted the exquisite and tissue-specific sensitivity to subtle reductions in Pten levels towards tumor initiation and progression with important implications for cancer prevention and therapy.
PTEN was originally discovered as the tumor suppressor gene frequently lost on chromosome 10q23 (1, 2). Soon after, the relevance of PTEN in cancer was addressed through the generation of germline knockout Pten mice by several independent laboratories (3–5). These studies revealed the requirement of PTEN for embryonic development. Importantly, heterozygous loss of this tumor suppressor in the mouse resulted in the development of cancer of multiple origins (3–6) as well as in a lethal lymphoproliferative disease (3–5). These studies provided the first evidence for a haploinsufficient tumor suppressive activity of PTEN, since a fraction of cancers arising in Pten heterozygous mice would not exhibit loss of the wild type allele (4, 5). In humans, germline loss and mutation of PTEN is observed in a group of autosomal dominant syndromes (PTEN hamartoma tumor syndromes or PHTS) which are characterized by neurological disorders, multiple hamartomas and cancer susceptibility (7). Studies from the mouse have recapitulated a fraction of the features observed in PHTS patients, however, the cooperative genetic or environmental factors contributing to the full symptomatic spectrum in this group of syndromes remain to be defined. In addition, analysis of cancer biopsies suggests that the notion that PTEN is a haploinsufficient tumor suppressor, formally proven in the mouse, can also be translated to humans (8).
How does PTEN exert its powerful tumor suppressive activity?
PTEN functions as a lipid phosphatase, dephosphorylating the 3’ position of phosphoinoisitde 3,4,5-triphosphate (PIP3). This lipid second messenger is the product of a potent proto-oncogenic kinase, Phosphoinositide 3-Kinase (PI3K), and the trigger for activation of the PI3K pathway. The relevance of the PI3K pathway in cancer is highlighted by the elevated number of components within the cascade whose level or activity is found altered, and represents one of the main targets for cancer therapy (9). However, despite the main role of PTEN as a negative regulator of the PI3K pathway, recent studies report a number of tumor suppressive activities for PTEN that are exerted from within the nucleus (10–14), where catalysis of PIP3 does not appear to represent a central function of this enzyme (15). These nuclear PTEN activities include the regulation of genomic stability, cell cycle progression, differentiation and gene expression (10–14). On the basis of these studies, it is apparent that we have only begun to understand the multiple other functions of PTEN that occur in the nucleus or other cellular compartments, and to elucidate the activities of PTEN for which the phosphatase domain might not be required.
What are the consequences of progressive PTEN dose reduction in tumorigenesis?
PTEN is one of the most mutated and deleted tumor suppressors in human cancer but, importantly, it also found partially down-regulated in cancer in the absence of genetic loss or mutation (10). Mouse genetic engineering has allowed us to study the impact of fractional PTEN level reduction in cancer. Pten heterozygous mice develop a number of cancers, including mammary, prostate, uterine and pheochromocytoma (3–5). However, a lethal lymphoproliferative disease in the Pten heterozygous model has limited the study of cancers that might develop with longer latencies. In this respect, the generation and use of conditional Pten knockout mice has contributed to a better understanding of its tumor suppressive activity in specific cell/tissues in vivo, in a partial or complete loss setting (16). Moreover, conditional Pten knockout mouse models have uncovered unexpected and seemingly paradoxical consequences of complete Pten-loss. These include the activation of potent failsafe mechanisms such as a novel cellular senescence program (17), or the exhaustion of the hematopoietic stem cell compartment prior to leukemia development (18, 19). These findings, however, did not address the impact of subtle or incremental reductions in PTEN levels.
In an effort to study the impact of subtle Pten reduction in a non-allelic manner (i.e. WT (100%) vs Heterozygous (50%) vs Null (0%)), our lab generated a hypomorphic Pten mouse allelic series taking advantage of genetic expression interference. These mice harbor a neomycin cassette in one allele of Pten, which interferes with Pten transcription and reduces mature mRNA levels (20). In this manner, we at first generated and characterized mice in which the levels of Pten lie between the heterozygous and knockout (by combining the hypomorphic allele with a knockout allele, which resulted in a 70–80% decrease in Pten levels, termed Pten “hypo” mice (Ptenhy/−)). While the Pten heterozygous mutants develop high-grade prostate intraepithelial neoplasia (HG-PIN), an in situ form of prostate cancer, at incomplete penetrance, the Pten hypo mice develop prostate cancer lesions at full penetrance. Importantly, a fraction of cancers in these mutants now progress towards invasive prostate carcinoma in the absence of loss of heterozygosity of the wild type allele (20). This study provided the first experimental evidence demonstrating that a subtle decrement in Pten expression can have dramatic consequences in cancer progression.
