Abstract
Aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor that regulates genes involved in drug/xenobiotic metabolism, cell cycle progression, cell fate determination, immune function, and inflammatory response. Increasing evidence that AHR plays a role in the pathophysiology of a number of human disease states is driving the need for improved pharmacological tools to be used for understanding the in vivo impact of AHR modulation. In this study, we have characterized and used structure-activity relationship analyses of a newly synthesized library of derivatives of the potent AHR antagonist 2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-o-tolylazo-phenyl)-amide (CH223191). Initial screening of these compounds revealed that those bearing groups with strong electronegativity at the R1 position (i.e., CHD-5, CHD-11, and CHD-12) versus those that are more electron-poor at this position (i.e., CHD-7 and CHD-8) elicited the most potent AHR antagonistic properties. The ability of these derivatives to inhibit agonist (2,3,7,8-tetrachlorodibenzo-p-dioxin) binding, nuclear translocation of AHR, and agonist-induced enzyme activity also were determined and support the initial findings. Furthermore, CH223191, but not CHD-5, CHD-11, or CHD-12, was found to exhibit AHR-independent proproliferative properties. These results contribute to our understanding of the structural requirements of potent AHR antagonists and the development of effective pharmacological tools to be used for studying the pathophysiological role of AHR.
Introduction
The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor that is known best historically for regulating cytochrome P450 genes such as CYP1A1, CYP1A2, and CYP1B1 that are involved in drug and xenobiotic metabolism (Okey, 2007; Beischlag et al., 2008). During the past decade, increasing evidence has indicated that AHR also can modulate a variety of cellular and physiological processes that can affect cell proliferation, cell fate determination, cell migration, immune and inflammatory responses, and morphogenesis. The mechanisms by which AHR ligands, both exogenous and endogenous, affect these processes are poorly understood but appear to involve multiple interactions between AHR and other signaling pathways. It also is apparent that modulation of AHR via certain agonists and antagonists likely impinges on the pathophysiology of human disease states, such as cardiovascular diseases (Zhang et al., 2010), diabetes (Kerkvliet et al., 2009), cancer (Dietrich and Kaina, 2010), and blood diseases (Casado et al., 2010). Thus, selective pharmacological tools are needed to identify the various roles of AHR ligands, to assess the impact of AHR modulation in preclinical animal models of human diseases, and to determine whether AHR is an appropriate target for novel therapies.
In its unliganded state, AHR exists as a cytosolic complex (Okey, 2007; Beischlag et al., 2008). Upon agonist binding, AHR translocates to the nucleus, dimerizes with its DNA-binding partner aryl hydrocarbon receptor nuclear translocator, and binds specific DNA recognition sites that alter the transcription of AHR/aryl hydrocarbon receptor nuclear translocator target genes. The most potent AHR agonist is the environmental contaminant 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), which binds AHR with high affinity and specificity. Additional agonists that are characterized poorly as yet with respect to their AHR binding specificities and in vivo characteristics include endogenous chemicals such as 6-formylindolo[3,2-b]carbazole and 2-(1′-H-indole-3′′-carbonyl)-thiazole-4-carboxylic acid methyl ester (Denison and Nagy, 2003). Although the toxicological/adverse effects of exogenous agonists such as TCDD are well described, the physiological effects of these endogenous AHR agonists are poorly understood. Recent clues that they may play important functional roles include observations that 6-formylindolo[3,2-b]carbazole is present within human subjects (Wincent et al., 2009), alters the differentiation of Th17 cells in vitro (Veldhoen et al., 2009), and exacerbates the severity of experimental autoimmune encephalomyelitis (Veldhoen et al., 2008).
With respect to AHR antagonists, the most potent include two flavonoid compounds, 3′-methoxy-4′-nitroflavone (MNF) (Lu et al., 1995; Gasiewicz et al., 1996) and 6,2′,4′-trimethoxyflavone (TMF) (Murray et al., 2010a), and 2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-o-tolylazo-phenyl)-amide (CH223191) (Kim et al., 2006). Although MNF exhibits acceptable antagonistic properties, its use is confounded by its partial agonistic activities and rapid in vivo metabolism. It is noteworthy that the ability of MNF to exert an agonistic versus antagonistic AHR response is species dependent and involves specific ligand/amino acid interactions that occur within the AHR ligand binding domain (Henry and Gasiewicz, 2008). A similar flavone AHR antagonist, TMF, appears to act in a species-independent manner and does not appear to exhibit partial agonistic properties (Murray et al., 2010a). CH223191 was identified as an AHR antagonist using a chemical library screen (Kim et al., 2006). The evidence available thus far indicates that CH223191 is a potent, pure AHR antagonist, does not exhibit AHR agonistic activities, and competitively binds AHR, thereby inhibiting AHR transformation (Kim et al., 2006; Zhao et al., 2010).
We sought to further our understanding of how the AHR antagonist CH223191 interacts with AHR with the goal of developing more potent and pharmacologically effective AHR antagonists than CH223191. With this in mind, we synthesized a chemical library composed of CH223191 derivatives and examined their abilities to interact with AHR and block the agonistic activities of TCDD. Our results have identified key structural features of the R1 position in the CH223191 molecule that play important roles in altering binding to AHR. The novel CH223191 derivatives characterized in this study add additional members to the diverse AHR ligand family that vary in their structures and pharmacological properties and provide additional tools to be used for exploring the potential therapeutic benefits of AHR-based drug therapies.
Materials and Methods
Materials.
