Abstract
Dose selection during antiparasitic drug development in animal models and humans traditionally has relied on correlations between plasma concentrations obtained at or below maximally tolerated doses that are efficacious. The objective of this study was to improve the understanding of the relationship between dose and plasma/tissue exposure of the model antiparasitic agent, pafuramidine, using a semiphysiologically based pharmacokinetic (semi-PBPK) modeling approach. Preclinical and clinical data generated during the development of pafuramidine, a prodrug of the active metabolite, furamidine, were used. A whole-body semi-PBPK model for rats was developed based on a whole-liver PBPK model using rat isolated perfused liver data. A whole-body semi-PBPK model for humans was developed on the basis of the whole-body rat model. Scaling factors were calculated using metabolic and transport clearance data generated from rat and human sandwich-cultured hepatocytes. Both whole-body models described pafuramidine and furamidine disposition in plasma and predicted furamidine tissue (liver and kidney) exposure and excretion profiles (biliary and renal). The whole-body models predicted that the intestine contributes significantly (30–40%) to presystemic furamidine formation in both rats and humans. The predicted terminal elimination half-life of furamidine in plasma was 3- to 4-fold longer than that of pafuramidine in rats (170 versus 47 h) and humans (64 versus 19 h). The dose-plasma/tissue exposure relationship for the prodrug/active metabolite pair was determined using the whole-body models. The human model proposed a dose regimen of pafuramidine (40 mg once daily) based on a predefined efficacy-safety index. A similar approach could be used to guide dose-ranging studies in humans for next-in-class compounds.
Introduction
The primary goal of preclinical drug development is to identify compounds with optimal efficacy and safety profiles and desirable pharmacokinetic properties to advance to clinical trials. The design of safe and effective dosage regimens that are compatible with the target patient population and disease remains a major challenge. Suboptimal dose selection can adversely influence progression of a drug development program, resulting in additional time and expense for the dose-ranging study (dose too low), or a poor understanding of risk/benefit, causing unnecessary early termination of promising drug candidates (dose too high).
Human African trypanosomiasis (HAT), a life-threatening parasitic disease, affects the world's poorest populations (Barrett, 2010). HAT is characterized by a first stage, when parasites proliferate in the hemolymphatic system, and a second stage, when parasites cross the blood-brain barrier and invade the central nervous system. The disease is fatal if untreated. All current chemotherapies are unsatisfactory because of toxicity and/or inconvenient parenteral administration regimens (Barrett, 2010). Pafuramidine, a prodrug of furamidine, is the only orally active agent that has shown efficacy in clinical trials for treatment of first-stage infection (Paine et al., 2010). However, clinical development of pafuramidine was placed on hold because of transiently elevated liver transaminases observed in an expanded phase I safety trial (http://www.immtechpharma.com/documents/news_022208.pdf; Paine et al., 2010).
Bioconversion of pafuramidine to furamidine is believed to occur primarily in the liver. The metabolic pathway involves sequential oxidative and reductive reactions, producing four intermediate metabolites (Zhou et al., 2004). After a single oral dose of [14C]pafuramidine (10 mg/kg) to rats, tissue retention of total radioactivity, predominantly as furamidine, was extensive (Midgley et al., 2007). The highest concentration of radioactivity was detected in liver and was 3 orders of magnitude higher than that measured in plasma 24 h after administration; radioactivity was still detectable in liver after 7 days. Unlike with animal models, collection of liver tissue from human subjects over a prolonged period of time is impossible for obvious ethical reasons. As an alternative, a semiphysiologically based pharmacokinetic (semi-PBPK) modeling approach could be used to predict the furamidine hepatic exposure-time profile in humans, permitting improved understanding of the relationship between the dose of pafuramidine and plasma/hepatic exposure of furamidine.
Hepatic clearance is a fundamental PBPK model parameter. Several approaches have been developed to predict human hepatic clearance, including 1) empirical allometric scaling, 2) physiologically based direct scaling of in vitro human clearance, and 3) normalized scaling of in vivo animal clearance based on in vitro animal and human data (Luttringer et al., 2003; Ito and Houston, 2005). Empirical allometric scaling, a conventional technique based on body weight, frequently fails when drug disposition demonstrates large species differences (Lavé et al., 1999). With physiologically based direct scaling, intrinsic clearance, determined from human hepatocytes or liver microsomes, is corrected by a physiologically based scaling factor, and subsequently scaled up on the basis of a liver model (well stirred or parallel tube) (Ito and Houston, 2004). Although preferred to empirical allometric scaling, physiologically based direct scaling consistently underestimates human clearance because of decreased enzyme activity or incomplete enzyme composition associated with in vitro systems (Ito and Houston, 2005). Normalized scaling, via integration of in vivo and in vitro data, represents an alternative approach to predict human pharmacokinetics (Lave et al., 1997; Luttringer et al., 2003). Application of these approaches has been limited primarily to the estimation of metabolic clearance of parent compounds; interspecies extrapolation of both metabolic and transport clearance values is reported rarely for metabolites (Pang et al., 2008).
The objective of this study was to improve the understanding of the relationship between pafuramidine dose and furamidine plasma/hepatic exposure via a semi-PBPK modeling approach. First, a rat whole-liver PBPK model was developed using rat isolated perfused liver (IPL) data. Second, a whole-body semi-PBPK model for rats was developed on the basis of the rat whole-liver PBPK model. Third, normalized scaling was applied to rat IPL and rat and human sandwich-cultured hepatocyte data to predict the metabolic/transport clearance of pafuramidine/furamidine in humans. Fourth, a whole-body semi-PBPK model for humans was developed using the whole-body rat semi-PBPK model and normalized scaling factors. The final whole-body human model was used to predict furamidine plasma and tissue exposure under various multiple-dose scenarios of pafuramidine. This approach could be used to guide dose-ranging human studies for next-in-class compounds.
Materials and Methods
Materials and Chemicals.
Dulbecco's modified Eagle's medium was purchased from Invitrogen (Carlsbad, CA). ITS+ (insulin-transferrin-selenium) culture supplement and Matrigel were obtained from BD Biosciences (San Jose, CA). Penicillin, streptomycin, nonessential amino acids, dexamethasone, Hanks' balanced salt solution (HBSS), modified HBSS (HBSS without Ca2+ and Mg2+, with 0.38 g/l EGTA), and phosphate-buffered saline (PBS) were purchased from Sigma-Aldrich (St. Louis, MO). Human plasma was obtained from Biological Specialty Corporation (Colmar, PA). Pafuramidine, furamidine, and internal standards (d8-pafuramidine and d8-furamidine) were synthesized in the laboratory of Dr. David W. Boykin (Georgia State University, Atlanta, GA) as described previously (Boykin et al., 1996). All other chemicals and reagents were of analytical grade and were used without further purification.
Animals.
Male Wistar and Sprague-Dawley rats (250–300 g) were purchased from Charles River Laboratories (Raleigh, NC). Animals had free access to water and food before surgery. All animal procedures were compliant with guidelines of the University of North Carolina Institutional Animal Care and Use Committee.
Disposition of Pafuramidine and Furamidine in IPLs from Rats.
Data were obtained from a previous recirculating rat IPL study, in which pafuramidine was added as a bolus to the perfusate reservoir to yield an initial concentration of 10 μM (Yan et al., 2011). In brief, perfusions were conducted ex vivo over designated times (up to 2 h) in a temperature-controlled chamber. Aliquots of perfusate (∼400 μl) were collected from the IPL reservoir at 5-min intervals from 0 to 40 min and at 10-min intervals thereafter, and bile was collected at 10-min intervals; the liver was harvested at the end of perfusion.
Determination of Unbound Fraction.
The unbound fractions of furamidine in liver and perfusate from rat IPL experiments and rat plasma, determined using rapid equilibrium dialysis devices (Thermo Fisher Scientific, Waltham, MA), were provided from a previous study (Yan et al., 2011). In the present study, the unbound fractions of pafuramidine in liver and perfusate from rat IPL experiments and rat plasma, as well as unbound fractions of pafuramidine and furamidine in human plasma, were determined using the same method. In brief, pafuramidine or furamidine was added to thawed rat liver homogenates/perfusate/plasma to yield a concentration of 1 μM. An aliquot (200 μl) and 0.1 M PBS (350 μl) were placed in tissue and buffer chambers, respectively, and incubated (37°C) on a Thermomixer (350 rpm) (Eppendorf AG, Hamburg, Germany). After 6 h, aliquots (100 μl) were collected from the sample and buffer chambers and analyzed for total (bound + unbound) and unbound pafuramidine or furamidine, respectively, by LC-MS/MS.
Determination of Blood/Plasma Ratio in Rats.
The blood/plasma (B/P) ratios of pafuramidine and furamidine in rats were determined using an in vitro method described previously (Berry et al., 2010). In brief, pafuramidine or furamidine was added to prewarmed fresh rat blood and reference (blank) plasma to yield a concentration of 0.1 μM. After incubation at 37°C for 1 h in a humidified and oxygenated incubator, compound-treated rat blood was centrifuged at 1500g for 10 min, and plasma was separated from blood cells. Plasma was analyzed for pafuramidine and furamidine by LC-MS/MS. The B/P ratios were calculated by dividing the peak area observed in the reference plasma (representing nominal blood concentration) by the peak area observed in the compound-treated plasma (representing plasma concentration).
Disposition of Pafuramidine and Furamidine in Rat and Human Sandwich-Cultured Hepatocytes.
Rat sandwich-cultured hepatocyte (SCH) data were obtained from a previous study (Yan et al., 2011). Freshly isolated suspended human hepatocytes, provided by Invitrogen, were seeded at 1.5 × 106 cells/well onto 6-well plates and overlaid with Matrigel in the same manner as described for rat hepatocytes (Yan et al., 2011). The liver donors were reported as white (two women and one man; 50, 56, and 57 years old, respectively). Culture medium [Dulbecco's modified Eagle's medium supplemented with 1% (v/v) ITS+, 1 μM dexamethasone, 2 mM l-glutamine, 1% (v/v) nonessential amino acids, 100 units penicillin G sodium, and 100 μg/ml streptomycin sulfate] was changed daily for 5–7 days until extensive canalicular networks were formed. On the day of experimentation, SCH were incubated with culture medium (1.5 ml) containing pafuramidine at the same concentration (10 μM) as that used in rat SCH experiments (Yan et al., 2011). At designated times up to 24 h, aliquots of medium (500 μl) were collected, and cells were washed twice and incubated at 37°C for 5 min with 2 ml of standard HBSS (to maintain bile canalicular networks; cells + bile) or Ca2+-free HBSS (to open bile canalicular spaces; cells) (Turncliff et al., 2006). After incubation, buffer was removed, and cells were washed three times with 2 ml of ice-cold standard HBSS and lysed with 1 ml of ice-cold methanol-water (7:1, v/v) containing 0.1% (v/v) trifluoroacetic acid (TFA). Media and cell lysates were stored at −80°C pending analysis for pafuramidine and furamidine by LC-MS/MS.
