Abstract
Background & objectives:
Multidrug resistant methicillin-resistant Staphylococcus aureus (MRSA) is a major cause of nosocomial and community acquired infections and is on the rise. The glycopeptide vancomycin has been proposed as the drug of choice for treating such infections. The present study aimed at identifying the vancomycin resistance both phenotypically and genotypically among the MRSA isolates from two tertiary care hospitals in Hyderabad, south India.
Methods:
MRSA were isolated and identified from different clinical samples collected from ICUs of tertiary care hospitals in Hyderabad using conventional methods. Antibiogram of the isolates and vancomycin MIC were determined following CLSI guidelines. vanA was amplified by PCR using standard primers.
Results:
All vancomycin resistant S. aureus (VRSA) isolates were MRSA. The VRSA isolates were positive for vanA gene, except one which was negative. All VRSA had a vancomycin MIC in the range of 16-64 mg/l.
Interpretation & conclusions:
The increase in vancomycin resistance among MRSA and excessive use of antimicrobial agents have worsened the sensitivity. Larger studies need to be done in various geographical regions of the country to better define the epidemiology, mechanism of vancomycin resistance in S. aureus and its clinical implications.
Keywords: MRSA, Staphylococcus aureus, vanA gene, vancomycin, VRSA
Staphylococcus aureus is one of the most common causes of nosocomial infections, especially pneumonia, surgical site infections and blood stream infections and continues to be a major cause of community-acquired infections. Methicillin-resistant S. aureus (MRSA) was first detected approximately 40 years ago and is still among the top three clinically important pathogens1,2. The emergence of high levels of penicillin resistance followed by the development and spread of strains resistant to the semisynthetic penicillins (methicillin, oxacillin, and nafcillin), macrolides, tetracycline, and aminoglycosides has made the therapy of staphylococcal disease a global challenge3.
The glycopeptide vancomycin was considered to be the best alternative for the treatment of multi drug resistant MRSA4. However, there are increasing numbers of reports indicating the emergence of vancomycin-resistant S. aureus (VRSA) strains exhibiting two different resistance mechanisms. Initially vancomycin-intermediate S. aureus (VISA) noted in Japan in 1996 and subsequently in United States in 1997, was believed to be due to the thickened cell wall5, where many vancomycin molecules were trapped within the cell wall. The trapped molecules clog the peptidoglycan meshwork and finally form a physical barrier towards further incoming vancomycin molecules6. The second, noted in United States in 20027 among S. aureus, was identical to the mechanism seen in vancomycin-resistant Enterococcus8. Vancomycin-resistant Enterococcus faecium harbours the vanA operon, which contains five genes, VanS, -R, -H, -A and -X8. But Tiwari and Sen have reported a VRSA which is van gene-negative9. Subsequent isolation of VISA and VRSA isolates from other countries including Brazil10, France11, United Kingdom12, Germany13, India9,14 and Belgium15 has confirmed that the emergence of these strains is a global issue.
The aim of the present study was to identify the emergence of vancomycin resistant MRSA among S. aureus isolates from tertiary care hospitals in Hyderabad, south India, and to determine the sensitivity of these isolates to different antimicrobial agents. Further search is also conducted for the vanA gene in VRSA strains.
Material & Methods
Bacterial isolates: A total of 358 S. aureus isolates were obtained randomly from clinical samples (blood, urine and throat swabs, wound and ear swabs) of ICUs of Osmania hospital (OH) and Durgabhai Deshmukh h0 ospital and r0 esearch c0 enter (DDHRC), Hyderabad, between March and October 2008. The study was done at department of microbiology, Gulberga University, Gulberga, Karnataka. S. aureus was identified by colony morphology, Gram stain, DNase, catalase and coagulase tests and fermentation of mannitol by conventional methods.