The synthesis and initial screening of the 50 compounds representing structural modifications of CH223191 are described elsewhere (H. Lee, E.-Y. Choi, H. Swanson, and K. B. Kim, submitted for publication). TCDD was a generous gift from Dr. Stephen H. Safe (Texas A&M University, College Station, TX). [3H]TCDD was obtained from Chemsyn (Lenexa, KS). Unless otherwise mentioned, all of the other chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Cell Culture.
Immortalized murine hepatocytes established from either AHR(−/−) or AHR(+/+) mice were obtained from Dr. Gary Perdew (Pennsylvania State University, University Park, PA) (Murray et al., 2005). The hepatocytes were cultured in α-minimal essential medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.1 μM dexamethasone. Human hepatoma (HepG2) cells that were transfected stably with the CYP1A1 promoter upstream of the luciferase reporter gene (HepG2-p450luc) were as described previously (Heid et al., 2001). T84 cells were cultured in Dulbecco's modified Eagle's medium/Ham's F-12 medium (Lonza, Basel, Switzerland) supplemented with 5% fetal bovine serum and 1× antibiotic and antimycotic mix (Invitrogen). All of the other cell lines were maintained in Dulbecco's modified Eagle's medium with glucose and glutamine (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (Invitrogen) at 37°C and 5% CO2.
Luciferase Reporter Assays.
HepG2-p450luc cells were aliquoted into 96-well plates and cultured until approximately 70% confluent. The cells then were pretreated for 1 h with the indicated chemicals at the indicated concentrations before the administration of either dimethyl sulfoxide (DMSO) (0.1%) or TCDD (1 nM). After incubation for 4 h, the cells were harvested, and the luminescence values were obtained according to the manufacturer's protocol (Promega, Madison, WI) using a microplate luminometer (Applied Biosystems, Foster City, CA).
Cell Viability Analyses.
HepG2 cells (3000 cells/well), AHR(+/+) and AHR(−/−) hepatocytes (1500 cells/well), and Hepa1 cells (2000 cells/well) were aliquoted into 96-well plates. After 24 h, the indicated chemicals were added, and cell viability was determined after an additional 24, 48, or 72 h using the WST-1 assay (Roche Applied Science, Indianapolis, IN).
Proliferation/Apoptosis Analyses.
HepG2 cells were aliquoted into a 96-well plate (3000 cells/well) and after 24 h were treated with either DMSO (0.1%), 3′,4′-dimethoxyflavone (DMF) (5 μM), CH223191 (10 μM), or the indicated CH223191 derivatives (10 μM). After incubation for 72 h, 5-bromo-2′-deoxyuridine (BrdU) was added, allowed to incorporate for 6 h, and analyzed as described in the manufacturer's protocol (Roche Applied Science). The luminescence values were obtained using a Microplate Luminometer (Applied Biosystems; Invitrogen).
For the analyses of apoptosis, caspase 3/7 activity was measured using the Caspase-Glo 3/7 reagent according to the manufacturer's protocol (Promega). In brief, HepG2 cells were aliquoted into a 96-well plate (3000 cells/well) and after 24 h were treated with either DMSO (0.1%), DMF (5 μM), CH223191 (10 μM), or the indicated CH223191 derivatives (10 μM). After incubation for 72 h, Caspase-Glo 3/7 was added, and the luminescence values were obtained using a Microplate Luminometer (Applied Biosystems; Invitrogen).
Ligand Binding Assays.
Competitive ligand binding assays were performed as described previously (Puppala et al., 2007) using varying concentrations of the indicated competitors. The IC50 values were determined using Prism 5.0 (GraphPad Software Inc., San Diego, CA).
Ethoxyresorufin O-Deethylase Assays.
Ethoxyresorufin O-deethylase (EROD) assays were performed essentially as described previously (Sanderson et al., 1996). In brief, the cells (15,000 cells/well) were plated into 96-well plates. After 24 h, the cells were cultured in the presence of the indicated chemicals and harvested, and fluorescence was determined using a plate-reading spectrofluorometer (HTS 7000 Plus BioAssay Reader; PerkinElmer Life and Analytical Sciences, Waltham, MA).
Western Blot Analyses.
The effect of the CH223191 derivatives on the nuclear translocation of AHR in cultured cells was determined by pretreating Hepa1 cells with the indicated derivatives for 1 h before the administration of either DMSO (0.1%) or TCDD (1 nM). After 1 h, the cells were harvested, and the nuclear extracts were prepared using the protocol described previously (Holden and Tacon, 2011), except that 1 mM rather than 10 mM dithiothreitol was used. The protein concentrations were determined using the BCA Protein Assay (Thermo Fisher Scientific, Rockford, IL). Aliquots of the extracts (20 μg) were loaded on a 7.5% SDS polyacrylamide gel electrophoresis gel. The AHR band was visualized with mouse monoclonal AHR antibody (Abcam Inc., Cambridge, MA), and histone deacetylase 1 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) was used as a loading control.
Results
Initial Observations.