In Vivo Studies.
Rats.
Data were provided from a previous study, in which four Sprague-Dawley rats were administered a single dose of pafuramidine (7.5 μmol/kg) by oral gavage (Generaux, 2010). Pafuramidine was prepared as a suspension in acidified water (pH 3)-70% Tween 80 in ethanol (7:3, v/v). Blood (0.2 ml) was collected via a jugular vein cannula over 24 h after pafuramidine administration. Plasma was separated from blood cells by centrifugation (1500g for 10 min) and analyzed for pafuramidine and furamidine by LC-MS/MS (see below).
Humans.
A phase I, open-label study was conducted by Hammersmith Medicines Research (London, UK). The primary objective was to assess the absorption, metabolism, and excretion of [14C]pafuramidine maleate in healthy male volunteers. The Medicines and Healthcare Products Regulatory Agency, Administration of Radioactive Substances Advisory Committee, and Huntingdon Research Ethics Committee reviewed the study protocol; the Brent Medical Ethics Committee conducted a site-specific assessment of the study. The study commenced upon authorization by the Medicines and Healthcare Products Regulatory Agency and approval by the Administration of Radioactive Substances Advisory Committee and ethics committees. Potentially eligible subjects provided written, informed consent before screening, which entailed a medical history, physical examination, vital signs (blood pressure and heart rate), 12-lead electrocardiogram (ECG), standard blood and urine laboratory analyses, and breath tests for alcohol and smoking (carbon monoxide). Exclusion criteria included radiation exposure (a radioactive substance or X-rays, with the exception of dental X-rays or X-rays of the chest, hands, or feet) during the 12 months before the study; abnormal history or physical observations, ECG, or laboratory values that could interfere with the study objectives or safety of the volunteer; acute or chronic illness that could preclude or render hazardous the volunteer's participation; severe adverse reaction to any drug or a history of sensitivity to dicationic compounds; prescription medication use during the 28 days before the study or use of over-the-counter preparations, including herbal/dietary supplements, during the 7 days before the study; presence or history of drug or alcohol abuse; tobacco product use within the previous 6 months; and blood pressure and heart rate in the seated position outside the ranges of 90 to 160 mm Hg systolic/40 to 95 mm Hg diastolic and 40 to 100 beats/min, respectively.
Healthy male white volunteers (n = 6), aged from 48 to 63 years and weighing from 63 to 108 kg, were enrolled in the study. All subjects reported to the hospital ward at 8:00 PM the evening of day −1 (the day before drug administration); urine was collected and tested for drugs of abuse, and breath was collected and tested for alcohol and smoking (carbon monoxide). After an overnight fast, a cannula was placed into an antecubital vein the next morning (day 1), and the following were undertaken within 60 min before drug administration: 12-lead ECG; measurement of vital signs; venous blood collection for laboratory safety tests and baseline radioactivity measurement; and urine collection for laboratory safety tests. Subjects finished eating a high-fat breakfast (2 eggs fried in butter, 2 bacon strips fried in butter, 2 slices of buttered toast, 4 ounces of hash brown potatoes, and 8 fluid ounces of whole milk), approximately 15 min before dosing to facilitate drug absorption.
Each volunteer was administered a single oral dose of [14C]pafuramidine maleate (nominal dose 131.9 mg, equivalent to 100 mg of pafuramidine free base) with 100 ml of water between 9:00 and 9:25 AM. The dose was prepared in capsule form and contained 0.7 MBq (19 μCi) of radioactivity. The dose of pafuramidine free base was consistent with clinical use of the drug; the dose of radioactivity was consistent with the upper limit for a category IIa study set by the International Commission of Radiological Protection for trials in male volunteers. After [14C]pafuramidine administration, blood was collected at designated times from 0.5 to 168 h; vital signs and 12-lead ECGs were recorded from 1 to 168 h; urine was collected continuously in 3-, 6-, 12-, and 24-h intervals from 0 to 168 h; and feces were collected as individual evacuations from 0 to 168 h. Within 30 min of blood collection, plasma was separated from blood cells by centrifugation (700–1500g for 10 min). Whole blood and the resultant plasma, as well as urine and feces, were stored at −20°C pending analysis for total radioactivity, pafuramidine, and furamidine. Approximately 168 h after drug administration, blood and urine were collected for laboratory safety tests, and a physical examination was conducted. Other than the high-fat breakfast on day 1, standard meals and beverages were provided to the subjects during their stay on the ward. Alcoholic and caffeinated beverages, grapefruit and grapefruit juice, smoking, and strenuous exercise were not allowed from 24 h before drug administration until after discharge.
Subjects were discharged on the morning of day 8 (∼168 h after drug administration). They returned to the ward as outpatients to provide complete urine and fecal collections from the previous 24 h. These outpatient visits, planned on the mornings of days 10, 14, 21, 28, 35, and 42, continued until there was no measurable radioactivity in urine and feces. At each visit, vital signs and 12-lead ECGs were recorded and a physical examination was conducted.
Total radioactivity was measured in whole blood, plasma, urine, and feces by high-performance liquid radiochromatography. Pafuramidine and furamidine concentrations in plasma were measured by LC-MS/MS (see below).
LC-MS/MS Analysis.
In vitro samples and rat plasma.
Pafuramidine and furamidine were quantified using an API 4000 triple quadrupole mass spectrometer (Applied Biosystems, Foster City, CA) equipped with a TurboIonSpray interface (Applied Biosystems/MDS Sciex, Foster City, CA). The sample preparation procedure and LC-MS/MS conditions for the quantification of pafuramidine and furamidine were detailed previously (Yan et al., 2011). In brief, pafuramidine and furamidine and internal standards (d8-pafuramidine and d8-furamidine) were separated on an Aquasil C18 HPLC column (2.1 mm × 50 mm, 5 μm) (Thermo Fisher Scientific) with a high-pressure linear gradient program. Calibration curves were prepared in appropriate matrices (0.05–5 μM in liver homogenates; 1–10,000 nM in perfusate, medium, cell lysates, human plasma, and PBS) and were linear over the respective ranges (R2 > 0.98). The limit of quantification was 5 nM for both compounds.
Human plasma.
Quantification of pafuramidine and furamidine was conducted by Tandem Labs (Salt Lake City, UT). The stock solutions of analytes (pafuramidine and furamidine) and internal standards (d8-pafuramidine and d8-furamidine) were prepared in 100% methanol to yield concentrations of 0.37, 0.41, 0.21, and 0.3 mg/ml, respectively. Stock solutions of pafuramidine and furamidine were diluted in blank human plasma to yield working concentrations of 5000 and 2000 ng/ml for preparation of calibration standards and quality controls, respectively. Calibration standards (0.25–250 ng/ml) and quality controls (0.75, 75, and 200 ng/ml) for pafuramidine and furamidine were prepared by serial dilution of each working solution with blank human plasma. The working internal standard solutions (100 ng/ml d8-pafuramidine and d8-furamidine) were prepared by diluting the respective stock solution with 0.05% TFA in water-methanol (5:5, v/v); 50 μl of the diluted stock solution and 500 μl of 50 nM ammonium acetate buffer (pH 3) were added to 100 μl of human plasma, standards, and quality controls. After vortex mixing, all samples were loaded onto Polycrom B1000 1cc, 20-mg extraction cartridges (Cera, Inc., Baldwin Park, CA), followed by serial washing of the solid phase with 300 μl of 50 mM ammonium acetate buffer (pH 3), followed by 300 μl of 50 mM ammonium acetate buffer (pH 3)-methanol (8:2, v/v). Analytes were eluted with 300 μl of acetonitrile, followed by 300 μl of 0.1% hydrochloric acid in methanol. After evaporation at 45°C (TurboVap; Zymark Corp., Hopkinton, MA), samples were reconstituted with 50 μl of 0.05% TFA in water-acetonitrile (9:1; v/v) and transferred to HPLC vials. Pafuramidine and furamidine were quantified on an API 3000 triple quadrupole mass spectrometer (PE Sciex, Concord, ON, Canada). Analytes were separated on a BDS Hypersil Phenyl HPLC column (2 mm × 50 mm, 5 μm) at 35°C with a high-pressure linear gradient program consisting of 0.05% TFA in HPLC-grade water (A) and HPLC-grade acetonitrile (B) delivered by a HP 1100 pumping system (Hewlett Packard, Palo Alto, CA) at a flow rate of 300 μl/min. Mobile phase composition was increased from 10 to 70% B from 0 to 3.5 min and then decreased to 10% B from 3.5 to 3.6 min; the column was reequilibrated for 2 min before the next injection. Calibration curves for pafuramidine and furamidine were linear from 0.25 to 250 ng/ml (R2 > 0.99). Intra- and interday precision (expressed as coefficient of variation percentage) and accuracy (expressed as bias percentage) of quality controls for both compounds were <15%. The mass spectrometers were operated in positive ion mode using multiple reaction monitoring: pafuramidine, 365.1 → 334.1 m/z; furamidine, 305.3 → 288.1 m/z; d8-pafuramidine, 373.1 → 242.0 m/z; d8-furamidine, 313.3 → 296.1 m/z.
Semi-PBPK Modeling: Model Approach.
Kinetic parameters associated with hepatic disposition of pafuramidine and furamidine were generated from the whole-liver semi-PBPK model and used to develop the whole-body rat semi-PBPK model. Preclinical data on preformed furamidine kidney-to-plasma partitioning and renal excretion were incorporated to predict formed furamidine disposition in rats. In vivo rat data were used to develop the semi-PBPK model structure and associated parameters. Kinetic parameters for humans (see Semi-PBPK Modeling: Model Parameterization) were used to predict the disposition of pafuramidine and furamidine and, ultimately, to predict the relationship between dose and plasma or tissue exposure.