Antibiotic susceptibility testing: The antibiotic-resistance profile was determined by the disc agar diffusion (DAD) technique using different antimicrobial agents; penicillin G (10 U), oxacillin (1 μg), methicillin (30 μg), ampicillin (10 μg), ceftazidime (30 μg), gentamicin (10 μg), clarithromycin (15 μg), erythromycin (15 μg), clindamycin (2 μg), co-trimoxazole (25 μg), chloramphenical (30 μg), rifampin (5 μg), tetracycline (30 μg) (Hi-media, Mumbai) according to the guidelines recommended by Clinical and Laboratory Standards Institute (CLSI)16. Resistance to the aminoglycoside streptomycin was assessed similarly but the results were interpreted as per the guidelines of the manufacturer (Hi-Media). The standard S. aureus strains NCTC 12493 and ATCC 29213 were used as reference strains (department of microbiology, University of Gulberga) for MRSA and VSRA, respectively.
Determination of MIC: Minimal inhibitory concentration (MIC) of vancomycin was determined by agar dilution method using CLSI guidelines16. Briefly, gradient plates of Mueller-Hinton agar (Hi-media) were prepared with vancomycin (0.5-256 mg/l, Sigma-Aldrich). 0.5 McFarland equivalent inoculum prepared using 18-24 h old culture was spotted on to gradient plates. Plates were incubated overnight at 35°C for 24 h before assessing the visible growth.
PCR amplification for van A: The primer synthesis and the PCR amplification of the vanA were done at Bioserve Biotechnologies, Hyderabad, using Gene Amp (Applied Biosystems, USA) thermocycler. The primers and the PCR conditions were as described17. The primers for amplification of vanA were; Forward ATGAATAGAATAAAAGTTGC and Reverse TCACCCCTTTAACGCTAATA. The amplification conditions were initial denaturation at 98°C for 10 sec, annealing at 50°C for 1 min, polymerization at 72°C for 1 min; and final extension at 72°C for 5 min.
Results
Among the 358 clinical isolates of S. aureus, 285 (79.6%) were identified as methicillin resistant S. aureus (MRSA) by disc diffusion method.
The MIC for 335 of 358 isolates (93.57%) for vancomycin was ≤2 mg/l indicating that all were sensitive to vancomycin. Sixteen isolates showed an MIC range between 4-8 mg/l, indicating vancomycin intermediate resistance. For the remaining seven isolates, the MIC was in the range of 16-64 mg/l indicating that these seven isolates were vancomycin-resistant (VRSA). These were all MRSA and resistant to majority of the other antibiotics tested. All these seven isolates showed resistance to a minimum of six other antibiotics including vancomycin and methicillin (Table).
Table.
Description of VRSA (n=7) including antibiotic susceptibility profile as determined by disc diffusion method
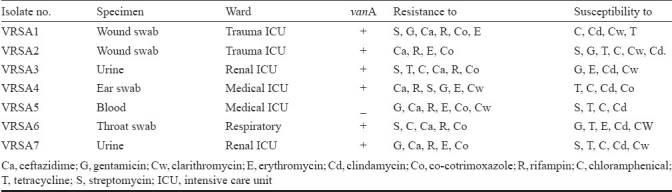
One isolate of S. aureus had vancomycin MIC of 4 mg/l and considered VISA according to recent CLSI breakpoints for vancomycin. However, this isolate was not distinguished from vancomycin susceptible S. aureus in the disc diffusion test because it has shown >20 mm zone of inhibition. All isolated VRSA were resistant to ceftazidime, rifampicin, and were inducible for clindamycin resistance. About 86 per cent of the isolates were susceptible to tetracycline and 71.4 per cent each were susceptible to chloramphenicol and clarithromycin.
PCR amplification for vanA among the seven VRSA showed that six contained vanA, whereas all the isolates expressed mecA (unpublished data). One VRSA isolate was negative for vanA by PCR (Fig.).
Fig.