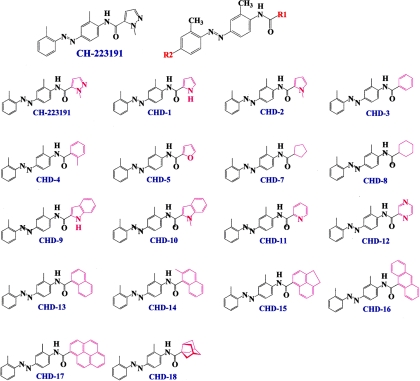
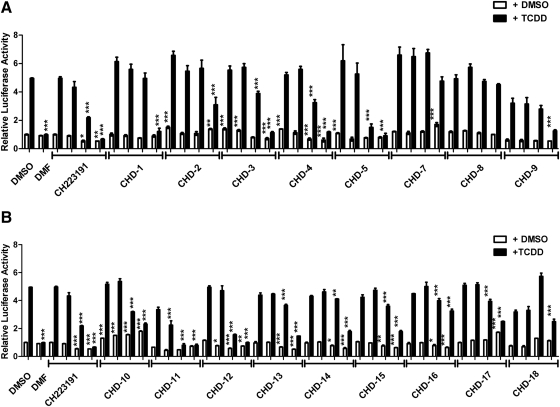
A privileged library consisting of 50 compounds that are structurally similar to CH223191 but vary with respect to their substituents, the sites of substitution, and number of rings was synthesized and subjected to an initial screening (Lee et al., manuscript submitted for publication). The structures of some of these compounds, CHD-1 to CHD-18, are shown in Fig. 1. This initial screening indicated that substitutions at the site depicted as R1 of CH223191 significantly altered the abilities of these compounds to block TCDD's induction of gene transcription. Additional reporter analyses of compounds CHD-1 to CHD-18 were performed using a range of doses (10−8 to 10−5 M) that were administered in the presence of either DMSO or TCDD (Fig. 2). The analyses included the parent compound CH223191 and the previously reported pure AHR antagonist (DMF) (Lee and Safe, 2000) for comparative purposes. Comparison of the characteristics of CHD-2, CHD-5, CHD-11, and CHD-12 versus those of CHD-7, CHD-8, CHD-16, and CHD-17 reveal that the first group contains an electron-rich π system with at least one heteroatom functioning as a Lewis donor. As shown in Fig. 2, CHD-5, CHD-11, and CHD-12 blocked TCDD-induced reporter activity in a dose-dependent manner to an extent that was at least as good as that of the parent CH223191 compound. In contrast, the antagonistic activities of CHD-7, CHD-8, CHD-16, and CHD-17 were among the poorest.
Fig. 1.
Structures of the CH223191 derivatives. Schematic representation of the CH223191 derivatives that vary with respect to their substituents at either the R1 or R2 (not shown) positions and were synthesized and characterized in this study.
Fig. 2.
Reporter analyses of the CH223191 derivatives. A, CHD-1 through CHD-9. B, CHD-10 through CHD-18 HepG2 cells that have been transfected stably with a luciferase reporter regulated by the human CYP1A1 promoter (HepG2-p450luc) were pretreated with either DMSO alone or the indicated compounds (10−5, 10−6, 10−7, or 10−8 M) for 1 h. DMF was used at a concentration of 5 × 10−6 M. After the addition of TCDD (1 nM), the cells were incubated for an additional 4 h and harvested, and luciferase activities were determined. The mean ratios of triplicate wells (±S.D.) are depicted. The results are representative of at least three independent experiments that were subjected to one-way analysis of variance and Tukey's post hoc test analyses. ***, p < 0.001; **, p < 0.01; *, p < 0.05.
A majority of the derivatives, when cultured in the presence of only the DMSO vehicle control, exhibited relatively low AHR agonistic activities. Those exhibiting the most potent, dose-responsive agonistic activities include CHD-7, CHD-10, and CHD-17. However, it should be noted that, even at the highest concentrations used (i.e., 10−5 M), their induction of reporter activity was less than 2-fold that of the DMSO vehicle control. This fold induction is considered to be relatively modest compared with that of TCDD (≥5-fold). With this data, we then selected five derivatives for more in-depth analyses, with CHD-7 and CHD-8 representing derivatives of CH223191 with the lowest and CHD-5, CHD-11, and CHD-12 representing those with the highest AHR antagonistic properties, respectively.
Impact of the CH223191 Derivatives on Cell Viability and Proliferation.
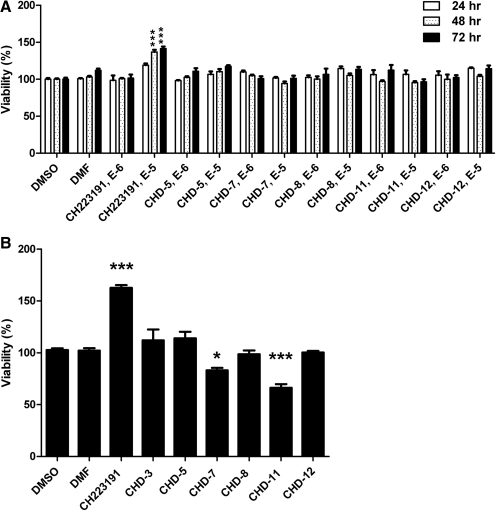
The impact of CH223191 and its derivatives, CHD-5, CHD-7, CHD-11, and CHD-12 on the viability of cells that were representative of human versus murine sources was determined using murine (Hepa1; Fig. 3A) and human (HepG2; Fig. 3B) hepatoma cell lines. In the Hepa1 cells, neither CH223191 nor its derivatives significantly decreased cell viability. In the HepG2 cells, the most significant loss of viability occurred after the treatment with CHD-7 and CHD-11 at the highest concentration (i.e., 10−5 M) and at the longest time point (i.e., 72 h). Because these negative effects on cell viability were not observed in Hepa1 cells, it is likely that the effects of the CH223191 derivatives are species and/or cell line specific. It is noteworthy that treatment of both cell lines with the parent CH223191 compound at the highest concentration (i.e., 10−5 M) increased the number of viable cells, indicating that at this concentration CH223191 may exert proliferative effects.