Semi-PBPK Modeling: Model Structure.
Rat whole liver.
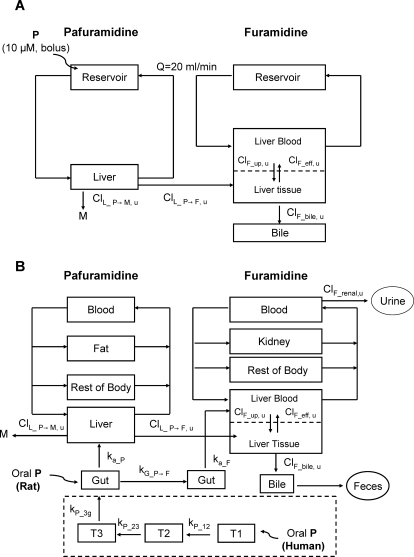
Pafuramidine and furamidine submodels were linked by pafuramidine metabolism in the liver (Fig. 1A). The pafuramidine submodel for rat IPLs was composed of two compartments: perfusate reservoir and liver (Fig. 1A). The furamidine submodel was composed of three compartments: perfusate reservoir, liver, and bile. Pafuramidine is highly lipophilic (log DpH_7 = 4.3) (Zhou et al., 2002) and poorly soluble, qualifying as a class II compound according to the Biopharmaceutics Classification System (Wu and Benet, 2005). As such, pafuramidine was assumed to diffuse passively through the hepatic basolateral membrane. In contrast to pafuramidine, furamidine is hydrophilic (log DpH_7 = −3) (Zhou et al., 2002). Furamidine basolateral reuptake and efflux clearances (ClF_up, u and ClF_eff, u) (Table 1) estimated by the whole-liver model (Fig. 1A) were at least 2-fold lower than the perfusate flow rate (4 l · h−1 · kg−1), suggesting that diffusional barriers exist for furamidine. Therefore, distribution of pafuramidine and furamidine in the liver was assumed to be flow- and diffusion-limited, respectively (Fig. 1A).
Fig. 1.
Semi-PBPK model schemes depicting disposition of pafuramidine and furamidine in rat IPLs (A) and in vivo in rats and humans (B). Distinct model structures were developed for the prodrug, pafuramidine, and the derived active metabolite, furamidine, and linked by liver (and gut) metabolism. Pafuramidine distribution in all tissue compartments was assumed to be flow-limited. Furamidine distribution in the liver was described as diffusion-limited, as depicted by the dashed line in the liver compartment; furamidine distribution in all other tissue compartments was assumed to be flow-limited. Compartments T1, T2, and T3 in the dashed square represent three consecutive transit compartments for pafuramidine before reaching the absorption site in the gut after oral administration in humans, where kP_12, kP_23, and kP_3g represent the first-order rate constants for pafuramidine distribution along the transit compartments and gut. kG_P→F represents the first-order rate constant for metabolic conversion from pafuramidine to furamidine in the gut; ka_P and ka_F represent the first-order rate constants for the movement from gut to liver of pafuramidine and formed furamidine, respectively. ClL_P→F, u represents hepatic unbound intrinsic clearance for the formation of furamidine from pafuramidine; ClL_P→M, u represents hepatic unbound intrinsic clearance for metabolic conversion from pafuramidine to other metabolites; ClF_up, u represents unbound intrinsic clearance for hepatic basolateral uptake of furamidine, ClF_eff, u represents unbound intrinsic clearance for hepatic basolateral efflux of furamidine, ClF_bile, u represents unbound intrinsic clearance for biliary excretion of furamidine, and ClF_renal, u represents unbound renal clearance of furamidine from plasma. P, pafuramidine; M, other metabolites; F, furamidine.
TABLE 1.
Semiphysiologically based pharmacokinetic model parameters associated with the disposition of pafuramidine and furamidine in rats and humans
| Parameters | Value |
Source | |
|---|---|---|---|
| Rat | Human | ||
| Physiological | |||
| Body weight (kg) | 0.3 | 70 | Measured |
| Cardiac output (l · h−1 · kg−1) | 14 | 5 | Brown et al., 1997 |
| Blood and tissue volumesa | |||
| Blood | 0.074 | 0.077 | Brown et al., 1997 |
| Fat | 0.07 | 0.2 | Brown et al., 1997 |
| Liver | 0.03 | 0.03 | Brown et al., 1997 |
| Kidney | 0.007 | 0.004 | Brown et al., 1997 |
| Tissue blood flowsa | |||
| Fat | 0.07 | 0.05 | Brown et al., 1997 |
| Liver | 0.17 | 0.25 | Brown et al., 1997 |
| Kidney | 0.14 | 0.19 | Brown et al., 1997 |
| Rest of body | 0.62 | 0.51 | Calculated |
| Chemical-specific | |||
| Pafuramidine | |||
| Unbound fractions (%) | |||
| Liver (fu, L_P)b | 0.07 ± 0.02 | Same as rats | Measured |
| Perfusate (fu, per_P) | 1.1 ± 0.1 | N.A. | Measured |
| Plasma (fu, p_P) | 0.2 ± 0.02 | 0.2 ± 0.01 | Measured |
| Liver-to-perfusate partition coefficient | 70 | N.A. | Fitted |
| Tissue-to-plasma partition coefficients | |||
| Liver | 14 | Same as rats | Calculated |
| Fat | 260 | Same as rats | Fitted |
| Rest of body | 2 | Same as rats | Fitted |
| B/P ratioc | 1.1 ± 0.1 | Same as rats | Measured |
| Fraction of dose absorbed into enterocytes (fa) | 0.3 | 1 | Fitted |
| Gastrointestinal rate constants (h−1)d | |||
| kP_12, kP_23, kP_3g | N.A. | 1.1 | Fitted |
| ka_P | 0.5 | 1.1 | Fitted |
| kG_P→F | 0.07 | 0.3 | Fitted |
| Hepatic clearance (l · h−1 · kg b.wt.−1)e | |||
| ClL_P→M, u | 70 | 124 | Fitted (rat); scaled (human) |
| ClL_P→F, u | 40 | 71 | Fitted (rat); scaled (human) |
| Furamidine | |||
| Unbound fractions (%) | |||
| Liver (fu, L_F)b | 0.3 ± 0.1 | Same as rats | Measured |
| Perfusate (fu, per_F) | 44 ± 5 | N.A. | Measured |
| Plasma (fu, p_F) | 24 ± 2 | 25 ± 3 | Measured |
| Tissue-to-plasma partition coefficients | |||
| Kidney | 4000 | Same as rats | Fitted |
| Rest of body | 1 | Same as rats | Fitted |
| B/P ratioc | 0.9 ± 0.1 | Same as rats | Measured |
| Gastrointestinal rate constants (h−1)d | |||
| ka_F | 2.4 | 0.2 | Fitted |
| Hepatic and renal clearances (l · h−1 · kg b.wt.−1)e | |||
| ClF_up, u | 1.9 | 1 | Fitted (rat); scaled (human) |
| ClF_eff, u | 0.08 | 0.05 | Fitted (rat); scaled (human) |
| ClF_bile, u | 0.08 | 0.05 | Fitted (rat); scaled (human) |
| ClF_renal, u | 0.12 | 0.024 | Calculated |
| GFRf | 0.4 | 0.08 | Lin, 1998 |
N.A., not applicable.
Tissue volumes and blood flows denote fractions of total body weight and cardiac output, respectively.
Liver unbound fractions in humans were assumed to be equal to those in rats.
B/P ratio of pafuramidine or furamidine at 0.1 μM in rat blood (see Materials and Methods).
Rate constants associated with pafuramidine and furamidine absorption and metabolism in the gut are defined in the legend to Fig. 1 and were optimized by fitting the semi-PBPK model (scheme depicted in Fig. 1B) to in vivo rat and human data.
Clearance values associated with the disposition of both pafuramidine and furamidine in rat livers are defined in the legend to Fig. 1 and were optimized by fitting the semi-PBPK model (scheme depicted in Fig. 1A) to rat IPL data, corresponding hepatic clearances in humans were calculated as described under Materials and Methods, furamidine renal clearance in rats was calculated on the basis of in vivo rat data, and human renal clearance was estimated using a “GFR ratio approach” proposed by Lin (1998) (see Materials and Methods).
GFR was obtained from Lin (1998).
Rat whole body.
The whole-liver rat model was expanded into a whole-body rat model by substituting the perfusion reservoir with the blood compartment (Fig. 1B). On the basis of the high lipophilicity of pafuramidine, fat was added as a storage organ in the pafuramidine submodel. The kidney was incorporated as an additional storage organ in the furamidine submodel on the basis of the significant kidney retention of furamidine (Midgley et al., 2007; Goldsmith, 2011); all other tissues were grouped together as “rest of body” in both submodels to maintain mass balance (Fig. 1B). Pafuramidine distribution into the liver was assumed to be flow-limited, whereas furamidine distribution into the liver was assumed to be diffusion-limited. To avoid overparameterization, distribution of both compounds into all other organs was assumed to be flow-limited (Fig. 1B). On the basis of the high hepatic extraction ratio calculated from IPL data (EH = 0.88) (Yan et al., 2011), pafuramidine was assumed to be cleared from the body primarily via hepatic metabolism. On the basis of in vivo data, furamidine was assumed to be eliminated via both biliary and renal excretion (Midgley et al., 2007; Goldsmith, 2011). To simulate pafuramidine and furamidine plasma concentration-time profiles, the model incorporated a single gut compartment for pafuramidine absorption after oral administration. Absorption of pafuramidine from gut to liver was assumed to be a first-order process. Because the initial model failed to describe the prompt appearance of furamidine in plasma, the model was modified to include the gut as a site of furamidine formation during pafuramidine absorption.
Human whole body.
