Abstract
The analysis of protein-protein-interactions is a key focus of proteomics efforts. The yeast two-hybrid system has been the most commonly used method in genome-wide searches for protein interaction partners. However, the throughput of the current yeast two-hybrid array approach is hampered by the involvement of the time-consuming LacZ assay and/or the incompatibility of liquid handling automation due to the requirement for selection of colonies/diploids on agar plates. To facilitate large-scale yeast two-hybrid assays, we report a novel array approach by coupling a GFP reporter based yeast two-hybrid system with high throughput flow cytometry that enables the processing of a 96 well plate in as little as 3 minutes. In this approach, the yEGFP reporter has been established in both AH109 (MATa) and Y187 (MATα) reporter cells. It not only allows the generation of two copies of GFP reporter genes in diploid cells, but also allows the convenient determination of self-activators generated from both bait and prey constructs by flow cytometry. We demonstrate a Y2H array assay procedure that is carried out completely in liquid media in 96-well plates by mating bait and prey cells in liquid YPD media, selecting the diploids containing positive interaction pairs in selective media and analyzing the GFP reporter directly by flow cytometry. We have evaluated this flow cytometry based array procedure by showing that the interaction of the positive control pair P53/T is able to be reproducibly detected at 72 hrs post-mating compared to the negative control pairs. We conclude that our flow cytometry based yeast two-hybrid approach is robust, convenient, quantitative, and is amenable to large-scale analysis using liquid-handling automation.
Key terms: HT flow cytometry, Protein-protein interaction, Yeast two-hybrid system, Array approach
Introduction
The identification and characterization of protein-protein interactions (PPIs) is a key topic in post-genome research. The yeast two-hybrid system (Y2H) is the most commonly used method for large-scale, high throughput (HT) identification of PPIs and has contributed the majority of the currently available data (1–6). The Y2H system, developed two decades ago, is based on the split and reconstitution of a transcription factor, such as yeast Gal4p (6). Two proteins, X and Y, one fused with the Gal4p DNA binding domain (BD-X), often called “Bait”, the other protein fused with the Gal4p activation domain (AD-Y), often called “Prey”, are co-expressed in yeast cells. The interaction between proteins X and Y brings the Gal4p BD and AD together to form a functional transcription factor and subsequently activates reporter gene expression, such as His3, URA3, LacZ, yEGFP and SdHA (6–12).
There are basically two Y2H approaches: library screen and array approaches (including both matrix and mass mating approaches) (4,5). Fishing in cDNA libraries has been the typical use of the Y2H system in academic labs aiming at the isolation of binding partners for a protein of interest. The standard time-consuming procedure, the complexity of the cDNA in the library and the cost of DNA sequencing for clone identification have limited its application in the current high-throughput, large-scale Y2H campaign (5). Therefore, the matrix approach, i.e., a one-toone test of a binary protein interaction has been employed. This pair-wise strategy requires cloning all individual cDNAs into the Y2H bait and prey vectors and arraying in 96-well or 384-well microtiter plates. This approach is likely to be more comprehensive and more sensitive than the library approach because the identity of each clone is defined, each cDNA is equally tested against the bait and the weak interactions can be more easily distinguished from the background (5). The matrix one-to-one testing approach affords the most definitive detection strategy.
However, for large sets of bait and prey, each individual bait has to be mated with all the prey plates, requiring a large number of time-consuming and labor-intensive manipulations, such as mating (13). To reduce the labor, pooling strategies, which pool multiple strains of bait or prey together followed by “mass mating” have been developed for large-scale purposes (13,14). Due to the requirement of a large number of time-consuming manipulations in the array approach, automation thus becomes the key to perform a large-scale Y2H assay. The array approach is often performed on agar plates by mating haploid cells on rich media plates and scoring the positive PPIs on selective plates (15–18). Using the matchmaker Y2H system as an example, Rajagopala and Uetz (18) summarized the high throughput Y2H array protocol as follows: 1) mating haploid cells on rich media plate; 2) selecting the diploid cells on plates lacking leucine and tryptophan; 3) determining the positive PPIs by scoring the His3 reporter on plates lacking histidine (Fig. 1A). This protocol is applicable to the other Y2H system using different selection markers and reporters.
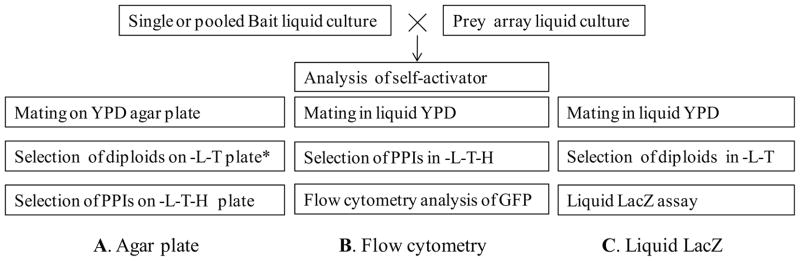
Fig. 1.
Schematic diagram of the flow cytometry based array approach (B) and the comparison with the current agar plate based (A) and liquid LacZ based (C) approaches. –L-T-H: yeast synthetic media without leucine (L), tryptophan (T) and histidine (H). * This step is optional in high throughput screens.
While this approach has been automated, it remains difficult because of the requirement for colony handling and replication. To avoid colony handling, an automated Y2H array approach in a liquid format was developed with the whole procedure performed in liquid media, including mating in YPD media, selection of diploid cells in SD-T-L media and measuring the LacZ reporter in a microtiter plate (Fig.1C) (19). However, measuring LacZ in the liquid format is time-consuming and cumbersome when processing large numbers of samples because of the requirement for lysing cells and adding substrates (19–22).