Fig. 3.
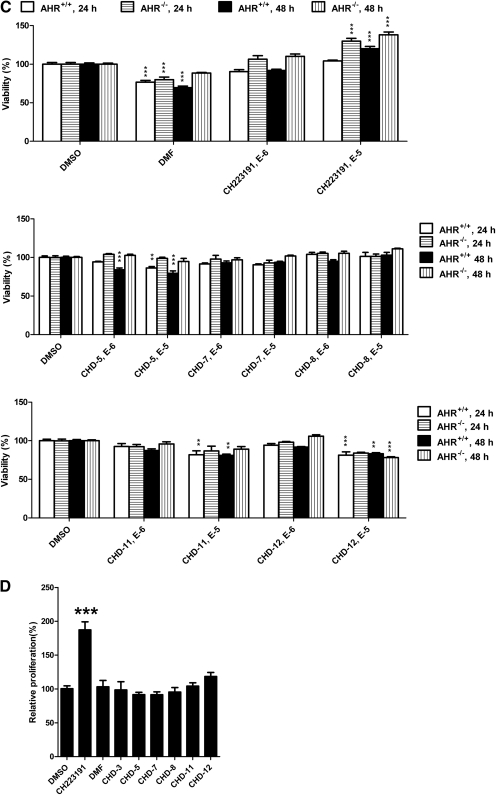
Impact of the CH223191 derivatives on viability and proliferation. A–C, viability analyses of Hepa1 (A), HepG2 (B), and immortalized murine AHR(+/+) versus AHR(−/−) hepatocytes (C). The cultured cells were incubated with the indicated compounds. After either 24, 48, or 72 h, the cells were harvested, and cell numbers were determined using the WST-1 assay. D, proliferation analyses of HepG2 cells. HepG2 cells were incubated with the indicated compounds for 72 h, and proliferation was addressed using the BrdU incorporation assay. The mean ratios of triplicate wells (±S.D.) are depicted. The results are representative of at least three independent experiments that were subjected to one-way analysis of variance and Tukey's post hoc test analyses. ***, p < 0.001; **, p < 0.01; *, p < 0.05.
Immortalized murine hepatocytes that varied with respect to their expression of AHR [i.e., AHR(+/+) and AHR(−/−)] also were used to determine whether the effects of these compounds on cell viability were AHR dependent. As shown in Fig. 3C, the most significant loss in viability in both AHR(+/+) and AHR(−/−) hepatocytes occurred after the treatment with DMF. The absence of AHR expression at least partially alleviated the DMF-induced loss of viability at the 48-h time point. It is noteworthy that treatment with the parent CH223191 (10−5 M) resulted in an increase in the number of viable cells that was accentuated by the lack of AHR expression. With respect to the CH223191 derivatives, the impact on viability was relatively modest. In AHR(+/+) cells, treatment with either CHD-5, CHD-11, or CHD-12 resulted in a maximal loss of 20%. Likewise, in AHR(−/−) cells, the greatest impact was observed after treatment for 48 h with CHD-12 (10−5 M), which resulted in an approximately 22% loss of viability. Only the CHD-5-induced loss of viability was altered (i.e., alleviated) by the presence of AHR.
To determine whether the CH223191-induced increase in the number of viable cells was due to an increase in proliferation, we performed BrdU incorporation analyses in the HepG2 cells (Fig. 3D). As shown, treatment with only CH223191 resulted in an approximately 2-fold increase in BrdU incorporation. Analyses of caspase 3/7 as a measure of apoptosis indicated that treatment with these chemicals did not reduce apoptosis (data not shown). Taken together, these results indicate that CH223191 harbors proproliferative activities and that these activities are not AHR dependent and are not observed with the CH223191 derivatives.
Competitive Ligand Binding Analyses.
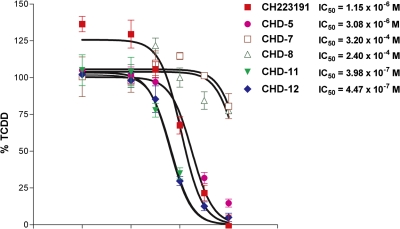
We then examined the relative AHR binding affinities of CHD-5, CHD-7, CHD-8, CHD-11, and CHD-12 using a competitive ligand binding assay. As shown in Fig. 4, the IC50 values determined from CHD-5, CHD-11, and CHD-12 ranged from 0.4 to 3 × 10−6 M and were similar to that of the parent CH223191 (10−6 M). In contrast, the relative AHR binding affinities of CHD-7 and CHD-8 were considerably lower (3.2 × 10−4 and 2.4 × 10−4 M, respectively).
Fig. 4.
CHD-5, CHD-11, and CHD-12, but not CHD-7 and CHD-8, competitively inhibit TCDD binding to AHR. [3H]TCDD, in the absence or presence of varying concentrations of CH223191 and its derivatives (10−10 to 10−5 M) was incubated with protein extracts prepared from Hepa1 cells. After incubation at room temperature for 2 h, nonspecific binding was removed using a 10% dextran-coated charcoal slurry and hydroxyapatite gel. A 200-fold molar excess of 2,3,7,8-tetrachlorodibenzofuran, an analog of TCDD, was used to estimate the nonspecific binding of TCDD. All of the values are expressed as the percentage of the value obtained using TCDD alone. The data are averages ± S.D. of three independent experiments. The IC50 values were determined using GraphPad Prism.
