Abstract
The fixation of atmospheric N2 by cyanobacteria is a major source of nitrogen in the biosphere. In Nostocales, such as Anabaena, this process is spatially separated from oxygenic photosynthesis and occurs in heterocysts. Upon nitrogen step-down, these specialized cells differentiate from vegetative cells in a process controlled by two major regulators: NtcA and HetR. However, the regulon controlled by these two factors is only partially defined, and several aspects of the differentiation process have remained enigmatic. Using differential RNA-seq, we experimentally define a genome-wide map of >10,000 transcriptional start sites (TSS) of Anabaena sp. PCC7120, a model organism for the study of prokaryotic cell differentiation and N2 fixation. By analyzing the adaptation to nitrogen stress, our global TSS map provides insight into the dynamic changes that modify the transcriptional organization at a critical step of the differentiation process. We identify >900 TSS with minimum fold change in response to nitrogen deficiency of eight. From these TSS, at least 209 were under control of HetR, whereas at least 158 other TSS were potentially directly controlled by NtcA. Our analysis of the promoters activated during the switch to N2 fixation adds hundreds of protein-coding genes and noncoding transcripts to the list of potentially involved factors. These data experimentally define the NtcA regulon and the DIF+ motif, a palindrome at or close to position −35 that seems essential for heterocyst-specific expression of certain genes.
Cyanobacteria are oxygen-producing, photosynthetic organisms that are responsible for approximately one-half of the global CO2 fixation. In addition, many cyanobacteria are able to perform N2 fixation, a process that is extremely sensitive to oxygen. To protect the nitrogenase complex from photosynthetically evolved oxygen, many filamentous strains, including Anabaena sp. PCC7120 (also known as Nostoc sp. PCC7120, from here on Anabaena 7120), differentiate heterocysts, a specialized cell type devoted to N2 fixation (1, 2). Heterocyst differentiation is integrated into a series of physiological responses that take place when a source of combined nitrogen is not available. Those responses are globally controlled by NtcA, a transcriptional regulator of the cAMP receptor protein (CAP) family (3–5). NtcA-mediated regulation involves the binding of NtcA to a consensus binding site with the sequence GTAN8TAC (3). In the absence of ammonium, the preferred nitrogen source, NtcA activates the expression of genes required for alternative assimilation pathways, such as those encoding nitrate and nitrite reductases (3). NtcA also acts as a transcriptional repressor of some genes, such as the gif gene, which encodes the glutamine synthetase inactivating factor (5). NtcA is also required for heterocyst differentiation and subsequent N2 fixation (6, 7). A key factor activated by NtcA is HetR, a master regulator of many genes involved in the differentiation process (8, 9). HetR binds to DNA (10) and folds into an unusual structure (11). Although HetR was hypothesized to control the expression of hundreds of genes, only a single 17-base pair palindrome has been identified as a binding site (12). Moreover, HetR exerts positive feedback on NtcA expression (13), but it is not known how the double-positive feedback between both factors is terminated at a later step in the differentiation cascade.
During the last three decades, <100 transcriptional start sites (TSS) have been mapped in Anabaena 7120 on a gene-by-gene basis, primarily associated with highly expressed genes or genes regulated by nitrogen availability. An analysis of the Anabaena 7120 transcriptome during vegetative cell growth and in response to nitrogen deprivation has been recently published (14).
To complement the scarce existing information, our study used a differential RNA-seq (dRNA-seq) approach, which is selective for the 5′ ends of primary transcripts (15, 16) and allows the comprehensive determination of the transcriptional organization of a genome. Pretreatment of bacterial RNA with Terminator 5′ phosphate-dependent exonuclease (TEX) specifically degraded transcripts with a 5′ P (processed RNAs) (16). Based on this approach, we present a genome-wide map of 13,705 candidate TSS that were experimentally mapped for the chromosome and the six plasmids of Anabaena 7120. Analyzing the transcriptional changes that occur upon nitrogen step-down in both the wild-type (WT) and a hetR mutant lead to the identification of all promoters controlled directly or indirectly by HetR, to a precise definition of the NtcA regulon and to the identification of the DIF+ motif, a sequence element involved in heterocyst-specific expression. The availability of its annotated primary transcriptome will greatly facilitate the use of this genetically tractable organism as a model for prokaryotic cell differentiation and N2 fixation.
Results
Large-Scale Mapping of Primary 5′ Ends.