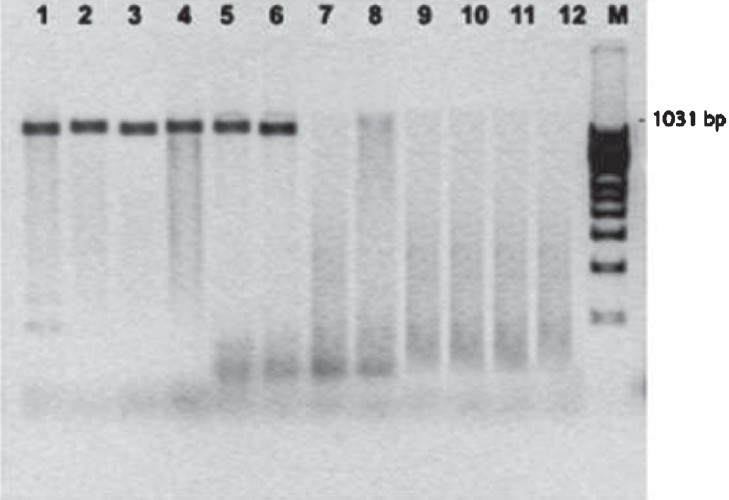
PCR amplification of the vanA gene for vancomycin resistance in methicillin resistant S. aureus. Lanes 1-6 vanA positive VRSA; Lane 7: vanA negative VRSA and Lanes 8-12: vanA negative VSSA, M-100 bp ladder.
Discussion
Infections caused by methicillin-resistant S. aureus have been associated with high morbidity and mortality rates. In Indian hospitals, MRSA is one of the common causes of hospital-acquired infections and 30 to 80 per cent methicillin resistance in S. aureus based on antibiotic sensitivity tests has been reported from different hospitals18. The present study showed 79.6 per cent of MRSA. Vancomycin is the main antimicrobial agent available to treat serious infections with MRSA but unfortunately, decrease in vancomycin susceptibility of S. aureus and isolation of vancomycin-intermediate and resistant S. aureus have recently been reported from many countries19. Many reports from north India also recorded the emergence of low level and intermediate vancomycin resistance9,14,20–23.
The MIC values indicated that only 1.9 per cent isolates were resistant to vancomycin. One isolate with intermediate resistance as per the MIC was recorded as sensitive in the disk diffusion test. There has been increasing evidence that isolates with a vancomycin MIC of 4 mg/l behave similarly in the clinical settings to VRSA and clinical failure generally results if treatment with vancomycin is continued24. Thus, the previous breakpoints for vancomycin [<4 mg/l (S); 8-16 mg/l (I); >32 mg/l (R)] have recently been revised [<2 mg/l (S); 4-8 mg/l (I); >16 mg/l (R)]16.
Vancomycin-resistant S. aureus tend to be multidrug resistant against a large number of currently available antimicrobial agents, compromising treatment options and increasing the likelihood of inadequate antimicrobial therapy and increase in morbidity and mortality24. In the present study, VRSA showed resistance to a wide range of antimicrobial agents. Though all the isolates were phenotypically sensitive to clindamycin, these showed inducible clindamycin resistance. This may be due to the increased use of antibiotics in the intensive care units. Linezolid and quinupristin/dalfopristin were recently approved by the Food and Drug Administration and are antimicrobials with activity against glycopeptides-resistant Gram positive microorganisms such as VRSA25.
The genetic mechanism of vancomycin resistance in VRSA is not well understood. Several genes have been proposed as being involved in certain clinical VRSA strains26–28. The experimental transfer of the vanA gene cluster from E. faecalis to S. aureus29 has raised fears about the occurrence of such genetic transfer in clinical isolates of methicillin resistant S. aureus. In this study, all the VRSA isolates contained mecA, but only six contained vanA. Tiwari & Sen8 have also reported a van gene-negative VRSA.
In conclusion, the results of this study showed the occurrence of vanA gene-negative VRSA in south India. The increase of vancomycin resistance among MRSA and excessive use of antimicrobial agents have worsened the sensitivity, which call for further epidemiological studies.