The final whole-body rat model was used initially to predict the disposition of pafuramidine/furamidine in humans. However, model predictions failed to describe the delayed absorption of pafuramidine observed in humans, which may be attributed to the capsule formulation, concurrent administration of a high-fat meal, and/or species differences in gut metabolism and/or transport. Therefore, three consecutive transit compartments were added between the site of administration (oral route) and site of absorption (gut) in the pafuramidine submodel (Fig. 1B) to represent dissolution from the dosage form (capsule), stomach emptying, and/or partitioning from the fat components of the concomitant high-fat meal. Species differences in gut metabolism and/or transport were considered as described under Semi-PBPK Modeling: Model Parameterization.
Semi-PBPK Modeling: Model Parameterization.
Absorption and metabolism in the gut.
Absorption and metabolism of pafuramidine/furamidine in the gut have not been characterized extensively in rats and humans. Thus, the relevant kinetic parameters, including the fraction of the dose absorbed into enterocytes (fa), the rate constants associated with absorption of pafuramidine or furamidine (ka_P or ka_F), and the rate constant associated with metabolic conversion from pafuramidine to furamidine in the gut (kG_P→F) were estimated by fitting the semi-PBPK models (Fig. 1B) to in vivo rat and human data.
Tissue distribution.
Tissue-to-perfusate/plasma partition coefficients were optimized by fitting relevant PBPK models (Fig. 1, A and B) to IPL and in vivo rat data. The pafuramidine liver-to-plasma partition coefficient in rats was calculated on the basis of the liver-to-perfusate partition coefficient generated from the rat IPL data after correction for the 5-fold difference in unbound fraction between plasma and perfusate (Table 1). Because of the lack of human tissue data, tissue partition coefficients and liver binding for pafuramidine/furamidine in humans were assumed to be equal to those in rats (Table 1).
Hepatic clearance.
Rat hepatic clearance (Clrat_liver) values were derived by fitting the whole-liver rat model to the rat IPL data. Clrat_liver, which represented metabolic and/or active transport capacities, was normalized by a scaling factor (SF) and corrected for liver weight (LW) to estimate the human hepatic clearance value: Clhuman_liver = Clrat_liver × SF × (LWhuman/LWrat), where SF is Clhuman SCH/Clrat SCH or khuman SCH/krat SCH and LWhuman and LWrat were normalized to body weight for humans and rats, respectively. Because biliary Cl of formed furamidine was too small to measure in either rat or human SCH, in vivo human biliary Cl was estimated by scaling in vivo rat biliary Cl on the basis of liver weight.
Renal clearance.
Furamidine unbound renal clearance from plasma in rat (ClR_rat, u) was calculated on the basis of preclinical data in rats administered furamidine intravenously (10 μmol/kg) (Goldsmith, 2011). In brief, rats (n = 3–7) were sacrificed at designated time points up to 16 days after furamidine administration, after which kidneys were harvested and homogenized; urine was collected at 0 to 3, 3 to 6, 6 to 12, and 12 to 24 h intervals after furamidine administration. ClR_rat, u was calculated by dividing the total amount of furamidine recovered in urine from 0 to 24 h by the area under the concentration-time curve (AUC) in plasma within the same time interval and was corrected by the unbound fraction of furamidine in plasma (Table 1). The unbound renal clearance of furamidine from plasma in humans (ClR_human, u; liters per hour per kilogram) was estimated by the “glomerular filtration rate (GFR) ratio approach” (Lin, 1998), assuming that active processes were not involved: ClR_human, u = ClR_rat, u/GFR ratio, where the GFR ratio between rats and humans is 4.8.
PBPK Modeling and Simulation.
Pharmacokinetic analysis.
PBPK modeling and simulation were performed with Berkeley Madonna (version 8.0.2; University of California, Berkeley, CA), a differential equation-based modeling software program used extensively in the development of PBPK models (Rowland et al., 2004). The goodness-of-fit of model simulations was assessed on the basis of a visual comparison of the predicted mass/concentration-time profiles and observed in in vivo rat and human data. Observed and predicted plasma/tissue concentration-time profiles were analyzed for plasma/tissue AUC values and terminal half-lives (t1/2, terminal) of pafuramidine/furamidine by noncompartmental analysis using WinNonlin (version 5.0.1; Pharsight, Mountain View, CA).
Prediction of dose-exposure relationship.
Furamidine plasma concentration-time profiles were simulated under different multiple-dose regimens on the basis of the single-dose human semi-PBPK model (Fig. 1B). The efficacy and safety indices of furamidine were defined by a minimum effective concentration (Ceff, min) and a hypothetical no observable adverse effect level (NOAEL), respectively. Selection of an optimal multiple-dose regimen of pafuramidine was based on the assumption that furamidine concentrations in plasma must be greater than Ceff, min at least 80% of the time during the dosing interval, whereas the average steady-state and maximum concentrations in plasma (Css, ave and Css, max) must be less than the NOAEL. Ceff, min was determined on the basis of an in vitro IC50 (1 ng/ml ≈ 3 nM) of furamidine against the Trypanosoma brucei rhodesiense strain, STIB900 (Wenzler et al., 2009). This trypanosome strain is used routinely at the Swiss Tropical and Public Health Institute to assess in vitro and in vivo activity of diamidine compounds (Ismail et al., 2005). The IC50, determined with culture medium containing 15% heat-inactivated horse serum, was converted to Ceff, min on the basis of the following assumptions: 1) no species differences in plasma binding (fu, p_F = 25%) (Table 1); and 2) binding in 15% horse serum (fu, ser_F) approximated that in rat perfusate (fu, per_F), which consisted of 20% rat blood; that is, fu, ser_F = fu, per_F = 44% (Table 1). Ceff, min for total (bound + unbound) furamidine in plasma was corrected to 5 nM (Ceff, min = IC50 × fu, ser_F/fu, p_F = 3 nM × 44%/25% = 5 nM). The NOAEL has not been determined for furamidine in humans. A concentration of 100 nM, which is 20% less than the predicted Css, ave under the dosage regimen used in the expanded phase I safety study (100 mg twice daily for 14 days), was selected to define the NOAEL. Dose safety was evaluated by comparing the furamidine concentrations in plasma with the hypothetical NOAEL and comparing furamidine exposure in liver between the clinically used and semi-PBPK model-predicted dosage regimens.
Results
Whole-Liver Rat PBPK Model Prediction.
The disposition of pafuramidine and furamidine in rat IPLs was characterized in a previous study (Yan et al., 2011). In summary, pafuramidine was taken up by IPLs and eliminated primarily by metabolism, with negligible biliary excretion; at the end of the 2-h perfusion, >98% of total formed furamidine was recovered in the liver. Pafuramidine distribution between perfusate and liver reached equilibrium after ∼20 min. The liver-to-perfusate partition coefficient for pafuramidine, generated on the basis of the whole-liver model (Fig. 1A), was 70 (Table 1). Pafuramidine was highly bound to proteins in plasma, perfusate (composed of 20% blood), and liver tissue (Table 1), whereas furamidine was highly bound only to liver tissue. The unbound fraction of furamidine in liver (fu, L_F) was at least 80-fold lower than that in plasma (fu, p_F) and perfusate (fu, per_F) (Table 1). Hepatic unbound intrinsic formation clearance of furamidine (ClL_P→F, u) accounted for approximately one-third of total hepatic unbound intrinsic clearance of pafuramidine (ClL_P→F, u + ClL_P→M, u) (Table 1). The unbound intrinsic clearance for furamidine hepatic basolateral reuptake (ClF_up, u) was 24-fold higher than that for basolateral efflux (ClF_eff, u) (Table 1), whereas the unbound intrinsic clearance for furamidine biliary excretion (ClF_bile, u) was similar to that of ClF_eff, u (Table 1).
Disposition of Pafuramidine and Furamidine in Rat and Human Sandwich-Cultured Hepatocytes.
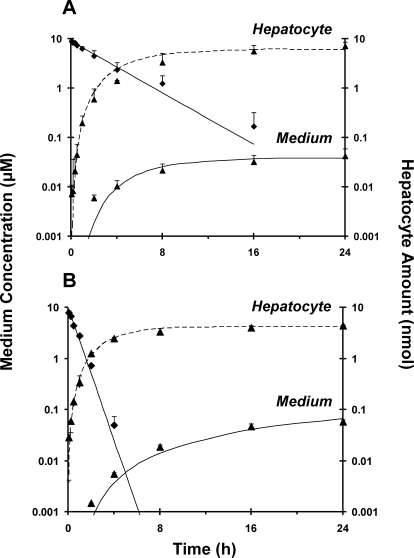
The disappearance of pafuramidine from medium was faster in human than in rat SCH (Fig. 2), as reflected by the 3-fold higher intrinsic clearance of pafuramidine in human than in rat SCH (Table 2). Disposition profiles of formed furamidine were similar between rat and human SCH (Fig. 2). The kinetic parameters associated with pafuramidine and furamidine hepatic disposition in rat and human SCH were derived from a previously developed compartmental model (Yan et al., 2011). The rate constants for furamidine basolateral efflux were similar between rats and humans, whereas the rate constant for furamidine basolateral reuptake in human SCH was three-fourths of that in rat SCH (Table 2). Furamidine was not detected in bile in either rat or human SCH, which could be due to the fact that biliary excretion of furamidine was so small that the difference in substrate accumulation between standard HBSS (cells + bile) and Ca2+-free HBSS (cells) was not measurable (Yan et al., 2011).
Fig. 2.
Disposition of pafuramidine (♦) and furamidine (▴) over 24 h in SCH from rats (A) and humans (B). Rat SCH data were obtained from a previous study (Yan et al., 2011); human SCH data were obtained using a similar study design. Pafuramidine (10 μM) was administered as a bolus to each well, which contained 1.5 ml of culture medium. Symbols and error bars denote mean values and SDs, respectively of n = 3 livers. Lines represent the computer-generated best fit of a previously developed pharmacokinetic model (Yan et al., 2011) to the data.
TABLE 2.
Kinetic parameters associated with disposition of pafuramidine and furamidine in rat and human SCH
| Parametera | Rat SCH | Human SCH | Scaling Factorb |
|---|---|---|---|
| ClL_P→M (ml · min−1 · 106 cells−1) | 0.0047 | 0.013 | 3 |
| ClL_P→F (ml · min−1 · 106 cells−1) | 0.0023 | 0.0067 | 3 |
| kF_up (h−1) | 0.8 | 0.6 | 0.75 |
| kF_eff (h−1) | 0.008 | 0.008 | 1 |
| kF_bile (h−1)c | N.A. | N.A. | N.A. |
N.A., not applicable.