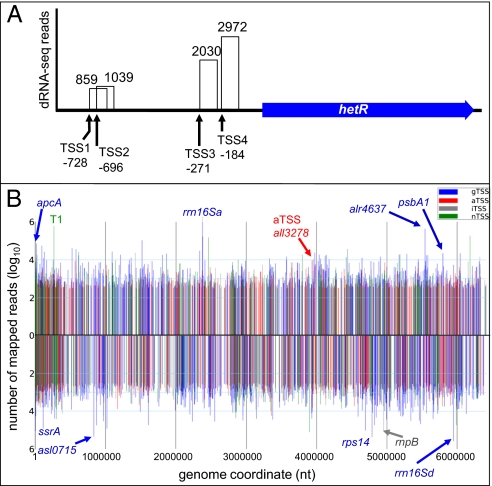
RNA samples were obtained from WT and hetR mutant strains that were grown in the presence of ammonia (WT-0 and hetR-0) or subjected to nitrogen step-down for 8 h (WT-8 and hetR-8). In total, 24,312,062 sequence reads (up to 75 nt long) were analyzed, and 1.6 billion bases of cDNA were mapped to the Anabaena 7120 chromosome and its six large plasmids. In the four samples, between 48.3% and 64.7% of the reads did not correspond to rRNAs. The complete dataset was taken to identify possible TSS, based on a minimum number of 50 sequencing reads associated with an RNA 5′ end. An example of the data obtained is shown for the gene encoding HetR in Fig. 1A. All four previously described TSS, including that at −271 whose induction is heterocyst-specific (13, 17, 18), were identified by 6,900 reads in our dataset.
Fig. 1.
Genome-wide identification of TSS in Anabaena 7120. (A) Differential RNA-seq identifies single TSS in complex promoter regions as exemplified by the hetR gene. The total number of reads mapped to each 5′ end is indicated for each of the four previously described TSS. (B) Distribution of 3401 TSS with ≥300 reads each along a linear plot of the Anabaena 7120 chromosome. TSS mapped for the forward strand are plotted above the x axis, and for the reverse strand below. The number of sequence reads is given on the y axis. The location of each TSS according to SI Appendix, Fig. S1A served for classification as gTSS (blue), nTSS (green), aTSS (red), or iTSS (gray). Selected TSS for each of the four classes are annotated.
We identified 12,797 putative chromosomal TSS and 908 putative TSS on the six plasmids (Table 1). From these TSS, 4,186 TSS were located within a distance of 200 nt upstream of an annotated gene (gTSS, mostly mRNAs); 4,172 TSS in inverse orientation (or ≤50 bp 5′ or 3′) to annotated genes (aTSS), suggesting antisense transcription; and 1,414 TSS of potential ncRNAs in intergenic spacers (nTSS). In addition, 3,933 TSS in sense orientation were located internally within annotated genes (iTSS). For consistency, this classification (SI Appendix, Fig. S1A), solely based on location, was used throughout. Therefore, some of the TSS here categorized as gTSS may actually give rise to ncRNAs, and some of the nTSS may rather drive the transcription of genes with long 5′UTRs (see also comments in Dataset S1, Table S1). Because for 704 TSS an association with more than one category was possible (SI Appendix, Fig. S1B), we prioritized gTSS over aTSS and iTSS, and all remaining TSS were automatically categorized as nTSS. A global overview of the distribution of TSS is given in Fig. 1B (chromosome) and SI Appendix, Fig. S2 (plasmids). The exact positions of all putative TSS are indicated in SI Appendix, Supplementary Data Files 1 (chromosome) and 2–7 (plasmids) and in Dataset S1, Table S1. To benchmark, we compared our data with a set of 93 TSS previously reported in 59 independent studies (SI Appendix, Table S2). From the previously reported TSS, 81 were confirmed with 69 of them being associated with 50 or more reads. In addition, TSS that had remained unnoticed were observed for several genes, including glnA, ntcA, patS, and rbcL.
Table 1.
Overview on the number and types of putative TSS mapped for the chromosome (Chr) and plasmids alpha, beta, gamma, delta, epsilon, and zeta
| Chr | Alpha | Beta | Gamma | Delta | Epsilon | Zeta | Total | |
| Length, nt | 6,413,771 | 408,101 | 186,614 | 101,965 | 55,414 | 40,340 | 5,584 | — |
| No. of genes | 5,430 | 386 | 186 | 90 | 85 | 31 | 5 | |
| gTSS | 3,955 | 145 | 41 | 24 | 14 | 6 | 1 | 4,186 |
| aTSS | 3,854 | 188 | 73 | 34 | 13 | 8 | 2 | 4,172 |
| iTSS | 3,722 | 113 | 50 | 18 | 21 | 6 | 3 | 3,933 |
| nTSS | 1,266 | 88 | 15 | 22 | 8 | 7 | 8 | 1,414 |
| Total | 12,797 | 534 | 179 | 98 | 56 | 27 | 14 | 13,705 |
Finally, 37 TSS were identified for 35 tRNA genes. Based on their alignment, a consensus for a constitutive promoter was defined (SI Appendix, Fig. S3). The length of the 5′ leaders varied from 5 to >200 nt, but most were between 10 and 20 nt (SI Appendix, Fig. S3 and Supplementary Data File 8).
Nitrogen Deficiency- Versus Differentiation-Related Promoters in the Nitrogen Stress Response of Anabaena 7120.