In this study, we sought to employ HT flow cytometry for large-scale analysis of PPIs through the use of the GFP reporter based Y2H system which we recently established (7). Flow cytometry is a versatile high speed cell analysis method that is playing key roles in proteomics and systems biology efforts (23). HT flow cytometry, such as HyperCyt®, enables the processing of a 96- and 384-well plate in as little as 3 and 12 min, respectively. It is therefore well-suited for large-scale cell screening and selection applications (24–27). To explore its utility for large-scale PPI analysis, we demonstrated an automated high throughput Y2H array procedure that was carried out in 96-well plate by mating yeast cells in YPD media, enriching positive PPIs in selective media and analyzing the GFP reporter by HyperCyt® flow cytometry (Fig. 1B).
Materials and Methods
Strains, Plasmids, and Molecular Cloning
Plasmid construction and molecular cloning were performed in the cloning host cell E.coli DH5a (Invitrogen) following standard protocols. The Y2H kit ‘‘Matchmaker system’’ was purchased from Clontech (San Jose, CA). The kit includes the bait vector pGBKT7 (containing the GAL4p DNA binding domain, BD) and the prey vector pGADT7 (containing the GAL4p activation domain, AD) along with the negative interaction control pair, pGBKT7-Lam/pGADT7-T (Lam/T) and positive interaction control pair, pGBKT7-P53/pGADT7-T (P53/T). PGBKT7-Lam encodes the Gal4p BD fused with Lamin. PGBKT7-53 encodes the Gal4p BD fused with murine p53. PGADT7-T encodes the Gal4 AD fused with the SV40 large T-antigen. The PCL1 plasmid (which encodes the full length Gal4p, served as the strongest positive control) as well as the AH109 and the Y187 reporter strains are also included in the kit. PGBKT7-VP16 (BD-VP16) was made by cloning the VP16 activation domain into the EcoRI/SalI sites of the pGBKT7 vector as a positive control for the self-activators. The AH109-yEGFP reporter strain was constructed as described (7). The Y187-yEGFP reporter strain was constructed as follows: the GAL1 promoter was amplified from the pYM_N22 plasmid (EUROSCARF) with OJC-201 (5’-AGgtcgacGAGCTCTAGTACGGATTAG-3’) and OJC-202 (5’-gggttaattaaATCCGGGGTTTTTTCTCCTT-3’) primers. The amplified GAL1 promoter fragment was inserted into the SalI/PacI sites of PKT127 plasmid (EUROSCARF) to make the GAL1-PKT127 plasmid which contains the yEGFP reporter driven by the Gal1 promoter. Two primers OJC-221 (5’-aagcttttgaccaggttattataaaagaaacttcatgctcgaaaaagCGAGCTCTAGTACGGATTAG-3’) and OJC-197 (5’-tttatattatttgctgtgcaagtatatcaataaacttatatattaTTAGAAAAACTCATCGAGCA-3’) were used to amplify the cassette containing the Gal1 promoter-yEGFP-Km from the GAL1-PKT127 plasmid. The resulting PCR products were purified and transformed into Y187 cells by the standard LiAC/PEG method. The selection of G418 resistant transformants was performed on YPD media supplemented with 400 μg/ml G418. After 3 days, 3 colonies were picked and transformed with the Lam/T, P53/T and PCL1 plasmid to evaluate GFP expression. The strains with the lowest GFP signal for the Lam/T pair and the highest signal for the P53/T pair or PCL1 were chosen for future use. Chromosomal integration of the yEGFP gene was confirmed by PCR and DNA sequencing.
Preparation of the bait and prey stock plates
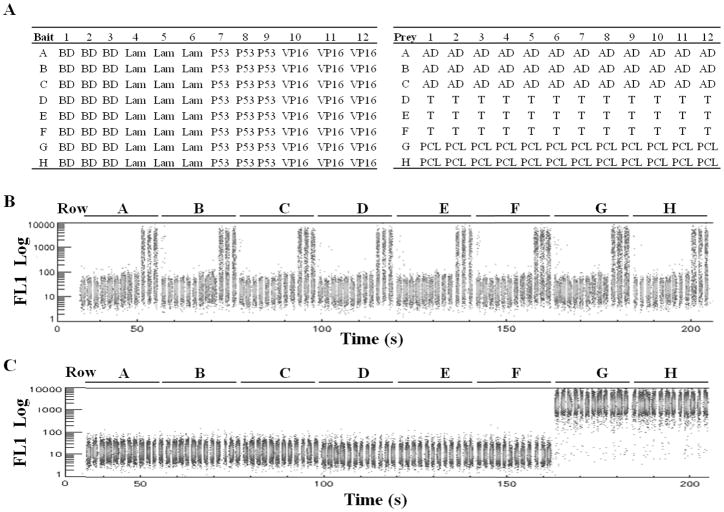
The bait constructs, BD, BD-Lam, BD-P53 and BD-VP16 were transformed into the AH109-yEGFP reporter cells by the standard LiAc method (28) and the transformants were selected on SD-T (Synthetic media lacking Tryptophan) plates. Colonies were streaked once and grown in SD-T liquid media overnight. Glycerol was added to the cultures (20% final concentration) and arrayed in a 96-well plate called the “bait plate” as follows: 200 μl AH109-yEGFP strains bearing BD, BD-Lam, BD-P53 and BD-VP16 bait constructs were added respectively into each well of 3 separate columns of the bait plate (BD: column 1–3, BD-Lam: column 4–6, etc). The prey constructs, AD, AD-T, and PCL1 were transformed into the Y187-yEGFP reporter cells and the transformants were selected on SD-L (Synthetic media lacking Leucine) plates. Colonies were streaked once and grown in SD-L liquid media overnight. Glycerol was added to the cultures to a final concentration of 20% and arrayed in a 96-well prey plate as follows: 200 μl Y187-yEGFP strain bearing AD and AD-T prey constructs were added respectively into each well of 3 rows of the prey plate while the last two rows were arrayed with the Y187-yEGFP bearing PCL1 plasmid. The bait and array plates were frozen at −80°C. The plate maps are shown in Figure 3.