References
- 1.Van Belkum A, Verbrugh H. 40 years of methicillin-resistant Staphylococcus aureus. MRSA is here to stay - but it can be controlled. BMJ. 2001;323:644–5. doi: 10.1136/bmj.323.7314.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deresinski S. Methicillin-resistant Staphylococcus aureus: an evolution, epidemiologic and therapeutic Odyssey. Clin Infect Dis. 2005;40:562–73. doi: 10.1086/427701. [DOI] [PubMed] [Google Scholar]
- 3.Maranan MC, Moreira B, Boyle-Vavra S, Daum RS. Antimicrobial resistance in Staphylococci. Epidemiology, molecular mechanisms, and clinical relevance. Infect Dis Clin North Am. 1997;11:813–49. doi: 10.1016/s0891-5520(05)70392-5. [DOI] [PubMed] [Google Scholar]
- 4.Wootton M, Howe RA, Hillman R, Walsh TR, Bennett PM, Mac-Gowan AP. A modified population analysis (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J Antimicrob Chemother. 2001;47:399–403. doi: 10.1093/jac/47.4.399. [DOI] [PubMed] [Google Scholar]
- 5.Hiramatsu K, Hanaki H, Ino T, Oguri YT, Tenover FC. Methicillin-resistant Staphylococcus aureus clinical strain with reduced susceptibility. J Antimicrob Chemother. 1997;40:135–6. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 6.Cui L, Iwamoto A, Lian JQ, Neoh HM, Maruyama T, Horikawa Y, et al. Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:428–38. doi: 10.1128/AAC.50.2.428-438.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bozdogan BÜ, Esel D, Whitener C, Browne FA, Appelbaum PC. Antibacterial susceptibility of a vancomycin-resistant Staphylococcus aureus strain isolated at the Hershey Medical Center. J Antimicro Chemother. 2003;52:864–8. doi: 10.1093/jac/dkg457. [DOI] [PubMed] [Google Scholar]
- 8.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–27. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiwari HK, Sen MR. Emergence of vancomycin resistant Staphylococcus aureus (VRSA) from a tertiary care hospital from northern part of India. Infect Dis. 2006;6:156. doi: 10.1186/1471-2334-6-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliveira GA, Dell’Aquila AM, Masiero RL, Levy CE, Gomes MS, Cui L, et al. Isolation in Brazil of nosocomial Staphylococcus aureus with reduced susceptibility to vancomycin. Infect Control Hosp Epidemiol. 2001;22:443–8. doi: 10.1086/501932. [DOI] [PubMed] [Google Scholar]
- 11.Poly MC, Grelaud C, Martin C, de Lumley L, Denis F. First clinical isolate of vancomycin-intermediate Staphylococcus aureus in a French hospital. Lancet. 1998;351:1212. doi: 10.1016/s0140-6736(05)79166-2. [DOI] [PubMed] [Google Scholar]
- 12.Howe RA, Bowker KE, Walsh TR, Feest TG, Mac-Gowan AP. Vancomycin-resistant Staphylococcus aureus. Lancet. 1998;351:602. doi: 10.1016/S0140-6736(05)78597-4. [DOI] [PubMed] [Google Scholar]
- 13.Bierbaum G, Fuchs K, Lenz W, Szekat C, Sahl HG. Presence of Staphylococcus aureus with reduced susceptibility to vancomycin in Germany. Eur J Clin Microbiol Infect Dis. 1999;18:691–6. doi: 10.1007/s100960050380. [DOI] [PubMed] [Google Scholar]
- 14.Assadullah S, Kakru DK, Thoker MA, Bhat FA, Hussain N, Shah A. Emergence of low level vancomycin resistance in MRSA. Indian J Med Microbiol. 2003;21:196–8. [PubMed] [Google Scholar]