EROD Analyses.
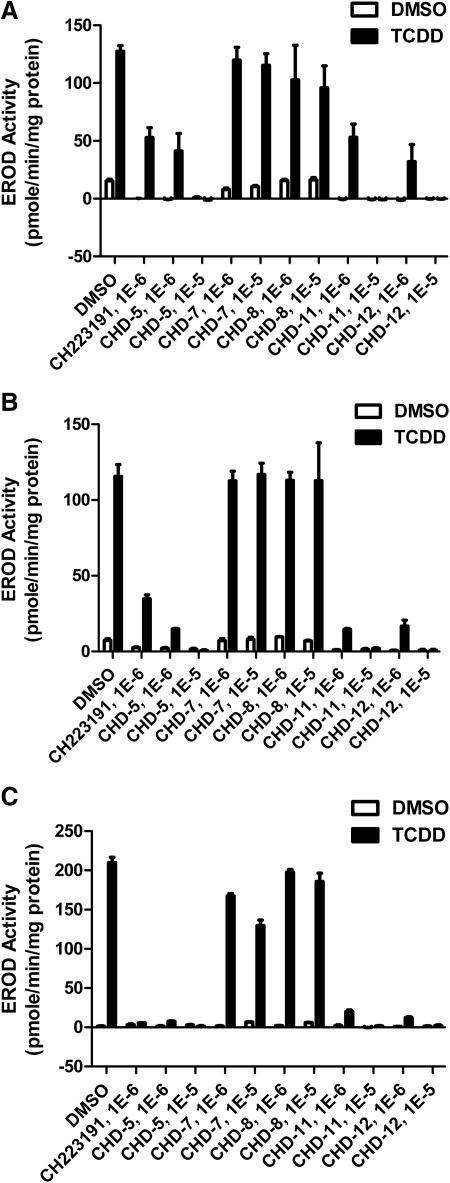
To further characterize the AHR antagonistic activities of the CH223191 derivatives, we cultured human cell lines that were representative of three different tissue types (hepatocytes, HepG2; keratinocytes, HaCaT; and intestinal epithelial, T84) with TCDD in the absence or presence of either CH223191 or the indicated derivatives. The analyses of EROD activity served as a measure of endogenous AHR target gene activity. These data (Fig. 5; Table 1) are consistent with those obtained using the reporter assays and again indicate that CHD-5, CHD-11, and CHD-12 are the most potent and CHD-7 and CHD-8 are the least potent AHR antagonists, respectively. It is noteworthy that the range of potencies appeared to be the most dramatic when the HaCaT and T84 cells were assayed.
Fig. 5.
CHD-5, CHD-11, and CHD-12 inhibit the ability of TCDD to induce CYP1A1 activity in EROD analyses. A–C, either HepG2 (A), HaCaT (B), or T84 (C) cells were aliquoted into 96-well plates. When either 90% (HepG2 and T84) or postconfluent (HaCaT), the indicated chemicals were added. After an additional 16 h of incubation, the cells were harvested, and EROD activities were determined as described under Materials and Methods. The mean ratios of triplicate wells (± S.D.) are depicted. The results are representative of at least three independent experiments. The statistical analyses of these experiments are shown in Table 1.
TABLE 1.
| Antagonist and Concentration | EROD Activity (Percentage of 10 nM TCDD) |
||
|---|---|---|---|
| HaCaT | HepG2 | T84 | |
| % | % | % | |
| CH223191 | |||
| 1 μM | 33 ± 6 | 58 ± 16 | 30 ± 8 |
| CHD-5 | |||
| 1 μM | 27 ± 6 | 58 ± 13 | 29 ± 8 |
| 10 μM | 21 ± 11 | 28 ± 9 | 28 ± 9 |
| CHD-7 | |||
| 1 μM | 104 ± 10*† | 82 ± 9‡ | 94 ± 9*† |
| 10 μM | 96 ± 4 | 98 ± 16‡ | 68 ± 3 |
| CHD-8 | |||
| 1 μM | 113 ± 4*† | 68 ± 7 | 103 + 9*† |
| 10 μM | 109 ± 9 | 79 ± 7 | 87 ± 2*† |
| CHD-11 | |||
| 1 μM | 36 ± 10 | 33 ± 10 | 32 ± 7 |
| 10 μM | 23 ± 3 | 29 ± 9 | 28 ± 9 |
| CHD-12 | |||
| 1 μM | 29 ± 7 | 34 + 8 | 31 ± 8 |
| 10 μM | 21 ± 2 | 29 + 9 | 24 ± 10 |
Statistically different (P < 0.05) from CH223191.
Statistically different (P < 0.05) from CHD-5, CHD-11, and CHD-12 at the corresponding concentrations.
Statistically different (P < 0.05) from CHD-11 and CHD-12 at the corresponding concentrations.
Impact of CH223191 Derivatives on AHR Nuclear Translocation.