ClL_P→M and ClL_P→F represent in vitro intrinsic clearance for the metabolic conversion of pafuramidine to other metabolites and furamidine, respectively. kF_up, kF_eff, and kF_bile represent first-order rate constants for furamidine hepatic basolateral uptake, efflux and biliary excretion, respectively. SCH values denote mean data from n = 3 separate livers.
Determined by the ratio of human to rat hepatic clearances or rate constants determined with SCH. The scaling factor was used to normalize in vivo clearance values from rats before scaling to human clearance values (see Materials and Methods).
Furamidine biliary excretion was undetectable in both rat and human SCH.
Disposition of Pafuramidine and Furamidine in Rats.
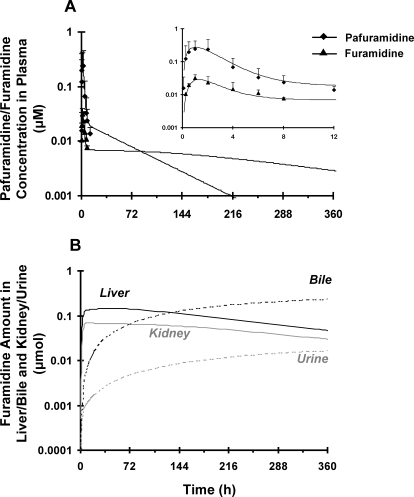
Pafuramidine was absorbed and metabolized to furamidine rapidly after oral administration; both compounds in plasma reached a maximum concentration at ∼1 h and declined approximately in parallel for up to 12 h (Fig. 3A, inset). The apparent terminal half-life (t1/2, app) of pafuramidine, based on the 0 to 12 h plasma concentration-time profile, was similar to that of furamidine (4 h).
Fig. 3.
Disposition of pafuramidine (♦) and furamidine (▴) in rats administered a single oral dose of pafuramidine (7.5 μmol/kg). A, comparison of observed (symbols) and semi-PBPK model-predicted (scheme depicted in Fig. 1B) (lines) plasma concentration-time profiles of pafuramidine and furamidine over 12 h (inset) and 360 h. Symbols and error bars denote mean values and SD, respectively, of four rats. B, semi-PBPK model-predicted (scheme depicted in Fig. 1B) amount-time profiles of furamidine in liver/kidney (solid lines) and bile/urine (dashed lines) over 360 h.
Whole-Body Rat Semi-PBPK Model Prediction.
Physiological and pafuramidine- and furamidine-specific parameters (Table 1) were used to develop the rat semi-PBPK model (Fig. 1B). Pafuramidine and furamidine hepatic disposition parameters were obtained from the rat whole-liver model (Fig. 1A). The partition coefficient of pafuramidine in fat was much higher than that in other tissues, which could be due to the high lipophilicity (Zhou et al., 2002; Andersen et al., 2008). On the basis of previous data from rats administered preformed furamidine intravenously (Goldsmith, 2011), the furamidine kidney-to-plasma partition coefficient was estimated to be as high as 4000 (Table 1); the renal clearance of furamidine from plasma (ClF_renal, u) in rats was approximately 3-fold less than the GFR (Table 1) (Goldsmith, 2011). The rate constant for pafuramidine absorption (ka_P) was 7-fold higher than that for metabolic conversion of pafuramidine to furamidine in the gut (kG_P→F). The overall fraction of furamidine formed from pafuramidine in the rat was approximately 40%, of which the liver contributed approximately twice as much as the gut (Table 3). The semi-PBPK model (Fig. 1B) adequately described pafuramidine/furamidine disposition observed in plasma up to 12 h (Fig. 3A, inset). On the basis of model predictions through 360 h, the t1/2, terminal values of both pafuramidine and furamidine were at least 10-fold longer than those measured from the 0 to 12 h observed in vivo data (t1/2, app); t1/2, terminal of furamidine was ∼4-fold longer than that of pafuramidine (170 versus 47 h) (Fig. 3A). The semi-PBPK model predicted that liver and kidney accumulation of furamidine was extensive, accounting for 63 and 32% of the total formed at 12 h, respectively; ∼1% of furamidine was recovered in plasma at 12 h. Furamidine exposure in liver and kidney reached a maximum at ∼24 h (Fig. 3B) and then declined in parallel with that in plasma (Fig. 3A). Furamidine was eliminated in rat primarily by biliary excretion; renal excretion was <10% (Table 3; Fig. 3B).
TABLE 3.
Semi-PBPK model prediction of furamidine presystemic formation and excretion in rats and humans after administration of a single oral dose of pafuramidine (7.5 μmol/kg) to rats or [14C]pafuramidine (100 mg) to healthy human volunteers
| Outcome |
||
|---|---|---|
| Rat | Human | |
| % | ||
| fma | 40 | 52 |
| Presystemic formation in gutb | 30 | 40 |
| Presystemic formation in liverb | 70 | 60 |
| Excretion into bilec | 93 | 96 |
| Excretion into urinec | 7 | 4 |
Total fraction of pafuramidine converted to furamidine.
Contribution of gut or liver to furamidine presystemic formation.
Calculated based on the cumulative amount of furamidine excreted in bile or urine, relative to total excreted, to infinite time.
Disposition of Total Radioactivity, Pafuramidine, and Furamidine in Humans.
All of the volunteers completed the study. All subjects reported 11 adverse events (3 moderate and 8 mild) after [14C]pafuramidine administration, the most common of which was headache. Only two events (mild headache) were considered to be possibly related to the study medication. There were no changes in vital signs, 12-lead ECG, or laboratory values that were clinically significant or that could reasonably be attributed to the study medication. Overall, a single oral dose of [14C]pafuramidine maleate was deemed safe and well tolerated.
Total radioactivity in whole blood was only measurable from 3 to 5 h after [14C]pafuramidine maleate administration. Maximum radioactivity concentrations in whole blood ranged between 490 and 890 ng equivalents/ml (mean Cmax = 570 ng equivalents/ml), and Tmax ranged from 2 to 4 h (median = 3 h). Total radioactivity in plasma was consistently measurable from 2.5 to 12 h; maximum radioactivity concentrations ranged between 650 and 980 ng equivalents/ml (mean Cmax = 760 ng equivalents/ml), and Tmax ranged from 2 to 5 h (median = 4.5 h). Radioactivity concentrations in whole blood were generally lower than corresponding concentrations in plasma.
After [14C]pafuramidine maleate administration, pafuramidine was measurable in plasma by 0.5 h in five of the six subjects. Pafuramidine plasma concentrations increased to a maximum value ranging between 98 and 644 nM (mean Cmax = 186 nM) from 1.5 to 4 h (median Tmax = 2.8 h). Pafuramidine plasma concentrations declined, with a t1/2, app ranging from 4 to 46 h (harmonic mean = 11.5 h). Furamidine was not consistently measurable in plasma until 2 h after [14C]pafuramidine administration and increased to a maximum concentration ranging between 39 and 114 nM (mean Cmax = 44 nM) from 4 to 10 h (median Tmax = 6 h). Furamidine plasma concentrations declined, with a t1/2, app ranging from ∼8 to 42 h (harmonic mean = 14.5 h). Both pafuramidine and furamidine were above the limit of quantification in plasma of all subjects up to 24 h after drug administration.
Within 168 h of [14C]pafuramidine administration (during which complete collections of excreta were available), the major route of elimination of radioactivity was via the feces. The average (range) percentage of the dose excreted into the feces (primarily as furamidine and one of the intermediate metabolites, M3) and urine (primarily as M3) was 36% (33–40%) and 13% (11–16%), respectively. Extrapolations of the data available after 168 h indicated that the average (range) percentage of the dose excreted into the feces and urine would be 39% (34–42%) and 13% (11–16%), respectively. There was considerable retention of radioactivity (∼50% of the dose), largely as furamidine, in the human body 7 days after [14C]pafuramidine administration.
Whole-Body Human Semi-PBPK Model Prediction.
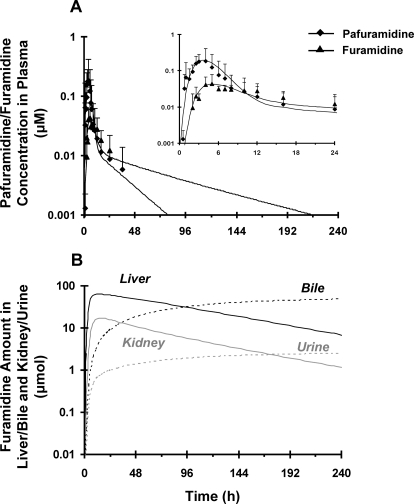
Physiological and pafuramidine/furamidine-specific parameters (Table 1) were used to develop the human semi-PBPK model. No significant difference in plasma binding of pafuramidine and furamidine was observed between rats and humans (Table 1). Furamidine unbound fraction in human plasma (fu, p_F) was consistent with previously reported values (∼24%) at concentrations ranging from 16 to 328 nM (Midgley et al., 2007), indicating concentration-independent binding. Pafuramidine/furamidine hepatic clearance values in humans were predicted by scaling the corresponding in vivo rat values normalized by scaling factors derived from rat and human SCH studies, as described under Materials and Methods. Similar to rats, the renal clearance of furamidine from plasma (ClF_renal, u), estimated by the GFR approach, was approximately 3-fold less than GFR (Table 1). Because a one-compartment oral absorption model failed to predict the delayed absorption of pafuramidine, transit compartments were added to the human semi-PBPK model (Fig. 1B). The rate constants for pafuramidine absorption along the transit compartments (kP_12, kP_23, and kP_3g) were similar (Table 1); the rate constant for pafuramidine absorption from gut to liver (ka_P) was comparable to kP_12, kP_23, and kP_3g but was 5-fold higher than that for furamidine absorption (ka_F) (Table 1); and the rate constant for metabolic conversion of pafuramidine to furamidine in the gut (kG_P→F) was approximately 4-fold lower than ka_P (Table 1). The overall fraction of furamidine generated from pafuramidine in the gut and liver was approximately 50%; similar to rats, the contribution by the liver was higher than that by the gut (Table 3). Model predictions, based on the scheme depicted in Fig. 1B, adequately described pafuramidine and furamidine disposition observed in plasma up to 24 h (Fig. 4A, inset). Prolonged (up to 240 h) predictions indicated that the t1/2, terminal values of both pafuramidine and furamidine were 1.5- and 4-fold longer, respectively, than those measured from the 24-h observed in vivo data; the t1/2, terminal of furamidine was ∼3-fold longer than that of pafuramidine (64 versus 19 h) (Fig. 4A). Similar to rats, the semi-PBPK model for human disposition predicted that the liver and kidney are the major organs for furamidine accumulation. Furamidine distribution between tissues (liver and kidney) and plasma reached equilibrium after ∼36 h, as reflected by the parallel terminal plasma concentration- and tissue mass-time profiles (Fig. 4, A and B). Furamidine was eliminated in humans primarily by biliary excretion; renal excretion was negligible (Table 3; Fig. 4B).