Since complete loss of Pten elicits the activation of a fail safe response in the prostate (17), and possibly in other tissues, tumors with loss of one copy of PTEN might benefit of “hypomorphing”, or further reducing PTEN levels, without losing the remaining functional allele, at least until the failsafe pathway (i.e. cellular senescence) has been bypassed (for instance through p53 mutation or loss (17)).
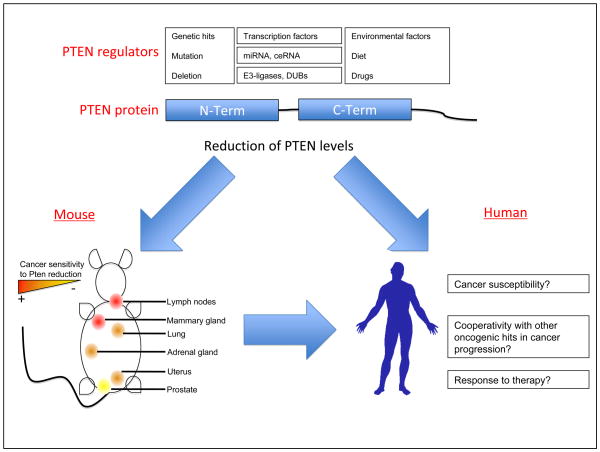
As for how PTEN reduction below heterozygosity can occur, it is plausible that environmental factors (diet and chemicals, such as Zinc (21)) or genetic events (i.e. microRNAs and loss of competitive endogenous RNAs or ceRNAs, such as the PTEN pseudogene (22)) might contribute leading to more aggressive cancer (Figure 1). Among the mechanisms that induce PTEN down-regulation in cancer as way to favor tumor progression, it is remarkable the effect of mitogen-activated protein kinase (MAPK) pathway on the transcription of this tumor suppressor. Expression of an oncogenic version of the small GTPase Ras (in which Glycine 12 is mutated to Valine, G12V) leads to the reduction of PTEN levels. PTEN down-regulation is transcriptional, and is driven by the induction of c-Jun downstream of MAPK signaling (23). On the other hand, a recent study from our lab has uncovered a novel type of regulatory mechanism based on the existence of competitive endogenous RNAs (ceRNAs). As proof of principle, we have shown that a non-coding RNA, the PTEN pseudogene, exerts a tumor suppressive activity based on the homology of its 3′UTR with PTEN, which allows it to quench PTEN-targeting miRNAs and in turn to regulate PTEN expression level (22). These studies exemplify the complexity of PTEN regulatory networks and therefore the importance of fine-tuning the expression of this tumor suppressor. Of interest, the promoter region of PTEN exhibits regulatory elements not only for c-Jun, but also for other survival signals (e.g. NFκB (24)), therefore suggesting that additional cancer promoting factors may act through PTEN down-regulation, leading to cancer progression or resistance to therapy.
Figure 1. Impact of PTEN regulatory cues in tumorigenesis.
Schematic representation of the tissue-specific cancer sensitivity to PTEN down-regulation and the regulatory factors that could lead to the reduction of PTEN levels. PTEN expression is regulated at multiple levels, genomic (mutation and deletions), transcriptional (transcription factors), post-transcriptional (miRNAs, competitive endogenous RNAs -ceRNAs- such as the PTEN pseudogene) and post-translational (drugs, E3-ligases, de-ubiquitinating enzymes or DUBs). In turn, the reduction of PTEN levels has gradual and tissue-specific impact on cancer initiation in the mouse, with the mammary tissue being among the most sensitive tissues to PTEN level reduction. This data suggests that, in humans, subtle reduction of PTEN levels may lead to cancer susceptibility, cooperate with other oncogenic events in cancer progression or impact on the response to therapy.
Is there evidence that a subtle reduction of PTEN expression level is sufficient to promote cancer susceptibility?
We have addressed this question by generating the Pten “hyper” mouse model. Taking advantage of the Pten hypomorphic allele, we generated mice with one Pten hypomorphic allele over a wild type allele (Ptenhy/+). These mice, as predicted, express 80% of normal Pten mRNA and protein levels (25). Surprisingly, hyper mice exhibit many of the features present in Pten heterozygous mice, including early lethality, signs of a lymphoproliferative disease and cancer susceptibility. More importantly, they develop tumors. Interestingly, the spectrum of tumors is distinct from that observed in heterozygous mice. Mammary tumors are most prevalent, whereas lung tumors (which were not observed in the Pten heterozygous cohort analyzed) are also detected. Interestingly, tumors arising in hyper mice retain the wild type Pten allele and gene expression, and do not harbor mutation at the remaining PTEN locus. In addition, Pten hyper mice do not show signs of lesions in the prostate epithelium, unlike heterozygous and hypo mice. This data suggests that there is an exquisite tissue-specific sensitivity to the reduction of Pten levels (Figure 1), with tissues that require a more profound down-regulation in Pten for cancer progression to occur (prostate) and others more sensitive to small decreases (lymph nodes, mammary epithelium). In the mammary gland, we can readily observe signs of hyperproliferation in the epithelium of 2 month-old females, indicating that the reduction of Pten has immediate cell autonomous consequences in the mammary tissue. The fact that 80% of total levels are not sufficient for Pten to suppress cancer revisits the idea that Pten is solely a haploinsufficent tumor suppressor, suggesting that levels above 50% still do not suffice to keep cancer development at bay, phenomenon that we have termed tumor suppressor “quasi-insufficiency”.