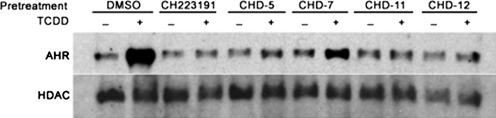
Finally, we determined whether the CH223191 derivatives could effectively block TCDD-induced AHR translocation into the nucleus. Previous studies indicate that CH223191 blocks TCDD-induced AHR nuclear translocation (Zhao et al., 2010). Given this, we pretreated cultured cells with either CH223191 or the indicated derivatives before their incubation with TCDD. The nuclear fractions of these cells then were subjected to Western blot analyses (Fig. 6). As shown, treatment with TCDD but not DMSO enriched the presence of AHR in the nuclear fraction. Pretreatment with CH223191, CHD-5, CHD-11, and CHD-12 but not CHD-7 effectively blocked the TCDD-induced translocation of AHR into the nucleus. These data are consistent with those obtained from the reporter (Fig. 2), ligand binding (Fig. 4), and EROD analyses (Fig. 5) and again indicate that CHD-5, CHD-11, and CHD-12 are the most potent whereas CHD-7 (as well as CHD-8) is the least potent AHR antagonists. The CH223191 derivatives that we synthesized for this study also appear to exert antagonistic activity by the same mechanism as CH223191, that is, blocking nuclear translocation of AHR.
Fig. 6.
CHD-5, CHD-11, and CHD-12, but not CHD-7, inhibit TCDD-induced AHR enrichment in the nuclei. Hepa1 cells were treated with the indicated derivatives (10 μM) for 1 h before the administration of either DMSO (0.1%) or TCDD (1 nM). After incubation for 1 h, the cells were harvested, and the nuclear extracts were prepared and subjected to Western blot analyses. HDAC, histone deacetylase.
Discussion
The work described in this study contributes to our toolbox of diverse AHR ligands that may be used for understanding the physiological and pharmacological properties of AHR. A wide variety of exogenous and endogenous substances has been identified that interact with AHR and display agonistic, partial agonistic, and/or antagonistic activities (Denison and Nagy, 2003). The differing pharmacological impact of these ligands is thought to arise from their distinct interactions with the amino acid residues that reside within the AHR ligand binding pocket and induce subtle changes in receptor conformation. Our current understanding of the molecular events involved in the interactions between the AHR ligand binding pocket and its ligands have been facilitated greatly by studies using site-directed mutagenesis and computational methodologies (Procopio et al., 2002; Henry and Gasiewicz, 2008; Bisson et al., 2009; Pandini et al., 2009; Petkov et al., 2009; Whelan et al., 2010). Within the AHR binding cavity are critical amino acid residues that ensure that the cavity is sufficiently sized to accommodate relatively large molecules such as TCDD. In addition, amino acid residues with aromatic properties are thought to interact with and stabilize the bound ligand, whereas those with relatively high polarity participate in hydrogen bonding and other interactions. The antagonistic versus agonistic properties of these ligands appear to be dictated by the localization of charge within the ligand molecules and the distinct interactions between the ligand and the amino acid residues that lie within the AHR ligand binding pocket (Petkov et al., 2009). For example, with respect to AHR agonists, molecular docking studies (Bisson et al., 2009) have revealed that 6-formylindolo[3,2-b]carbazole can form two hydrogen bonds with glutamine residues, whereas 2-(1′-H-indole-3′′-carbonyl)-thiazole-4-carboxylic acid methyl ester forms a single hydrogen bond with a serine residue. Along the same lines, the recently identified selective AHR modulator 3′,4′-dimethoxy-α-naphthoflavone is thought to form a hydrogen bond with serine residues as well as a novel bond with a threonine residue of AHR (Murray et al., 2010b). Finally, with respect to AHR antagonists, the best characterized are flavones that share an electronegative center that hypothetically can interact with a positively charged residue such as arginine (Gasiewicz et al., 1996; Henry et al., 1999; Henry and Gasiewicz, 2008). It has been hypothesized (Henry and Gasiewicz, 2008) that these positively charged amino acid residues within AHR may constrain the positioning of the flavone antagonists via an interaction with the electronegative center of the antagonist, such that agonist-induced conformational changes are inhibited. Thus, like other nuclear receptors, such as estrogen receptor α and estrogen receptor β (Moore et al., 2010), ligands of AHR contact distinct sets of amino acids within the ligand binding pocket that presumably induce differential receptor conformations and subsequently elicit distinct pharmacological properties.
CH223191 has been described previously both in vitro and in vivo as a pure and potent antagonist of AHR (Kim et al., 2006) that appears to preferentially antagonize halogenated aromatic hydrocarbons (Henry et al., 1999; Zhao et al., 2010. The preferential antagonism of CH223191 has been hypothesized to arise either from its binding outside the AHR ligand binding pocket or via a binding mechanism that allows for a conformational change of the receptor that can exclude the binding of only a subgroup of AHR agonists. The work presented herein furthers our understanding of how CH223191-like antagonists may interact with AHR. Here, we report that, in general, increasing the hydrophobicity at the R1 site of CH223191 results in a corresponding decrease in antagonistic activity toward AHR. For example, CHD-7 and CHD-8, which contain either a cyclopentane or a cyclohexane substitution, were found to exhibit low antagonistic properties (Figs. 1 and 2). A comparison of CHD-7 with CHD-5 indicates that the addition of the reactive oxygen within the ring structure enhances AHR antagonistic activities. Likewise, CHD-3, which harbors a phenyl moiety at the R1 position, exhibits more potent AHR antagonistic activities than CHD-8, where the substituent is the aforementioned cyclohexane. Thus, CHD-5, CHD-11, and CHD-12, which contain a heterocyclic moiety with either an oxygen (CHD-5) or a nitrogen (CHD-11 and CHD-12), appear to exhibit the highest antagonistic properties toward AHR. These findings are consistent with previous studies of other AHR ligands where 1) electron-rich rich substituents within flavone-based AHR antagonists were found to be necessary for optimal AHR ligand binding (Henry et al., 1999) and 2) within the AHR ligand binding pocket polar amino acid residues such as histidine and threonine residues were found to contribute to ligand binding, presumably via a network of weak interactions (Pandini et al., 2009). We hypothesize that electronegative, heterocyclic aromatic substituents at the R1 position of CH223191-based AHR antagonists may be involved in molecular interactions (such as hydrogen bonding or π-π interaction) within the AHR ligand binding pocket in a manner that is similar to that of the nitro substituents of the flavone antagonists.