Fig. 4.
Disposition of pafuramidine (♦) and furamidine (▴) in healthy male subjects administered a single oral dose of [14C]pafuramidine maleate [nominal dose 131.9 mg, equivalent to 100 mg (274 μmol) pafuramidine free base] in capsule form. A, comparison of observed (symbols) and semi-PBPK model-predicted (scheme depicted in Fig. 1B) (lines) plasma concentration-time profiles of pafuramidine and furamidine over 24 h (inset) and 240 h. Symbols and error bars denote mean values and SD, respectively, of six subjects. B, semi-PBPK model-predicted (scheme depicted in Fig. 1B) amount versus time profiles of furamidine in liver/kidney (solid lines) and bile/urine (dashed lines) over 240 h.
Prediction of Dose-Exposure Relationship.
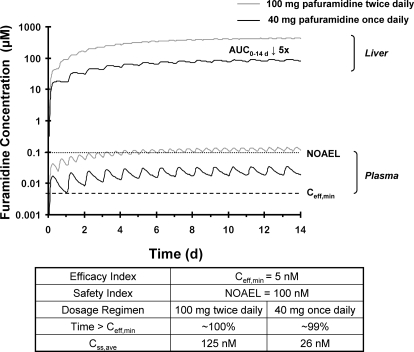
The human semi-PBPK model (Fig. 1B) was used to simulate plasma and liver time profiles associated with various pafuramidine oral dosing regimens. On the basis of the predefined hypothetical efficacy and safety indices, a regimen (40 mg once daily for 14 days) that maintained furamidine plasma concentrations above the Ceff, min for ∼99% of the time throughout the 14 days was selected (Fig. 5). On the basis of the predicted plasma t1/2, terminal of furamidine, plasma concentrations of furamidine reached steady-state at approximately 12 to 13 days; Css, ave was approximately 4-fold higher than the Ceff, min and was 4-fold lower than the hypothetical NOAEL (Fig. 5). Predicted furamidine liver exposure, expressed as the area under the liver concentration-time curve (AUC0–14 d), was 5-fold lower with this alternate dosage regimen compared with the dosage regimen administered in the expanded phase I safety study (100 mg twice daily for 14 days) (Fig. 5); a similar trend also was observed with furamidine kidney exposure between the two dosage regimens (data not shown).
Fig. 5.
Semi-PBPK model-predicted (scheme depicted in Fig. 1B) plasma and liver concentration-time profiles of furamidine in humans after oral administration of 100 mg of pafuramidine free base twice daily (gray lines) and 40 mg of pafuramidine free base once daily (black lines). Dotted and dashed lines represent safety and efficacy indices defined by a hypothetic NOAEL and a minimum effective concentration (Ceff, min) of furamidine in plasma, respectively. NOAEL and Ceff, min were determined as described under Materials and Methods. Time > Ceff, min denotes the time that furamidine concentrations in plasma were above the Ceff, min as a percentage of the entire dosing period (14 days), Css, ave denotes average plasma concentration of furamidine at steady state, and “AUC0–14 d ↓ 5x” denotes liver exposure of furamidine, expressed as the area under the liver concentration-time curve (AUC0–14 d), which was 5-fold lower with the modified pafuramidine dosage regimen (40 mg once daily) compared with the dosage regimen administered in the expanded phase I safety study (100 mg twice daily). The fold-difference in furamidine exposure in the kidney between the two dosage regimens was similar to that in the liver. The kidney profile is not shown because of near overlap with the liver profile, reducing visual clarity. Note that furamidine plasma concentrations under the modified dosage regimen reach a plateau in approximately 12 to 13 days; the time to reach steady state appears shorter because of the log scale applied to the y-axis.
Discussion
Accurate prediction of the pharmacokinetics of generated active metabolites in humans is essential in the selection of appropriate prodrug doses before clinical trials. Pafuramidine is a prodrug of the antiparasitic agent, furamidine, which demonstrated efficacy in animal models of first-stage human African trypanosomiasis (Mdachi et al., 2009; Wenzler et al., 2009). Dose selection for antiparasitic drug development traditionally has relied on achieving efficacious plasma drug concentrations in humans that correlate with those in animal models. A similar strategy was applied to pafuramidine, only plasma concentrations of furamidine were correlated between species. The current work represents the first attempt to optimize this prodrug dose-selection strategy using whole-body semi-PBPK models for rats and humans to predict plasma and tissue exposure and excretion profiles of the active metabolite. Preclinical and clinical data generated during pafuramidine development were used as a training set to demonstrate the utility of PBPK modeling to examine the relationship between pafuramidine dose and furamidine plasma/tissue exposure, which could be used to guide clinical dose-ranging studies of next-in-class compounds.
The final model structure and pafuramidine absorption characteristics in rats were used initially to predict pafuramidine and furamidine disposition in humans. However, the model prediction underestimated the Cmax and Tmax of pafuramidine and furamidine, which possibly could be attributed to differences in dosage formulation (suspension versus capsule) or concomitant diet (standard versus high-fat meal) between rats and humans. Either of these factors could increase the Cmax and Tmax of pafuramidine/furamidine in humans. As such, transit compartments were added incrementally to the pafuramidine human submodel; three transit compartments best described pafuramidine absorption. Pafuramidine is a Biopharmaceutics Classification System class II compound (Zhou et al., 2002); the high-fat meal was administered intentionally to facilitate pafuramidine absorption (Wu and Benet, 2005). This food effect was substantiated by the enhanced fa estimated from the human compared with that from the rat semi-PBPK model (Table 1). These observations emphasized that formulation/diet may be a significant determinant of pafuramidine/furamidine disposition.
Tissue-to-plasma partitioning (Kp) is another factor that influences the disposition of compounds in the body. The in vivo Kp values for pafuramidine and furamidine were not available for human tissues. Unbound Kp values in rats and humans were assumed to be similar (Kp, u, rat = Kp, u, human) (Arundel, 1997). Because no significant species difference in plasma binding of pafuramidine/furamidine was observed (Table 1), Kp values derived from the rat model were applied to describe the distribution of pafuramidine/furamidine in humans.
Extrapolation of metabolite kinetics from animals to humans remains a major challenge in PBPK modeling. Disposition of furamidine in the liver involves formation from pafuramidine, hepatocellular binding, basolateral efflux/reuptake, and biliary excretion. Empirical allometric scaling and physiologically based direct scaling were not used because of 1) marked species differences in pafuramidine metabolism (Table 2), 2) physiologically based direct scaling using rat SCH underestimated metabolic Cl of pafuramidine in rat IPLs, and 3) in vitro biliary Cl of furamidine was too small to measure in rat and human SCH (Table 2) (Yan et al., 2011). Normalized scaling via integration of IPL and SCH data was used in the current work, because this method successfully predicted hepatic metabolic Cl of 10 extensively metabolized drugs in humans (Lave et al., 1997). In addition, incorporation of Cl, predicted by normalized scaling using conventionally cultured hepatocytes, into the PBPK model of an antimalarial drug, epiroprim, provided more accurate predictions of epiroprim disposition in humans (Luttringer et al., 2003). SCH, rather than conventionally cultured hepatocytes, were used in the current study because of their ability to characterize both sinusoidal/biliary transport and metabolism (Swift et al., 2010). The human semi-PBPK model, based on these scaled parameters, adequately described pafuramidine/furamidine concentration-time profiles in human plasma and predicted the dose-plasma/tissue exposure relationship and excretion profiles (Fig. 4). To the authors' knowledge, the current work represents the first effort to extrapolate metabolism/transport clearance from rats to humans for a prodrug/active metabolite pair.
Previous studies in rats and monkeys indicated that pafuramidine has a low oral bioavailability (10–20%) (Midgley et al., 2007), suggesting that pafuramidine could undergo extensive first-pass biotransformation in the gut, as well as in the liver. One report examined pafuramidine metabolism in the gut; the intrinsic formation clearance of the first intermediate metabolite (M1) from pafuramidine in human intestinal microsomes was at least 10-fold lower than that in human liver microsomes (Wang et al., 2007). As such, the liver was assumed initially to be the sole site of furamidine formation. However, in vivo studies showed a near-simultaneous appearance of furamidine with pafuramidine in plasma in both rats and humans (Figs. 3A and 4A, insets). Initial model predictions showed a marked delay in the appearance of furamidine relative to pafuramidine. This discrepancy suggested that furamidine may be formed during pafuramidine absorption through the gut before entering the liver. Bioconversion of pafuramidine to furamidine involves sequential oxidative and reductive reactions mediated by cytochrome P450 enzymes and cytochrome b5/NADH cytochrome b5 reductases (Saulter et al., 2005; Wang et al., 2006). These enzymes are expressed in both the liver and gut. Previous studies demonstrated that CYP4F, a major catalyst of M1 formation, represented a significant portion of the human intestinal P450 “pie” (Wang et al., 2007). These observations prompted incorporation of a gut compartment, representing furamidine formation during pafuramidine absorption, in the furamidine rat/human submodel (Fig. 1B). The model described furamidine plasma disposition adequately (Figs. 3A and 4A, insets). Concordant with in vivo observations, the model predicted that, once absorbed, pafuramidine was converted efficiently to furamidine in rats and humans, as reflected by nearly 50% conversion from pafuramidine; the gut contributed approximately 30 to 40% to furamidine formation in both species (Table 3). These data suggested that the gut contributes significantly to furamidine formation after pafuramidine administration and substantiated the value of PBPK modeling to uncover potentially important biological determinants of drug disposition. In addition to formulation/diet differences, the delayed appearance of furamidine in humans may also reveal species differences in gut metabolism/transport. Further studies are warranted to confirm that the gut is a major presystemic site of furamidine formation after oral administration of pafuramidine and to elucidate species differences in furamidine disposition in the gut.