Fig. 3.
Determination of the self-activators by flow cytometry. A) The plate map for AH109-yEGFP cells bearing various bait constructs (bait plate) are shown in the left panel. The plate map for the Y187-yEGFP cells bearing prey constructs (prey plate) are shown in the right panel. B, C) Flow cytometry analysis of the GFP signal (FL1 Log) in each well of bait (B) and prey (c) plates. 2 μl cells from the stock bait and prey plates were transferred to 100 μl SD-T and SD-L media respectively to grow over night. PBS/BSA buffer was added to the each well and measured by HyperCyt flow cytometry row by row. The GFP signal (FL1 Log) of the whole plates (96 samples resolved by time) is shown.
Flow cytometry Array approach for analysis of protein-protein interaction
The bait and prey plates were thawed and 2 μl cells were transferred into new 96-well plates filled with 200 μl SD-T and SD-L respectively to grow overnight to 0.6–1.5 OD595. 50 μl bait cells and 50 μl prey cells (~105 cells) were mated for 18–20 hrs in a 96-well deep well plate filled with 100 μl 2xYPD media supplemented with 50 μg/ml kanamycin (to prevent bacterial contamination). The rest of cells in the bait and prey plates were added to 100 μl PBS/BSA solution and the GFP signal was analyzed by HyperCyt/Cyan HT flow cytometry at UNMCMD to determine the self-activators. After mating, the deep-well plates were spun at 1000 xg for 5 minutes and the supernatants were poured off. The wells were washed with 200 μl SD-T-L-H media (synthetic media lacking Tryptophan, Leucine and Histidine) once and resuspended in 1 ml SD-T-L-H supplemented with 50 μg/ml kanamycin to select the positive interaction pairs. 100 μl cells from wells A2, A5, A7, A11, E2, E5, E7, E11, H2, H5, H7, and H11 were spread to SD-T-L plates to determine the mating efficiency for each bait/prey combination. The mating efficiency was determined by the number of colonies grown on the SD-T-L plate divided by the number of bait or prey cells (whichever is less) used for mating. At 0, 24, 30, 48, 56, 72, and 96 hrs post-mating, 100 μl cells from each well were transferred to a clear round bottom 96-well plate to measure OD595 for scoring the His3 reporter. Subsequently, 100 μl PBS with 0.1%BSA was added to the same plate and the GFP signal was analyzed by HyperCyt® flow cytometry.
HT flow cytometry analysis of yeast cells
A HT flow cytometry platform containing HyperCyt® auto sampler (IntelliCyt) and CyAn ADP cytometer (Beckman Coulter) was used to analyze GFP expression in yeast cells grown in 96-well plates (29). Briefly, 100 μl cells were transferred to a new 96-well plate and 100 μl PBS/BSA buffer was added to each well by Nanoquot liquid dispenser (Biotek). The plate was mixed and read by HyperCyt® flow cytometry. 1,000–5,000 events were sampled from each well by using the following settings on the Cyan cytometer: FSC (Peak, Gain 2.5), SSC (Log, 360V), FL1 (Log, 570V). FITC MESF beads were run daily for calibration. HyperView software was used for data analysis. The mean GFP fluorescence and the percentage of positive cells in each well were calculated.
Measurement of OD595
100 μl cells were transferred to a clear round bottom 96-well plate and measured by plate reader (Biotek) for absorbance at 595 nm.
Results
Establishment of GFP reporters in both mating type reporter yeast cells: AH109 and Y187 strain
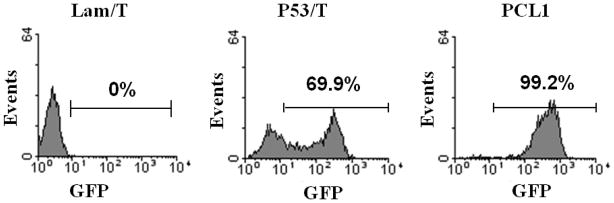
We are using AH109 (MATa) and Y187 (MATα) Y2H reporter strains for mating in both the array and library screen approaches. The bait construct is often transformed in AH109 cells and the prey constructs or cDNA library is transformed in Y187 cells. We previously established the AH109-yEGFP reporter strain in which the GFP reporter was demonstrated to have equal or greater sensitivity than the His3 and LacZ/MEL1 reporters (7). In order to generate two copies of GFP reporters in the mated diploid cells, we established a novel Y187-yEGFP reporter strain by integrating the yEGFP reporter at the ADE2 locus in Y187 cells using PCR based gene replacement. The Y187-yEGFP reporter strain was evaluated with Lam/T (negative control pair), P53/T (positive control pair) and PCL1 (the strongest positive control, encoding Gal4p). Both the P53/T interaction pair and the full-length Gal4p generated significant GFP expression in the Y187-yEGFP strain compared to the negative control Lam/T pair (Fig. 2). The Y187-yEGFP strain bearing the P53/T pair showed 69.9% GFP positive cells with signal/background ratio of ~10, which is very similar to the AH109-yEGFP strain bearing the P53/T pair (7). Gal4p generated ~100% GFP positive cells with signal/background ratio of ~100. These data indicate that the yEGFP reporter has been successfully established in the Y187 reporter cells. Thus, the chromosomally integrated yEGFP reporter in both the AH109 and Y187 strains allows the generation of two copies of the GFP reporter gene in mated diploid cells, which may increase the detection sensitivity. Moreover, it allows the easy determination of the self-activators originating from both bait and prey constructs (described below).