Although the antagonistic activities of most potent AHR antagonists identified herein (i.e., CHD-5, CHD-11, and CHD-12) appear to be similar to those of the parent compound, they may exert as yet unrealized pharmacokinetic/pharmacodynamic properties that affect their in vivo efficacies. Like the parent CH223191 compound, the three derivatives exhibit minimal agonistic activities. Unlike CH223191, however, they do not appear to exert proproliferative activities (Fig. 3). Although short-term, in vivo treatment with CH223191 did not appear to be toxic (Kim et al., 2006), the proproliferative, AHR-independent activities observed in this study have not been reported previously. DMF, however, has been observed previously to exhibit growth inhibitory properties in breast cancer cells (i.e., MCF7 and T47D), an effect attributed to its either estrogen receptor or protein kinase activities (Lee and Safe, 2000). Our observations that DMF treatment resulted in a loss of viability in both AHR(+/+) and AHR(−/−) cell lines (Fig. 3, C and D) are consistent with these previous findings.
In summary, we have identified and partially characterized a novel group of CH223191-based antagonists. These studies also have lent insights into the molecular requirements of potent AHR antagonists. The compounds characterized within this study, with their unique activities toward AHR, are potentially valuable pharmacological tools to be used to understand the pathophysiological role of AHR in human disease states.
Acknowledgments
We thank Dr. Gary Perdew for the AHR(−/−) cell lines, Dr. Stephen H. Safe for TCDD, and Dr. David Watt for critical review of this manuscript.
This work was supported by the National Institutes of Health National Institute of Environmental Health [Grant R01 ES014849].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.111.073643.
- AHR
- aryl hydrocarbon receptor
- BrdU
- 5-bromo-2′-deoxyuridine
- CH223191
- 2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-o-tolylazo-phenyl)-amide
- DMF
- 3′,4′-dimethoxyflavone
- DMSO
- dimethyl sulfoxide
- EROD
- ethoxyresorufin O-deethylase
- MNF
- 3′-methoxy-4′-nitroflavone
- TCDD
- 2,3,7,8-tetrachlorodibenzo-p-dioxin
- TMF
- 6,2′,4′-trimethoxyflavone.
Authorship Contributions
Participated in research design: Choi, Lee, Dingle, Kim, and Swanson.
Conducted experiments: Choi, Lee, and Swanson.
Contributed new reagents or analytic tools: Lee.
Performed data analysis: Choi and Swanson.
Wrote or contributed to the writing of the manuscript: Choi, Dingle, Kim, and Swanson.
References
- Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. (2008) The aryl hydrocarbon receptor complex and the control of gene expression. Crit Rev Eukaryot Gene Expr 18:207–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson WH, Koch DC, O'Donnell EF, Khalil SM, Kerkvliet NI, Tanguay RL, Abagyan R, Kolluri SK. (2009) Modeling of the aryl hydrocarbon receptor (AhR) ligand binding domain and its utility in virtual ligand screening to predict new AhR ligands. J Med Chem 52:5635–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casado FL, Singh KP, Gasiewicz TA. (2010) The aryl hydrocarbon receptor: regulation of hematopoiesis and involvement in the progression of blood diseases. Blood Cells Mol Dis 44:199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison MS, Nagy SR. (2003) Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol 43:309–334 [DOI] [PubMed] [Google Scholar]
- Dietrich C, Kaina B. (2010) The aryl hydrocarbon receptor (AhR) in the regulation of cell-cell contact and tumor growth. Carcinogenesis 31:1319–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiewicz TA, Kende AS, Rucci G, Whitney B, Willey JJ. (1996) Analysis of structural requirements for Ah receptor antagonist activity: ellipticines, flavones, and related compounds. Biochem Pharmacol 52:1787–1803 [DOI] [PubMed] [Google Scholar]
- Heid SE, Walker MK, Swanson HI. (2001) Correlation of cardiotoxicity mediated by halogenated aromatic hydrocarbons to aryl hydrocarbon receptor activation. Toxicol Sci 61:187–196 [DOI] [PubMed] [Google Scholar]
- Henry EC, Gasiewicz TA. (2008) Molecular determinants of species-specific agonist and antagonist activity of a substituted flavone towards the aryl hydrocarbon receptor. Arch Biochem Biophys 472:77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry EC, Kende AS, Rucci G, Totleben MJ, Willey JJ, Dertinger SD, Pollenz RS, Jones JP, Gasiewicz TA. (1999) Flavone antagonists bind competitively with 2,3,7, 8-tetrachlorodibenzo-p-dioxin (TCDD) to the aryl hydrocarbon receptor but inhibit nuclear uptake and transformation. Mol Pharmacol 55:716–725 [PubMed] [Google Scholar]
- Holden NS, Tacon CE. (2011) Principles and problems of the electrophoretic mobility shift assay. J Pharmacol Toxicol Methods 63:7–14 [DOI] [PubMed] [Google Scholar]