In rats and humans administered a single oral dose of pafuramidine, the t1/2, app of furamidine was approximately 4 and 14.5 h, respectively, similar to that of pafuramidine (Figs. 3A and 4A, insets). However, 1 week after [14C]pafuramidine administration, a considerable amount of radioactivity was retained in rats (Midgley et al., 2007) and humans, largely as furamidine. According to pharmacokinetic principles, the t1/2, terminal of a drug in plasma will equal that in the tissues once the drug reaches distributional equilibrium. If furamidine is assumed to comply with classic pharmacokinetic behavior at the pafuramidine doses examined, the observed plasma profile of furamidine (Figs. 3A and 4A, insets) may represent the distribution, rather than terminal, phase. Because of LC-MS/MS assay sensitivity limitations in the current study, furamidine was below the limit of quantification in plasma beyond 8 and 24 h after pafuramidine administration to rats and humans, respectively. In the absence of a more sensitive assay, the “true” t1/2, terminal of furamidine was predicted using semi-PBPK modeling. The model predicted that furamidine t1/2, terminal in rats was approximately 40-fold longer than t1/2, app (7 days versus 4 h). After intravenous administration of preformed furamidine (10 μmol/kg) to rats, furamidine was detected in the kidney for up to 16 days. The corresponding kidney t1/2, terminal was estimated to be 7 days (Goldsmith, 2011). The human plasma and tissue t1/2, terminal of furamidine was predicted to be 64 h (∼2.5 days), demonstrating the utility of PBPK modeling to estimate long-term plasma and tissue exposure, which may not be possible to measure directly in vivo because of analytical sensitivity and/or inaccessibility to sampling sites such as the liver/kidney.
A semi-PBPK modeling approach was used in the current work to examine the relationship between dose and plasma/tissue exposure for an antiparasitic prodrug/active metabolite pair in humans. The model predicted that the Css, ave of the active metabolite, furamidine, was 25-fold higher than the estimated Ceff, min in plasma (Fig. 5) based on the dose administered (100 mg twice daily) in the expanded phase I trial. This clinical dosage regimen triggered elevated liver transaminases in 25% of the subjects (Paine et al., 2010). An alternate dosage regimen (40 mg once daily) was predicted to maintain furamidine plasma concentrations half-way between the predefined hypothetical efficacy and safety indices ∼99% of the time throughout the entire 14-day dosing period (Fig. 5), while reducing furamidine hepatic exposure (Fig. 5). Model predictions suggested that if a patient were to inadvertently miss or double the projected dose, only modest fluctuations in plasma furamidine concentrations within the efficacy safety range would result. Next-in-class compounds in development for both stages of HAT are under investigation (Wenzler et al., 2009). This semi-PBPK modeling-based approach, which requires in vitro/in vivo data on metabolism, transport, and plasma/tissue binding, as well as estimated/known efficacy (e.g., Ceff, min) and safety (e.g., NOAEL) indices, could be applied to next-in-class compounds to predict plasma/tissue disposition and guide dose-ranging studies in humans.
Acknowledgments
We thank Dr. Gary M. Pollack for helpful discussions.
Appendix
Model Parameter Abbreviations
- Doral
oral dose
- fa
fraction absorbed
- Atissue/compartment
amount in whole organ or compartment
- AA
amount in arterial blood
- CA
arterial blood concentration
- CV
venous blood concentration
- CVP
venous plasma concentration
- B/P
blood/plasma ratio
- CVtissue
vascular tissue concentration
- Ctissue
tissue concentration
- CO
cardiac output
- Qtissue
tissue blood flow
- Vtissue
tissue volume
- Kp, tissue
tissue-to-plasma partition coefficient
- kP_12, kP_23, kP_3g
first-order rate constants for pafuramidine distribution along the transit compartments of gut
- ka_P, ka_F
first-order rate constants for the movement from gut to liver of pafuramidine and formed furamidine, respectively
- kG_P→F
first-order rate constant for metabolic conversion from pafuramidine to furamidine in the gut
- fu, L, fu, p
unbound fractions in liver and plasma, respectively
- ClL_P→F, u
hepatic unbound intrinsic clearance for the formation of furamidine from pafuramidine
- ClL_P→M, u
hepatic unbound intrinsic clearance for metabolic conversion from pafuramidine to other metabolites
- ClF_up, u
unbound intrinsic clearance for hepatic basolateral uptake of furamidine
- ClF_eff, u
unbound intrinsic clearance for hepatic basolateral efflux of furamidine
- ClF_bile, u
unbound intrinsic clearance for biliary excretion of furamidine
- ClF_renal, u
unbound renal clearance of furamidine from plasma
- Rabs_P
rate of pafuramidine movement from gut to liver
- Rabs_F
rate of intestinally formed furamidine movement from gut to liver
- Rgut formation_F
rate of furamidine formation in the gut
- Rliver formation_F
rate of furamidine formation in the liver
- Rliver formation_M
rate of other metabolite formation in the liver
- Rrenal_F
rate of furamidine renal excretion
- Rbile_F
rate of furamidine biliary excretion
Note: The volume of tissue and vascular blood in the liver represents 95 and 5%, respectively, of total liver volume (Nong et al., 2008). P, pafuramidine; F, furamidine; M, other metabolites.
Human Semi-PBPK Model Equations
Pafuramidine
Disposition in the gut
Amount in transit compartment 1 (T1): Single dose: dAT1/dt = −(kp_12 × AT1) + fa × Doral (t = 0); Multiple dose: dAT1/dt = −(kp_12 × AT1) + fa × Doral (at the beginning of each dosing interval)
Amount in transit compartment 2 (T2): dAT2/dt = (kp_12 × AT1) − (kp_23 × AT2)
Amount in transit compartment 3 (T3): dAT3/dt = (kp_23 × AT2) − (kp_3g × AT3)
Amount in the gut: dAgut_P/dt = (kp_3g × AT3) − Rabs_P − Rgut formation_F Rabs_P = ka_P × Agut_P Rgut formation_F = kG_P→F × Agut_P
Disposition in the liver (flow-limited)
dAliver_P/dt = Qliver × (CAP − CVliver_P) − Rliver formation_F − Rliver formation_M + Rabs_P
Cliver_P = Aliver_P/Vliver
CVliver_P = Cliver_P/Kp_liver_P
Rliver formation_F = Cliver_P × fu,L_P × ClL_P→F, u
Rliver formation_M = Cliver_P × fu,L_P × ClL_P→M, u
Disposition in the fat (flow-limited)
dAfat_P/dt = Qfat × (CAP − CVfat_P)
Cfat_P = Afat_P/Vfat
CVfat_P = Cfat_P/Kp_fat_P
Disposition in the rest of body (rest) (flow-limited)
dArest_P/dt = Qrest × (CAP − CVrest_P)
Crest_P = Arest_P/Vrest
CVrest_P = Crest_P/Kp_rest_P
Blood and plasma concentration
Venous blood concentration: CVP = (Qliver × CVliver_P + Qfat × CVfat_P + Qrest × CVrest_P)/CO
Venous plasma concentration: CVPP = CVP/(B/PP)
Arterial blood concentration: dAAP/dt = QC × (CVP − CAP) CAP = AAP/Vblood
Furamidine
Disposition in the gut
dAgut_F/dt = Rgut formation_F − Rabs_F
Rabs_F = ka_F × Agut_F
Disposition in the liver (diffusion-limited)
Liver blood: dAVliver_F/dt = Qliver × (CAF − CVliver_F) + Rabs_P − Ruptake_F + Refflux_F CVliver_F = AVliver_F/(0.05 × Vliver)
Liver tissue: dAliver_F/dt = Rliver formation_F + Ruptake_F − Refflux_F − Rbile_F Cliver_F = Aliver_F/(0.95 × Vliver) Ruptake_F = CVliver_F × fu,p_F × ClF_up, u Refflux_F = Cliver_F × fu,L_F × ClF_eff, u Rbile_F = Cliver_F × fu,L_F × ClF_bile, u
Disposition in the kidney (flow-limited)
dAkidney_F/dt = Qkidney × (CAF − CVkidney_F)
Ckidney_F = Akidney_F/Vkidney
CVkidney_F = Ckidney_F/Kp,kidney_F
Disposition in the rest of body (rest) (flow-limited)
dArest_F/dt = Qrest × (CAF − CVrest_F)
Crest_F = Arest_F/Vrest
CVrest_F = Crest_F/Kp,rest_F
Blood and plasma concentration
Venous blood concentration: CVF = (Qliver × CVliver_F + Qkidney × CVkidney_F + Qrest × CVrest_F)/CO
Venous plasma concentration: CVPF = CVF/(B/PF)
Arterial blood concentration: dAAF/dt = QC × (CVF − CAF) − Rrenal_F Rrenal_F = CAF × fu,p_F × ClF_renal, u CAF = AAF/Vblood
Urine
dAurine_F/dt = Rrenal_F
Feces
dAfeces_F/dt = Rbile_F
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [R01-GM41935, R25-GM74088]; and the Consortium for Parasitic Drug Development. G.Z.Y. was supported by an Eli Lilly Predoctoral Fellowship in Pharmacokinetics and Drug Disposition.
Dr. Kim Brouwer is a cofounder and Chair of the Scientific Advisory Board for Qualyst, Inc., which has exclusively licensed the sandwich-cultured hepatocytes technology for quantification of biliary excretion (B-CLEAR®).
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
- HAT
- human African trypanosomiasis
- PBPK
- physiologically based pharmacokinetic
- semi-PBPK
- semiphysiologically based pharmacokinetic
- IPL
- isolated perfused liver
- SCH
- sandwich-cultured hepatocyte(s)
- HBSS
- Hanks' balanced salt solution
- PBS
- phosphate-buffered saline
- LC
- liquid chromatography
- MS/MS
- tandem mass spectrometry
- B/P
- blood/plasma
- TFA
- trifluoroacetic acid
- ECG
- electrocardiogram
- HPLC
- high-performance liquid chromatography
- Cl
- clearance
- AUC
- area under the concentration-time curve
- GFR
- glomerular filtration rate
- NOAEL
- no observable adverse effect level.