Fig. 2.
Flow cytometry analysis of chromosomally integrated yEGFP reporter fluorescence in the Y187 strain. Y187-yEGFP cells were transformed with several protein pairs and the colonies grown on SD-L-T plates were suspended in PBS buffer and used for GFP fluorescence analysis by flow cytometry. The percentage of positive GFP cells triggered by the P53/T and BD/PCL1 constructs were shown by using the Lam/T pair as the negative control.
Analysis of self-activating bait and prey constructs by HT flow cytometry
Self-activating constructs refer to the bait or prey constructs in Y2H assays that activate Y2H reporters on their own without the occurrence of protein-protein interactions. This is observed more often for bait constructs and occasionally for prey constructs. For example, eukaryotic transcription factors, when fused with BD (bait constructs), often self-activate Y2H reporters. Other BD-fused proteins with features that can induce transcription in yeast also activate Y2H reporters (13). These self-activators in Y2H assay are often required to be determined prior to the HT screen and removed from the bait pools or prey arrays. We tested whether the flow cytometry based approach can be readily applied to determine the self-activators in a high throughput manner. Two self-activators, AH109-yEGFP bearing the BD-VP16 plasmid encoding the fusion protein of the Gal4BD and the VP16AD as well as Y187-yEGFP bearing the PCL1 plasmid were arrayed respectively in the bait and prey plate as shown in Fig. 3A. The GFP signal of cells in each well of the bait and prey plates was measured by HT flow cytometry (Fig. 3B, C). Compared to the wells containing the non-self-activating baits, such as BD, BD-Lam and BD-P53, all 24 wells in columns 10, 11, and 12 containing BD-VP16 showed clear GFP signal (Mean GFP fluorescence = 635.4±21.5) with 33.5% GFP positive cells (33.5±0.6). Compared to the wells containing non self-activating prey constructs, all 24 wells in row G and row H containing PCL1 triggered extremely high GFP expression (Mean GFP fluorescence = 1962.9±93.2) with almost 100% GFP positive cells (99.8±0.1). This indicates that the GFP reporter in both strains is suitable for detection of self-activators in a high throughput manner. Therefore, this HT flow cytometry based Y2H system affords unique advantages for analysis of self-activators.
Evaluation of an integrated liquid handing array approach for analysis of PPIs
To eliminate the agar plating and the lacZ assay procedures in the current Y2H array approach, we evaluated an integrated liquid handling Y2H array approach carried out in a 96-well plate by employing HT flow cytometry analysis of the yEGFP reporter (Fig. 1B).
To mimic the standard Y2H array approach, we made frozen stocks of the bait and prey cells in 96-well plates. The bait plate was arrayed with the AH109-yEGFP cells bearing the bait constructs BD, BD-Lam, BD-T and BD-VP16. The prey plate was arrayed with the Y187-yEGFP cells bearing the prey constructs AD, AD-T and PCL1. Plate maps are shown in Fig. 3. The cells in each well of the bait plate were mated with cells in the corresponding well of the prey plate (for example, bait A1 mated with prey A1). This mating strategy generated 6–9 replicates of 12 bait/prey pairs including: 1) 9 replicates of one positive control pair, P53/T; 2) 9 replicates of five negative control pairs, BD/AD, BD/T, Lam/AD, Lam/T and P53/AD and 3) 6–9 replicates of the self-activators, BD-VP16 or PCL1 pairing with any other constructs (in rows G and H and columns 10–12).
Next, we optimized the mating conditions in 96-well deep well plates as follows: 1) the bait and prey cells were grown to 0.6–1.2 OD595 prior to mating; 2) 50 μl bait cells (~105 cells) and 50 μl prey cells were mated in 100 μl 2xYPD media for 18–20 hrs. Under these conditions, the cell density of the post-mating culture is ~10 7 cells/ml. The mating efficiency of 12 wells representing the 12 bait/prey pairs was determined as follows: 1) negative controls: 0.48% (BD/AD, Well A2), 0.47% (Lam/AD, well A5), 0.26% (P53/AD, well A7), 0.44% (BD/T, well E2) and 0.62% (Lam/T, well, E5); 2) positive control: 0.23% (P53/T, well E7); 3) self-activators: 0.13% (BD-VP16/AD, well A11), 0.10% (BD-VP16/T, well, E11), 0.28% (BD-VP16/PCL1, well H11), 0.53% (BD/PCL1, H2), 0.55% (Lam/PCL1, well H5), and 0.25% (P53/PCL1, well H7). This indicates that the mating efficiency in each well is between 0.1–1%, and the mating efficiency of the positive control P53/T is equal or lower than the negative controls.