Can we find evidence of PTEN quasi-insufficiency in human cancer?
We define “quasi-insufficiency” as the inability to sustain adequate biological functions upon a subtle reduction in the protein levels of a given gene (below homozygosity but above heterozygosity). Microarray analysis of normal breast tissue allowed us to define the normal average PTEN mRNA level, and in turn to evaluate the level of PTEN in breast tumors (25). PTEN levels in cancer biopsies followed a linear distribution in the cohort analyzed, ranking from levels of expression equivalent to those observed in tissue from normal subjects to levels of expression below 20%. Notably, 20% of tumors displayed levels of PTEN slightly below the average levels of the normal tissue (above 65% of the normal averagePTEN levels: hyper tumors), and protein levels by immunohistochemistry that excluded a robust decrease in PTEN expression. Interestingly, these tumors presented a gene expression profile similar to the one detected in cells from our hyper mouse, suggesting that PTEN levels associate with a defined gene signature related to cell proliferation. We find particularly interesting that PTEN levels in the normal breast tissue also show a linear distribution, with a 15% standard deviation in our small cohort. This data suggests that PTEN levels may vary by a small fraction of expression among healthy individuals, and according to our data, these patients might present cancer susceptibility, and should therefore be followed up to closely monitor the development of tumors. Moreover, The idea that subtle changes in the expression of a gene can have profound effects on tumorigenesis adds complexity to the “two-hit hypothesis” postulated by Alfred Knudson (26). On the basis of the information obtained from our hyper mice, alterations in the protein levels of a TSG even in the absence of a “genetic hit”, could be sufficient to trigger a cellular response resulting in cellular transformation and cancer. Further studies will identify “cancer quasi-insufficient genes” and the deregulated pathways relevant for cancer development.
The other important implication derived from years of study on the role of PTEN in tumor suppression is the tissue-specific sensitivity to PTEN level reduction and its tissue-specific cooperativity with various oncogenes and tumor suppressors. In fact, mouse modeling has uncovered that loss of Pten cooperates with a diverse array of genetic hits in tumor development and progression. An exemplifying case is the study of cellular senescence evoked by Pten inactivation (17), Upon PTEN loss in the prostate epithelium, the tumor suppressor p53 is potently induced to elicit senescence and oppose cancer progression. In turn, compound loss of Pten and Tp53 in this tissue results in aggressive and lethal prostate cancer development. Additionally, Pten loss in the prostate cooperates with Tsc2 and p27-loss as well as Erg over-expression (16), among others. In the breast, data showing that BRCA1-loss results in increased mutations and micro-deletions in the PTEN locus (27) also suggest a possible cooperativity between PTEN and BRCA1.
On the basis of our data, it is tempting to speculate that the sensitivity to PTEN dosage reduction of a given tissue could be determined by its gene expression signature. Specifically, the sensitivity of the mammary gland to subtle PTEN down-regulation could be due to a gene expression network that makes it less tolerant to variations in PTEN expression. Therefore, identifying the PTEN loss-cooperating events (or susceptibility gene signatures) in specific tissues could contribute to define the network of functions exerted by PTEN in tumor suppression and the relevance of each PTEN function in a given tissue.
Future directions
More than ten years of PTEN research has led to a new perspective regarding this critical tumor suppressor. From the phoshatase that opposes PI3K signaling in the cytoplasm, PTEN has evolved into a protein that exerts a variety of tumor suppressive activities in the nucleus, beyond the dephosphorylation of PIP3. These studies have contributed to the understanding of how sensitive tissues are to even subtle reduction of PTEN levels, and in turn have raised further questions on how PTEN functions. Does the subtle reduction of PTEN drive cancer susceptibility due to the reduction of PIP3 hydrolysis, or due to loss of nuclear functions? Conversely, is elevation of PTEN level(s) a safe and effective approach to prevent or treat cancer? Can we quantitatively monitor PTEN levels in patients in order to predict cancer susceptibility? Our lab and others are focusing on providing definitive answers to these questions. Addressing these questions will in turn bring to a new exciting level our understanding of the rules and mechanisms of tumor suppression and oncogenesis. On the basis of this new notions, a journey of discovery that started at the bedside through the discovery of PTEN in human cancer specimen, which subsequently was translated to cell culture and faithful mouse models of cancer, will next be brought back to the bedside in the form of effective diagnostic, prognostic and therapeutic tools for cancer prevention and therapy.