Authorship Contributions
Participated in research design: Yan, Hall, Tidwell, Brouwer, and Paine.
Conducted experiments: Yan, Generaux, and Goldsmith.
Contributed new reagents or analytic tools: Yan, Yoon, Clewell, Brouwer, and Paine.
Wrote or contributed to the writing of the manuscript: Yan, Olson, Brouwer, and Paine.
References
- Andersen ME, Reddy MB, Plotzke KP. (2008) Are highly lipophilic volatile compounds expected to bioaccumulate with repeated exposures? Toxicol Lett 179:85–92 [DOI] [PubMed] [Google Scholar]
- Arundel PH. (1997) A multi-compartmental model generally applicable to physiologically-based pharmacokinetics. 3rd IFAC Symposium: Modelling and Control in Biomedical Systems; 1997 23–26 March; University of Warwick, Coventry UK: AstraZeneca, London, UK [Google Scholar]
- Barrett MP. (2010) Potential new drugs for human African trypanosomiasis: some progress at last. Curr Opin Infect Dis 23:603–608 [DOI] [PubMed] [Google Scholar]
- Berry LM, Roberts J, Be X, Zhao Z, Lin MH. (2010) Prediction of Vss from in vitro tissue-binding studies. Drug Metab Dispos 38:115–121 [DOI] [PubMed] [Google Scholar]
- Boykin DW, Kumar A, Hall JE, Bender BC, Tidwell RR. (1996) Anti-Pneumocystis carinii activity of bis-amidoximes and bis-O-alkylamidoxime prodrugs. Bioorg Med Chem 6:3017–3020 [Google Scholar]
- Brown RP, Delp MD, Lindstedt SL, Rhomberg LR, Beliles RP. (1997) Physiological parameter values for physiologically based pharmacokinetic models. Toxicol Ind Health 13:407–484 [DOI] [PubMed] [Google Scholar]
- Generaux CN. (2010) Effects of Parasitic Infection on the Pharmacokinetics and Disposition of Pentamidine Analogs. Ph.D. thesis, UNC Eshelman School of Pharmacy, University of North Carolina, Chapel Hill, NC [Google Scholar]
- Goldsmith RB. (2011) Detection and Mechanistic Comparison of Two Anti-Trypanosomal Diamidines in a Rat Renal Model. Ph.D. thesis, School of Medicine, University of North Carolina, Chapel Hill, NC [Google Scholar]
- Ismail MA, Batista-Parra A, Miao Y, Wilson WD, Wenzler T, Brun R, Boykin DW. (2005) Dicationic near-linear biphenyl benzimidazole derivatives as DNA-targeted antiprotozoal agents. Bioorg Med Chem 13:6718–6726 [DOI] [PubMed] [Google Scholar]
- Ito K, Houston JB. (2004) Comparison of the use of liver models for predicting drug clearance using in vitro kinetic data from hepatic microsomes and isolated hepatocytes. Pharm Res 21:785–792 [DOI] [PubMed] [Google Scholar]
- Ito K, Houston JB. (2005) Prediction of human drug clearance from in vitro and preclinical data using physiologically based and empirical approaches. Pharm Res 22:103–112 [DOI] [PubMed] [Google Scholar]
- Lalonde RL, Kowalski KG, Hutmacher MM, Ewy W, Nichols DJ, Milligan PA, Corrigan BW, Lockwood PA, Marshall SA, Benincosa LJ, et al. (2007) Model-based drug development. Clin Pharmacol Ther 82:21–32 [DOI] [PubMed] [Google Scholar]
- Lave T, Dupin S, Schmitt C, Chou RC, Jaeck D, Coassolo P. (1997) Integration of in vitro data into allometric scaling to predict hepatic metabolic clearance in man: application to 10 extensively metabolized drugs. J Pharm Sci 86:584–590 [DOI] [PubMed] [Google Scholar]
- Lavé T, Portmann R, Schenker G, Gianni A, Guenzi A, Girometta MA, Schmitt M. (1999) Interspecies pharmacokinetic comparisons and allometric scaling of napsagatran, a low molecular weight thrombin inhibitor. J Pharm Pharmacol 51:85–91 [DOI] [PubMed] [Google Scholar]
- Lin JH. (1998) Applications and limitations of interspecies scaling and in vitro extrapolation in pharmacokinetics. Drug Metab Dispos 26:1202–1212 [PubMed] [Google Scholar]
- Luttringer O, Theil FP, Poulin P, Schmitt-Hoffmann AH, Guentert TW, Lavé T. (2003) Physiologically based pharmacokinetic (PBPK) modeling of disposition of epiroprim in humans. J Pharm Sci 92:1990–2007 [DOI] [PubMed] [Google Scholar]
- Mdachi RE, Thuita JK, Kagira JM, Ngotho JM, Murilla GA, Ndung'u JM, Tidwell RR, Hall JE, Brun R. (2009) Efficacy of the novel diamidine compound 2,5-bis(4-amidinophenyl)-furan-bis-O-methlylamidoxime (Pafuramidine, DB289) against Trypanosoma brucei rhodesiense infection in vervet monkeys after oral administration. Antimicrob Agents Chemother 53:953–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midgley I, Fitzpatrick K, Taylor LM, Houchen TL, Henderson SJ, Wright SJ, Cybulski ZR, John BA, McBurney A, Boykin DW, et al. (2007) Pharmacokinetics and metabolism of the prodrug DB289 (2,5-bis[4-(N-methoxyamidino)phenyl]furan monomaleate) in rat and monkey and its conversion to the antiprotozoal/antifungal drug DB75 (2,5-bis(4-guanylphenyl)furan dihydrochloride). Drug Metab Dispos 35:955–967 [DOI] [PubMed] [Google Scholar]
- Nong A, Tan YM, Krolski ME, Wang J, Lunchick C, Conolly RB, Clewell HJ., 3rd (2008) Bayesian calibration of a physiologically based pharmacokinetic/pharmacodynamic model of carbaryl cholinesterase inhibition. J Toxicol Environ Health A 71:1363–1381 [DOI] [PubMed] [Google Scholar]
- Paine MF, Wang MZ, Generaux CN, Boykin DW, Wilson WD, De Koning HP, Olson CA, Pohlig G, Burri C, Brun R, et al. (2010) Diamidines for human African trypanosomiasis. Curr Opin Investig Drugs 11:876–883 [PubMed] [Google Scholar]
- Pang KS, Morris ME, Sun H. (2008) Formed and preformed metabolites: facts and comparisons. J Pharm Pharmacol 60:1247–1275 [DOI] [PubMed] [Google Scholar]
- Rowland M, Balant L, Peck C. (2004) Physiologically based pharmacokinetics in drug development and regulatory science: a workshop report (Georgetown University, Washington, DC, May 29–30, 2002). AAPS Pharm Sci 6:E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulter JY, Kurian JR, Trepanier LA, Tidwell RR, Bridges AS, Boykin DW, Stephens CE, Anbazhagan M, Hall JE. (2005) Unusual dehydroxylation of antimicrobial amidoxime prodrugs by cytochrome b5 and NADH cytochrome b5 reductase. Drug Metab Dispos 33:1886–1893 [DOI] [PubMed] [Google Scholar]
- Swift B, Pfeifer ND, Brouwer KL. (2010) Sandwich-cultured hepatocytes: an in vitro model to evaluate hepatobiliary transporter-based drug interactions and hepatotoxicity. Drug Metab Rev 42:446–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turncliff RZ, Hoffmaster KA, Kalvass JC, Pollack GM, Brouwer KL. (2006) Hepatobiliary disposition of a drug/metabolite pair: comprehensive pharmacokinetic modeling in sandwich-cultured rat hepatocytes. J Pharmacol Exp Ther 318:881–889 [DOI] [PubMed] [Google Scholar]
- Wang MZ, Saulter JY, Usuki E, Cheung YL, Hall M, Bridges AS, Loewen G, Parkinson OT, Stephens CE, Allen JL, et al. (2006) CYP4F enzymes are the major enzymes in human liver microsomes that catalyze the O-demethylation of the antiparasitic prodrug DB289 [2,5-bis(4-amidinophenyl)furan-bis-O-methylamidoxime]. Drug Metab Dispos 34:1985–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MZ, Wu JQ, Bridges AS, Zeldin DC, Kornbluth S, Tidwell RR, Hall JE, Paine MF. (2007) Human enteric microsomal CYP4F enzymes O-demethylate the antiparasitic prodrug pafuramidine. Drug Metab Dispos 35:2067–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzler T, Boykin DW, Ismail MA, Hall JE, Tidwell RR, Brun R. (2009) New treatment option for second-stage African sleeping sickness: in vitro and in vivo efficacy of aza analogs of DB289. Antimicrob Agents Chemother 53:4185–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CY, Benet LZ. (2005) Predicting drug disposition via application of BCS: transport/absorption/ elimination interplay and development of a biopharmaceutics drug disposition classification system. Pharm Res 22:11–23 [DOI] [PubMed] [Google Scholar]
- Yan GZ, Brouwer KL, Pollack GM, Wang MZ, Tidwell RR, Hall JE, Paine MF. (2011) Mechanisms underlying differences in systemic exposure of structurally similar active metabolites: comparison of two preclinical hepatic models. J Pharmacol Exp Ther 337:503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Lee K, Thakker DR, Boykin DW, Tidwell RR, Hall JE. (2002) Enhanced permeability of the antimicrobial agent 2,5-bis(4-amidinophenyl)furan across Caco-2 cell monolayers via its methylamidoxime prodrug. Pharm Res 19:1689–1695 [DOI] [PubMed] [Google Scholar]
- Zhou L, Thakker DR, Voyksner RD, Anbazhagan M, Boykin DW, Hall JE, Tidwell RR. (2004) Metabolites of an orally active antimicrobial prodrug, 2,5-bis(4-amidinophenyl)furan-bis-O-methylamidoxime, identified by liquid chromatography/tandem mass spectrometry. J Mass Spectrom 39:351–360 [DOI] [PubMed] [Google Scholar]