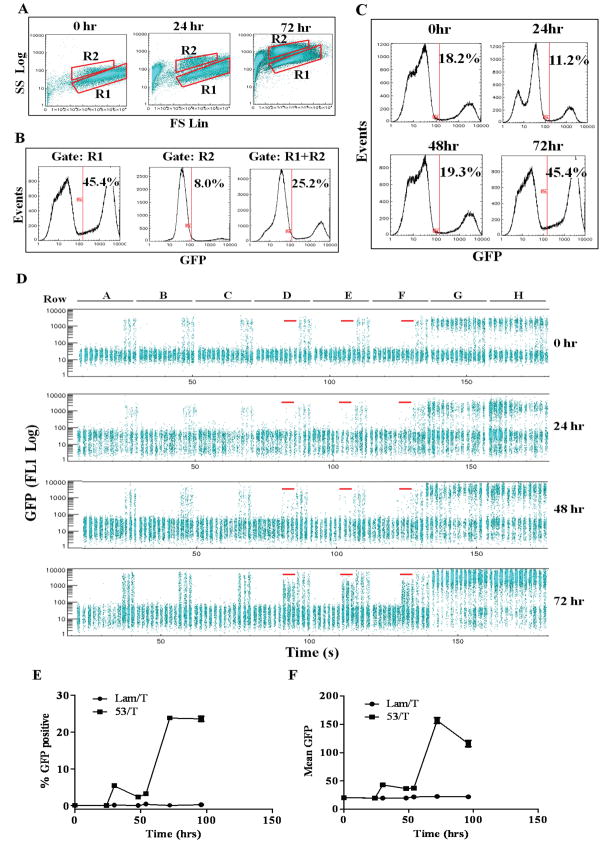
Then, we evaluated the GFP reporter and determined the optimal assay time for identification of P53/T interaction by measuring the GFP signal at 0, 24, 30, 48, 54, 72, and 96 hrs post-mating (Fig. 4). We first determined the appropriate region (gate) for data analysis by displaying the cell populations from the whole 96-well plate in a forward scatter versus side scatter dot plot at 0, 24 and 72 hrs time points (Fig. 4A). At the zero time point, ~95% cells were in the R1 region. A distinct yeast cell population in the R2 region with similar FSC but larger SSC values appeared after 24 hrs. The percentage of cells in the R2 region reached ~20% at 24 hrs and increased to ~50% at 72 hrs and 96 hrs (Fig. 4A and data not shown). To determine if the R2 region needs to be included for data analysis, we compared the GFP histogram of the cells in the R1, R2 and R1 plus R2 regions from the whole 96-well plate at 72 hrs (Fig. 4B). 45.4% cells were GFP positive in the R1 region whereas only 8.0% cells were GFP positive in the R2 region. The overall percentage of GFP positive cells decreased to 25.2%. These data indicate that cells in the R2 region contain a minor portion of GFP positive cells. We also displayed the GFP histogram of the R1 and R2 regions in individual wells at 72 hrs. For example, in well 43 containing the P53/T pair, the percentages of GFP positive cells were 23%, 7% and 16% in the R1, R2 and R1 plus R2 regions, respectively. In well 90 containing the Lam/PCL1 pair, the percentages of GFP positive cells were 91%, 26% and 60% in the R1, R2 and R1 plus R2 regions, respectively (data not shown). These results also showed that the R2 region contains much fewer GFP positive cells than the R1 region. Cells in the R2 region were likely to be dying haploid or diploid cells that couldn’t grow in the SD-T-L-H media. This agrees with the observation that cells in the R2 region showed similar FSC but larger SSC as well as higher background FL1 fluorescence compared to cells in the R1 region (Fig. 4A, 72hrs). Therefore, only cells in the R1 region were used in the subsequent data analysis. We then evaluated GFP expression at different time points by analyzing the GFP histogram of the cells on the whole plate (Fig. 4C) and by displaying and quantifying the GFP signal (Log FL1) of cells in each well (Fig. 4D–F). Because 51 wells would be anticipated to express GFP in the 96-well plate format, including 42 wells containing the self-activators and 9 wells containing the P53/T positive control, the GFP histogram of the whole plate would define the optimal assay time point. At the zero time point, 18.2% cells from the whole plate were positive (Fig. 4C). These positive cells were contributed by the haploid cells bearing self-activators, AH109-GFP/BD-VP16 (columns 10–12) and Y187-GFP/PCL1 (rows G and H), as shown in Fig. 4D (Fig. 4D, 0 hr). The percentage of GFP positive cells decreased to 11.2% at 24 hrs, indicating that the percentage of GFP positive haploid cells was decreased and that the diploid cells were growing. The percentage of GFP positive cells increased to 19.3% at 48 hrs and reached a plateau of 45.4% at 72 hrs and remained 43.6% at 96 hrs (Fig. 4C and data not shown). These data show that the diploid cells bearing the positive triggers (positive interactions or self-activators) started to overgrow after 48 hrs and dominated after 72 hrs incubation, suggesting that 72 hrs is the optimal time to assay PPIs.
Fig. 4.
Evaluation of the flow cytometry based array approach by analysis of the GFP signal at different time points post-mating. The bait plate and the prey plate were grown over night and mated with each other in YPD media. Positive PPIs were selected in SD-T-L-H media and cells were sampled at 0, 24, 30, 48, 54, 72 and 96 hrs post-mating to measure the GFP signal by HyperCyt/CyAn flow cytometer. (A) FSC/SSC dot plot analysis of two populations of cells in regions R1 and R2 from the whole 96-well plate at 0, 24 and 72 hrs post mating. (B) Histogram analysis of GFP signals of the cells in regions R1, R2 and R1 plus R2 from the whole 96-well plate at 72 hrs post-mating. The percentage of GFP positive cells in each region is shown. (C) Histogram analysis of the GFP signal of the cells in the region R1 at 0, 24, 48, 72 hrs post mating. The percentage of GFP positive cells on the whole plate at each time point is shown. (D) The GFP signal of the cells in each well at 0, 24, 48, 72 hrs post mating are shown in alignment. The 9 wells containing the P53/T mating pairs at each time point are highlighted by red line. (E) The mean value of the percentage of GFP positive cells (mean± SD) in all 9 wells containing the P53/T or Lam/T mating pairs are shown as a function of time. (F) The mean value of the GFP fluorescence of the cells (mean± SD) in all 9 wells containing the P53/T or Lam/T mating pairs are shown as a function of time.