Acknowledgments
We thank L. Salmena, J.G. Clohessy, I. Garcia-Cao and A.H. Berger for insightful discussions. A.C. is supported by the Ramón y Cajal program (Spanish Ministry of Education and Science). A.A. is supported by ESMO and Marie Curie Reintegration Grant.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 2.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–62. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki H, Freije D, Nusskern DR, Okami K, Cairns P, Sidransky D, et al. Interfocal heterogeneity of PTEN/MMAC1 gene alterations in multiple metastatic prostate cancer tissues. Cancer research. 1998;58:204–9. [PubMed] [Google Scholar]
- 4.Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, Yamada KM, et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci U S A. 1999;96:1563–8. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–55. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 6.Shen-Li H, Koujak S, Szablocs M, Parsons R. Reduction of Pten dose leads to neoplastic development in multiple organs of Pten(shRNA) mice. Cancer Biol Ther. :10. doi: 10.4161/cbt.10.11.13814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hobert JA, Eng C. PTEN hamartoma tumor syndrome: an overview. Genet Med. 2009;11:687–94. doi: 10.1097/GIM.0b013e3181ac9aea. [DOI] [PubMed] [Google Scholar]
- 8.Berger AH, Pandolfi PP. Haplo-insufficiency: a driving force in cancer. J Pathol. doi: 10.1002/path.2800. [DOI] [PubMed] [Google Scholar]
- 9.Wong KK, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Curr Opin Genet Dev. 20:87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–14. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 11.Semba S, Satake S, Matsushita M, Yokozaki H. Phosphatase activity of nuclear PTEN is required for CDX2-mediated intestinal differentiation of gastric carcinoma. Cancer Lett. 2009;274:143–50. doi: 10.1016/j.canlet.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 12.Rankin SL, Guy CS, Mearow KM. PTEN downregulates p75NTR expression by decreasing DNA-binding activity of Sp1. Biochem Biophys Res Commun. 2009;379:721–5. doi: 10.1016/j.bbrc.2008.12.075. [DOI] [PubMed] [Google Scholar]
- 13.Jacob AI, Romigh T, Waite KA, Eng C. Nuclear PTEN levels and G2 progression in melanoma cells. Melanoma Res. 2009;19:203–10. doi: 10.1097/CMR.0b013e32832ccd6e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosessian S, Avliyakulov NK, Mulholland DJ, Boontheung P, Loo JA, Wu H. Analysis of PTEN complex assembly and identification of heterogeneous nuclear ribonucleoprotein C as a component of the PTEN-associated complex. J Biol Chem. 2009;284:30159–66. doi: 10.1074/jbc.M109.027995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindsay Y, McCoull D, Davidson L, Leslie NR, Fairservice A, Gray A, et al. Localization of agonist-sensitive PtdIns(3,4,5)P3 reveals a nuclear pool that is insensitive to PTEN expression. J Cell Sci. 2006;119:5160–8. doi: 10.1242/jcs.000133. [DOI] [PubMed] [Google Scholar]
- 16.Nardella C, Carracedo A, Salmena L, Pandolfi PP. Faithfull Modeling of PTEN Loss Driven Diseases in the Mouse. Curr Top Microbiol Immunol. doi: 10.1007/82_2010_62. [DOI] [PubMed] [Google Scholar]
- 17.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–82. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–22. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- 20.Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu W, Wang X, Zhang W, Reed W, Samet JM, Whang YE, et al. Zinc-induced PTEN protein degradation through the proteasome pathway in human airway epithelial cells. J Biol Chem. 2003;278:28258–63. doi: 10.1074/jbc.M303318200. [DOI] [PubMed] [Google Scholar]
- 22.Poliseno L, Salmena L, Zhang J, Carver BSJHW, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010:1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vasudevan KM, Burikhanov R, Goswami A, Rangnekar VM. Suppression of PTEN expression is essential for antiapoptosis and cellular transformation by oncogenic Ras. Cancer research. 2007;67:10343–50. doi: 10.1158/0008-5472.CAN-07-1827. [DOI] [PubMed] [Google Scholar]
- 24.Vasudevan KM, Gurumurthy S, Rangnekar VM. Suppression of PTEN expression by NF-kappa B prevents apoptosis. Mol Cell Biol. 2004;24:1007–21. doi: 10.1128/MCB.24.3.1007-1021.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alimonti A, Carracedo A, Clohessy JG, Trotman LC, Nardella C, Egia A, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42:454–8. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proceedings of the National Academy of Sciences of the United States of America. 1971;68:820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saal LH, Gruvberger-Saal SK, Persson C, Lovgren K, Jumppanen M, Staaf J, et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet. 2008;40:102–7. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]