- 15.Pierard D, Vandenbussche H, Verschraegen I, Lauwers S. Screening for Staphylococcus aureus with a reduced susceptibility to vancomycin in a Belgian hospital. Pathologie Biologie. 2004;52:486–8. doi: 10.1016/j.patbio.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 16.Performance standards for antimicrobial susceptibility testing, 17th informational supplement (M100-517) Wayne, Pa: Clinical and Laboratory Standards Institute; 2007. Clinical and Laboratory Standards Institute. [Google Scholar]
- 17.Biswajit S, Anil KS, Abhrajyoti G, Manjusri B. Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia) J Med Microbiol. 2008;57:72–9. doi: 10.1099/jmm.0.47144-0. [DOI] [PubMed] [Google Scholar]
- 18.Anupurba S, Sen MR, Nath G, Sharma BM, Gulati AK, Mohapatra TM. Prevalence of methicillin resistant Staphylococcus aureus in a tertiary referral hospital in eastern Uttar Pradesh. Indian J Med Microbiol. 2003;21:49–51. [PubMed] [Google Scholar]
- 19.Benjamin PH, John KD, Paul DR. J, Timothy PS, Grayson ML.Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin- intermediate strains: Resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev. 2010;23:99–139. doi: 10.1128/CMR.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menezes GA, Harish BN, Sujatha S, Vinothini K, Parija SC. Emergence of vancomycin-intermediate Staphylococcus species in southern India. J Med Microbiol. 2008;57:911–2. doi: 10.1099/jmm.0.47829-0. [DOI] [PubMed] [Google Scholar]
- 21.Bhateja P, Mathur T, Pandya M, Fatma T, Rattan A Detection of vancomycin resistance Staphylococcus.aureus: A comparative study of three different phenotyic screening mehods. Indian J Med Microbiol. 2005;23:52–5. doi: 10.4103/0255-0857.13875. [DOI] [PubMed] [Google Scholar]
- 22.Bijiyani B, Purva M. Erroneous reporting vancomycine susceptibility for Staphylococcus spp.Vitek software version 2.01. Jpn J Infect Dis. 2009;62:298–9. [PubMed] [Google Scholar]
- 23.Veer P, Chande C, Chavan S, Wabale V, Chopdekar K, Bade J, et al. Increasing levels of minimum inhibitory concentration vancomycin in methicillin resistant Staphylococcus aureus alarming bell for vancomycin abusers.? Indian J Med Microbiol. 2010;28:413–4. doi: 10.4103/0255-0857.71810. [DOI] [PubMed] [Google Scholar]
- 24.De Lassence A, Hidri N, Timsit JF, Joly-Guillou ML, Thiery G, Boyer A, et al. Control and outcome of a large outbreak of colonization and infection with glycopeptide-intermediate Staphylococcus aureus in an intensive care unit. Clin Infect Dis. 2006;42:170–8. doi: 10.1086/498898. [DOI] [PubMed] [Google Scholar]
- 25.Schweiger ES, Scheinfeld NS, Tischler HR, Weinberg JM. Linezolid and quinupristin/dalfopristin: novel antibiotics for Gram-positive infections of the skin. J Drugs Dermatol. 2003;2:378–83. [PubMed] [Google Scholar]
- 26.Jansen A, Turck M, Szekat C, Nagel M, Clever I, Bierbaum G. Role of insertion elements and yycFG in the development of decreased susceptibility to vancomycin in Staphylococcus aureus. Int J Med Microbiol. 2007;297:205–15. doi: 10.1016/j.ijmm.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 27.Kuroda M, Kuwahara-Arai K, Hiramatsu K. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem Biophys Res Commun. 2000;269:485–90. doi: 10.1006/bbrc.2000.2277. [DOI] [PubMed] [Google Scholar]
- 28.Maki H, McCallum N, Bischoff M, Wada A, Berger-Bachi B. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2004;48:1953–9. doi: 10.1128/AAC.48.6.1953-1959.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noble WC, Virani Z, Cree RGA. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol Lett. 1992;93:195–8. doi: 10.1016/0378-1097(92)90528-v. [DOI] [PubMed] [Google Scholar]