To evaluate the GFP signal in the wells containing positive control P53/T mating, we displayed the FL1 signal of the cells in each well and quantified the GFP signal to compare the P53/T pair and the negative control pairs using Lam/T as an example (Fig. 4D–F). At 0 and 24 hr time points, only the wells containing self-activators (columns 10–12 as well as rows G and H) showed GFP signal due to the GFP positive haploid cells whereas the wells containing all five negative controls and the positive control P53/T did not show any GFP positive cells. A small population of GFP positive cells in the P53/T wells appeared at 48 hrs (Fig. 4D) while a larger population of GFP positive cells was shown clearly in all the P53/T wells at the 72 hr time point and remained at the same level at 96 hrs (Fig. 4D and data not shown). No GFP positive cells were detected in the wells containing 5 negative control pairs (45 wells) at any time points (Fig. 4D). The percentage of GFP positive cells in the wells containing the self-activators BD-VP16 (columns 10–12) or PCL1 (rows G and H) also increased significantly at 72 hrs compared to that at 24–48 hrs (Fig. 4D). These data further confirm the whole-plate histogram analysis (Fig. 4C) and show that 72 hrs post-mating is an optimal time point to detect the P53/T interaction. The percentage of GFP positive cells in the P53/T and Lam/T wells were quantified in Fig. 4E, showing that the percentage of GFP positive cells in the P53/T wells increased to 3–5% at 30–48 hrs and reached a plateau (23.8% ±3.1) at 72 hrs, which remained at the same level (23.5%± 3.4) at 96 hrs. The mean GFP fluorescence in the P53/T wells also reached the peak at 72 hrs (156.4±16.2) and decreased slightly (116±19.2) at 96 hrs (Fig. 4F). Both the percentage of GFP positive cells and the mean GFP fluorescence in the Lam/T wells and the other negative control wells remained at the same low level at different time points (Fig. 4E, 4F and data not shown). It is noteworthy that the mating efficiency of the negative control pairs was equal to or greater than the P53/T pair, ruling out the possibility that the negative results from these wells were due to the failure of mating. These data demonstrate that the interaction of the P53/T pair can be consistently detected in all 9 wells by flow cytometry at 72 hrs post-mating and the longer incubation time (96 hrs) still generated a significant GFP signal. They clearly show that GFP is a robust and reproducible reporter and that the flow cytometry array approach can be utilized for identification of PPIs in a high throughput manner.
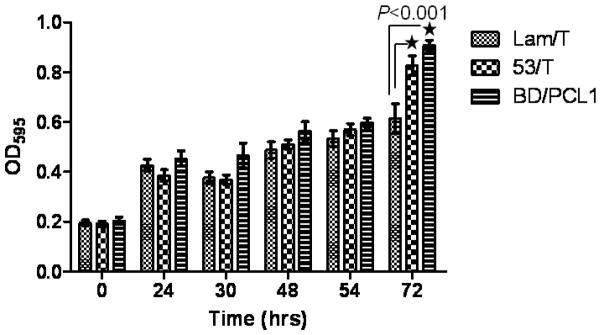
Finally, we evaluated the His3 reporter gene in the liquid array approach. The cell growth in the selective media lacking histidine (SD-T-L-H) was scored by measuring OD595 at different time points. The wells containing PCL1 (0.91 OD) and the P53/T pair (0.83 OD) showed statistically significant faster cell growth than that containing the Lam/T pair (0.61 OD) at 72 hrs post mating, suggesting that His3 could be used as a reporter gene in the array approach (Fig. 5). However, weobserved a significant and continuous cell growth (from 0.2 OD to 0.6 OD) in wells containing the Lam/T pair and other negative controls after mating (Fig. 5 and data not shown). The background cell growth may be due to the leaky expression and moderate stringency of the His3 reporter genes since we removed the YPD media by washing the wells with SD-T-L-H media after mating. This is commonly observed and typically overcome through the use of the His3 competitor 3-aminotriazole (3-AT) in the medium in the plate-based array approach. Addition of an appropriate amount of 3-AT may increase selection stringency in our assay, but the His3 reporter in our approach is not necessary as we are measuring GFP expression in the diploid cells which are selected by the Trp and Leu markers.”
Fig. 5.
Evaluation of the His3 Y2H reporter by measuring cell growth in the selective medium. The bait and prey cells were mated as described in Fig. 4 and the mated cells were grown in SD-T-L-H media. Cells were sampled at 0, 24, 30, 48, 54 and 72 hrs post-mating to measure the optical density at 595 nm by plate reader. Statistical analysis of OD595 of Lam/T wells (n=9), P53/T wells (n=9) and BD/PCL1 wells (n=6) was performed by the unpaired t-test using GraphPad Prism 5.