- Kerkvliet NI, Steppan LB, Vorachek W, Oda S, Farrer D, Wong CP, Pham D, Mourich DV. (2009) Activation of aryl hydrocarbon receptor by TCDD prevents diabetes in NOD mice and increases Foxp3+ T cells in pancreatic lymph nodes. Immunotherapy 1:539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Henry EC, Kim DK, Kim YH, Shin KJ, Han MS, Lee TG, Kang JK, Gasiewicz TA, Ryu SH, et al. (2006) Novel compound 2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-o-tolylazo-phenyl)-amide (CH-223191) prevents 2,3,7,8-TCDD-induced toxicity by antagonizing the aryl hydrocarbon receptor. Mol Pharmacol 69:1871–1878 [DOI] [PubMed] [Google Scholar]
- Lee JE, Safe S. (2000) 3′,4′-Dimethoxyflavone as an aryl hydrocarbon receptor antagonist in human breast cancer cells. Toxicol Sci 58:235–242 [DOI] [PubMed] [Google Scholar]
- Lu YF, Santostefano M, Cunningham BD, Threadgill MD, Safe S. (1995) Identification of 3′-methoxy-4′-nitroflavone as a pure aryl hydrocarbon (Ah) receptor antagonist and evidence for more than one form of the nuclear Ah receptor in MCF-7 human breast cancer cells. Arch Biochem Biophys 316:470–477 [DOI] [PubMed] [Google Scholar]
- Moore TW, Mayne CG, Katzenellenbogen JA. (2010) Minireview: Not picking pockets: nuclear receptor alternate-site modulators (NRAMs). Mol Endocrinol 24:683–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray IA, Flaveny CA, DiNatale BC, Chairo CR, Schroeder JC, Kusnadi A, Perdew GH. (2010a) Antagonism of aryl hydrocarbon receptor signaling by 6,2′,4′-trimethoxyflavone. J Pharmacol Exp Ther 332:135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray IA, Morales JL, Flaveny CA, Dinatale BC, Chiaro C, Gowdahalli K, Amin S, Perdew GH. (2010b) Evidence for ligand-mediated selective modulation of aryl hydrocarbon receptor activity. Mol Pharmacol 77:247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray IA, Reen RK, Leathery N, Ramadoss P, Bonati L, Gonzalez FJ, Peters JM, Perdew GH. (2005) Evidence that ligand binding is a key determinant of Ah receptor-mediated transcriptional activity. Arch Biochem Biophys 442:59–71 [DOI] [PubMed] [Google Scholar]
- Okey AB. (2007) An aryl hydrocarbon receptor odyssey to the shores of toxicology: the Deichmann Lecture, International Congress of Toxicology-XI. Toxicol Sci 98:5–38 [DOI] [PubMed] [Google Scholar]
- Pandini A, Soshilov AA, Song Y, Zhao J, Bonati L, Denison MS. (2009) Detection of the TCDD binding-fingerprint within the Ah receptor ligand binding domain by structurally driven mutagenesis and functional analysis. Biochemistry 48:5972–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov PI, Temelkov S, Villeneuve DL, Ankley GT, Mekenyan OG. (2009) Mechanism-based categorization of aromatase inhibitors: a potential discovery and screening tool. SAR QSAR Environ Res 20:657–678 [DOI] [PubMed] [Google Scholar]
- Procopio M, Lahm A, Tramontano A, Bonati L, Pitea D. (2002) A model for recognition of polychlorinated dibenzo-p-dioxins by the aryl hydrocarbon receptor. Eur J Biochem 269:13–18 [DOI] [PubMed] [Google Scholar]
- Puppala D, Gairola CG, Swanson HI. (2007) Identification of kaempferol as an inhibitor of cigarette smoke-induced activation of the aryl hydrocarbon receptor and cell transformation. Carcinogenesis 28:639–647 [DOI] [PubMed] [Google Scholar]
- Sanderson JT, Aarts JM, Brouwer A, Froese KL, Denison MS, Giesy JP. (1996) Comparison of Ah receptor-mediated luciferase and ethoxyresorufin-O-deethylase induction in H4IIE cells: implications for their use as bioanalytical tools for the detection of polyhalogenated aromatic hydrocarbons. Toxicol Appl Pharmacol 137:316–325 [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Hirota K, Christensen J, O'Garra A, Stockinger B. (2009) Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med 206:43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. (2008) The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453:106–109 [DOI] [PubMed] [Google Scholar]
- Whelan F, Hao N, Furness SG, Whitelaw ML, Chapman-Smith A. (2010) Amino acid substitutions in the aryl hydrocarbon receptor ligand binding domain reveal YH439 as an atypical AhR activator. Mol Pharmacol 77:1037–1046 [DOI] [PubMed] [Google Scholar]
- Wincent E, Amini N, Luecke S, Glatt H, Bergman J, Crescenzi C, Rannug A, Rannug U. (2009) The suggested physiologic aryl hydrocarbon receptor activator and cytochrome P4501 substrate 6-formylindolo[3,2-b]carbazole is present in humans. J Biol Chem 284:2690–2696 [DOI] [PubMed] [Google Scholar]
- Zhang N, Agbor LN, Scott JA, Zalobowski T, Elased KM, Trujillo A, Duke MS, Wolf V, Walsh MT, Born JL, et al. (2010) An activated renin-angiotensin system maintains normal blood pressure in aryl hydrocarbon receptor heterozygous mice but not in null mice. Biochem Pharmacol 80:197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Degroot DE, Hayashi A, He G, Denison MS. (2010) CH223191 is a ligand-selective antagonist of the Ah (Dioxin) receptor. Toxicol Sci 117:393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]