Discussion
Flow cytometry has been widely used in numerous aspects of biological research because of its unique capability for high content, multiplex and single cell analysis (24,25). We have employed flow cytometry in Y2H assays to improve the automation and throughput of both library screening and array approaches. We created a surface-display reporter in the Y2H system for HT library screening and proposed employing next-generation sequencing technology to determine clone identity (11). The robust flow cytometry based array approach presented in this study adds an alternative method for large-scale analysis of PPIs. The Y2H array approach is currently only used in a limited number of labs due to the unavailability of cDNA clone arrays. However, with the completion of interactome mapping projects in human, worm, yeast and other model organisms (30–33), the array format of cDNA clones in these species would be available and allow academic labs to identify the PPIs from defined arrays in the very near future. The flow cytometry based Y2H array approach demonstrated in this study is amenable to large-scale assays and affords these unique advantages. First, it allows easy determination of self-activators originating from both bait and prey constructs in a HT manner, which is difficult to achieve with the relatively laborious LacZ assay. Furthermore, quantitative analysis of GFP expression by flow cytometry allows the identification of binding partners of weak self-activators. We previously showed that the influenza NS1 protein is a weak self-activator in the Y2H system as evident from the fact that NS1 itself could trigger HIS3, MEL1 and GFP reporter expression (7). However, much stronger GFP expression could be detected by flow cytometry when BD-NS1 was co-expressed with AD-NS1 or AD-NS1BP (NS1BP: a known NS1 binding protein, our unpublished data). This feature allows us to include the self-activators in the array and to identify their binding partners, which is difficult to achieve in the other Y2H system. Thus, our system provides a more convenient and quantitative method than the current standard array approach to analyze the self-activators. Second, it allows easy automation with liquid handling. The whole liquid assay process we demonstrated can be easily automated by a liquid handling robot except for a washing step to replace the YPD media with the selective media, which requires simple manual manipulation. This step can also be automated via the utilization of commercially available plate washers that can aspirate supernatant from plates and wash with buffer. In addition, it has been reported that the 100-fold dilution of YPD media with selective media is able to minimize the effect of YPD (19). This strategy could be applied to our approach. In this study, we intentionally washed the wells to eliminate the effect of YPD media on cell growth in order to evaluate the His3 reporter properly. Third, it allows prompt reading of the plates by flow cytometry without additional treatment, such as lysing cells, and adding substrate required for the LacZ assay in the microtiter plate. Based on our screening experience, we can process ~120 96-well plates (more than 10,000 samples) per cytometer per day. Fourth, the GFP reporter produces much lower background signal than the His3 reporter does, which is evident from the result that the GFP signal remains at a very low level in all 45 wells containing negative control pairs whereas the cells in these wells showed significant growth in media lacking histidine. This makes the GFP reporter more suitable than the His3 reporter in the liquid Y2H array approach. Finally, it allows a large assay time window. The evaluation of the P53/T interaction showed that the optimal assay time point was 72 hrs post-mating and that the longer incubation time (96 hrs) did not affect the GFP signal significantly. The assay time is dependent on the time required for diploid cells containing the positive interaction pairs to overgrow in the culture, which is determined by the mating efficiency and the doubling time of diploid cells. Since the mating efficiency of 0.1% in the microtiter plates is routinely achievable, the assay time is largely dependent on the doubling time of diploid cells. The diploid cells bearing positive interaction pairs generally divide every 3–5 hrs in selective media. We observed that the doubling time for diploid cells bearing the BD/PCL1 or P53/T pairs is approximately 2 hrs and 3 hrs, respectively. Therefore, the diploid cells containing the P53/T pair will dominate the culture after 72 hrs growth. In the worst case, in principle, if 105 bait and prey cells were used to mate and only one diploid cell was produced (where the mating efficiency is 0.001%), and the total cell count in the after-mating culture is ~1 million, the ratio of diploid cells in the post-mating culture is 1/106. If the doubling time is 3–4 hrs, the diploid cell count will be ~1 million (220) after 72 hrs. In addition, the haploid cells may die in the selective media after 72 hrs, and can be gated out by flow cytometry as we observed (Fig. 4A). Thus, after 72 hrs, the diploid cells will dominate the culture and allow detection of the GFP signal by flow cytometry. We therefore believe that 72 hrs growth post-mating may be sufficient for the detection of most PPIs. A longer incubation time, such as 96 hrs, may be used for extremely slowly growing diploid cells. Taken together, these advantages make the flow cytometric GFP array approach a robust and attractive method for large-scale analysis of PPIs.
The multiplexing capability of flow cytometry affords another potentially unique advantage to analyze multiple bait proteins simultaneously. Multiple bait strains can be genetically color-coded with red fluorescent proteins, mixed and distinguished by flow cytometry, allowing the analysis of multiple bait proteins against prey arrays in a pair-wise manner simultaneously. This is difficult to achieve with the other platform. By pooling the same color-coded strains, our approach also allows analysis of multiple bait pools without increasing the assay complexity. We are currently developing strategies to genetically color-code the AH109-yEGFP cells with fluorescent proteins that don’t interfere with the GFP reporter signal.
In summary, this flow cytometry based Y2H array approach affords an alternative method for HT Y2H assays and adds a powerful new assay to the toolbox of flow cytometry-associated proteomics technologies. With the availability of arrayed cDNA libraries, we speculate that this unique flow cytometry based approach will become a very useful tool in large-scale analysis of PPIs, particularly, in the identification of host-pathogen PPIs.
Acknowledgments
We thank members of the University of New Mexico Center for Molecular Discovery and the National Flow Cytometry Resources at Los Alamos National Laboratory for very helpful discussion on this work. This work was supported by NIH grants 2P41RR01315 and 1U54MH084690.
References
- 1.Fields S. High-throughput two-hybrid analysis. The promise and the peril. Febs J. 2005;272:5391–9. doi: 10.1111/j.1742-4658.2005.04973.x. [DOI] [PubMed] [Google Scholar]
- 2.Parrish JR, Gulyas KD, Finley RL., Jr Yeast two-hybrid contributions to interactome mapping. Curr Opin Biotechnol. 2006;17:387–93. doi: 10.1016/j.copbio.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Bruckner A, Polge C, Lentze N, Auerbach D, Schlattner U. Yeast two-hybrid, a powerful tool for systems biology. Int J Mol Sci. 2009;10:2763–88. doi: 10.3390/ijms10062763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walhout AJ, Boulton SJ, Vidal M. Yeast two-hybrid systems and protein interaction mapping projects for yeast and worm. Yeast. 2000;17:88–94. doi: 10.1002/1097-0061(20000630)17:2<88::AID-YEA20>3.0.CO;2-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koegl M, Uetz P. Improving yeast two-hybrid screening systems. Brief Funct Genomic Proteomic. 2007;6:302–12. doi: 10.1093/bfgp/elm035. [DOI] [PubMed] [Google Scholar]
- 6.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–6. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Zhou J, Bae W, Sanders CK, Nolan JP, Cai H. A yEGFP-based reporter system for high-throughput yeast two-hybrid assay by flow cytometry. Cytometry A. 2008;73A:312–20. doi: 10.1002/cyto.a.20525. [DOI] [PubMed] [Google Scholar]
- 8.Serebriiskii IEJ, Berman M, Golemis EA. Approaches to detecting false positives in yeast two-hybrid systems. Biotechniques. 2000;28:328–330. 332–336. doi: 10.2144/00282rr03. [DOI] [PubMed] [Google Scholar]
- 9.Vidal M, Braun P, Chen E, Boeke JD, Harlow E. Genetic characterization of a mammalian protein-protein interaction domain by using a yeast reverse two-hybrid system. Proc Natl Acad Sci U S A. 1996;93:10321–6. doi: 10.1073/pnas.93.19.10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serebriiskii IG, Toby GG, Finley RL, Jr, Golemis EA. Genomic analysis utilizing the yeast two-hybrid system. Methods Mol Biol. 2001;175:415–54. doi: 10.1385/1-59259-235-X:415. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Zhou J, Sanders CK, Nolan JP, Cai H. A surface display yeast two-hybrid screening system for high-throughput protein interactome mapping. Anal Biochem. 2009;390:29–37. doi: 10.1016/j.ab.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Cormack BP, Bertram G, Egerton M, Gow NA, Falkow S, Brown AJ. Yeast-enhanced green fluorescent protein (yEGFP)a reporter of gene expression in Candida albicans. Microbiology. 1997;143 ( Pt 2):303–11. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 13.Zhong J, Zhang H, Stanyon CA, Tromp G, Finley RL., Jr A strategy for constructing large protein interaction maps using the yeast two-hybrid system: regulated expression arrays and two-phase mating. Genome Res. 2003;13:2691–9. doi: 10.1101/gr.1134603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin F, Avramova L, Huang J, Hazbun T. A yeast two-hybrid smart-pool-array system for protein-interaction mapping. Nat Methods. 2007;4:405–7. doi: 10.1038/nmeth1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cagney G, Uetz P, Fields S. High-throughput screening for protein-protein interactions using two-hybrid assay. Methods Enzymol. 2000;328:3–14. doi: 10.1016/s0076-6879(00)28386-9. [DOI] [PubMed] [Google Scholar]
- 16.Walhout AJ, Vidal M. High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods. 2001;24:297–306. doi: 10.1006/meth.2001.1190. [DOI] [PubMed] [Google Scholar]
- 17.Chern M, Richter T, Ronald PC. Yeast two-hybrid approaches to dissecting the plant defense response. Methods Mol Biol. 2007;354:79–83. doi: 10.1385/1-59259-966-4:79. [DOI] [PubMed] [Google Scholar]
- 18.Rajagopala SV, Uetz P. Analysis of protein-protein interactions using array-based yeast two-hybrid screens. Methods Mol Biol. 2009;548:223–45. doi: 10.1007/978-1-59745-540-4_13. [DOI] [PubMed] [Google Scholar]
- 19.Buckholz RG, Simmons CA, Stuart JM, Weiner MP. Automation of yeast two-hybrid screening. J Mol Microbiol Biotechnol. 1999;1:135–40. [PubMed] [Google Scholar]
- 20.Mockli N, Auerbach D. Quantitative beta-galactosidase assay suitable for high-throughput applications in the yeast two-hybrid system. Biotechniques. 2004;36:872–6. doi: 10.2144/04365PT03. [DOI] [PubMed] [Google Scholar]
- 21.Diaz-Camino C, Risseeuw EP, Liu E, Crosby WL. A high-throughput system for two-hybrid screening based on growth curve analysis in microtiter plates. Anal Biochem. 2003;316:171–4. doi: 10.1016/s0003-2697(02)00706-6. [DOI] [PubMed] [Google Scholar]
- 22.Serebriiskii IG, Golemis EA. Uses of lacZ to study gene function: evaluation of beta-galactosidase assays employed in the yeast two-hybrid system. Anal Biochem. 2000;285:1–15. doi: 10.1006/abio.2000.4672. [DOI] [PubMed] [Google Scholar]
- 23.Nolan JP, Yang L. The flow of cytometry into systems biology. Brief Funct Genomic Proteomic. 2007;6:81–90. doi: 10.1093/bfgp/elm011. [DOI] [PubMed] [Google Scholar]
- 24.Sklar LA, Carter MB, Edwards BS. Flow cytometry for drug discovery, receptor pharmacology and high-throughput screening. Curr Opin Pharmacol. 2007;7:527–34. doi: 10.1016/j.coph.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards BS, Oprea T, Prossnitz ER, Sklar LA. Flow cytometry for high-throughput, high-content screening. Curr Opin Chem Biol. 2004;8:392–8. doi: 10.1016/j.cbpa.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Young SM, Bologa C, Prossnitz ER, Oprea TI, Sklar LA, Edwards BS. High-throughput screening with HyperCyt flow cytometry to detect small molecule formylpeptide receptor ligands. J Biomol Screen. 2005;10:374–82. doi: 10.1177/1087057105274532. [DOI] [PubMed] [Google Scholar]
- 27.Black CB, Duensing TD, Trinkle LS, Dunlay RT. Cell-Based Screening Using High- Throughput Flow Cytometry. Assay Drug Dev Technol. 2010 doi: 10.1089/adt.2010.0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:31–4. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 29.Young SM, Curry MS, Ransom JT, Ballesteros JA, Prossnitz ER, Sklar LA, Edwards BS. High-throughput microfluidic mixing and multiparametric cell sorting for bioactive compound screening. J Biomol Screen. 2004;9:103–11. doi: 10.1177/1087057103262335. [DOI] [PubMed] [Google Scholar]
- 30.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–74. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–36. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 32.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–3. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–8. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]