To analyze the transcriptional changes induced by nitrogen stress and those specifically leading to the differentiation of heterocysts, we individually compared the numbers of dRNA-seq reads of all TSS identified in RNA isolated from the four samples. Normalized ratios (fold changes) in the number of reads between the different samples were determined as described (19). The comparison with previously described nitrogen stress-induced transcriptional responses, e.g., for the nirA-nrtABCD-narB cluster (NtcA-activated), the gifA gene (NtcA-repressed), and genes in the heterocyst envelope polysaccharide (HEP) island, which are involved in heterocyst maturation, revealed a high degree of consistency (SI Appendix, Fig. S4). We confirmed the TSS for nirA (−460 with respect to the translational start) (20), with a number of reads significantly higher in the samples collected 8 h after nitrogen step-down from both the WT and the hetR mutant (SI Appendix, Fig. S4A). We also confirmed the TSS for gifA at position −43 (21), associated with a high number of reads in the presence of NH4+ from both the WT and the hetR mutant (SI Appendix, Fig. S4B). SI Appendix, Fig. S4C shows that transcription of several genes involved in the synthesis of HEPs was almost completely restricted to WT under nitrogen stress.
Two main groups of TSS were defined that differed in their response to nitrogen step-down. The DEF category (deficiency-related changes) includes TSS showing transcriptional changes common to both strains (TSS for nirA or gifA above are paradigms for this category), whereas the DIF category (differentiation-related changes) includes TSS with transcriptional changes observed exclusively in the WT (e.g., TSS for genes in the HEP island; SI Appendix, Fig. S4C). The DIF group includes all transcriptional changes that depend on HetR and are thus likely involved in the process of heterocyst differentiation. Additionally, although most changes in the DEF category involved activation (DEF+), we also identified some TSS associated with a decrease in the number of reads upon nitrogen step-down (DEF−) (Dataset S1, Tables S3–S5). With a minimum fold change of eight, we identified 129, 28, and 209 TSS in the DEF+, DEF−, and DIF+ categories, respectively. Our dataset identifies strongly regulated TSS for many genes with previously described nitrogen-dependent regulation or role in heterocyst differentiation, including amt1 and amt4, heterocyst differentiation-related genes hepA, hepB, hepN, and hepS, regulatory genes nirB and patB, or nblA involved in phycobilisome degradation (SI Appendix, Table S6).
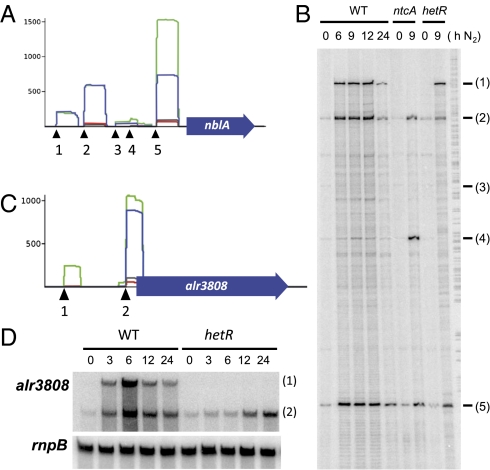
Our dataset allows the identification of multiple TSS in complex promoters (Fig. 1A), providing a powerful approach to the analysis of genes with complex regulation. Two such promoter regions containing several TSS were chosen for further validation by primer extension or Northern blot hybridization. Five putative TSS were identified for nblA (SI Appendix, Table S6 and Fig. 2A), a nitrogen stress-inducible gene required for phycobilisome degradation but not essential for heterocyst differentiation in Anabaena 7120 (22). The results presented in Fig. 2B confirmed all 5′ ends identified by dRNA-seq and their activity: TSS1 was not active in the presence of ammonium and NtcA-dependent, TSS2 and TSS5 were also inducible but NtcA-independent, TSS3 was barely detected, and TSS4 was mostly detected in the ntcA mutant. Two TSS were identified for alr3808 (SI Appendix, Table S6 and Fig. 2C) encoding a DpsA homolog with known nitrogen-dependent regulation (23, 24). Consistent with the dRNA-seq data, two transcripts covering alr3808 were identified by Northern blot (Fig. 2D). The longer transcript, probably originating at position 4601709f, is induced upon nitrogen step-down but was not expressed in the hetR mutant (therefore categorized as DIF+), whereas the shorter transcript, probably originating at position 4601982f, was also induced in the hetR mutant, although at later time points. Thus, not only the positions of TSS but also the regulation observed by dRNA-seq, were confirmed.
Fig. 2.
Analysis of genes with multiple TSS identified by dRNA-seq. RNA was isolated from ammonium-grown cells (lanes labeled 0) or from ammonium-grown cells incubated in the absence of combined nitrogen for the number of hours indicated. (A) Graphical representation of reads mapped to the promoter of nblA. The histograms correspond to the WT-0 (red), WT-8 (green), hetR-0 (black), and hetR-8 (blue) samples. (B) Primer extension analysis of the nblA mRNA in WT and mutant strains CSE2 (ntcA) and 216 (hetR). Samples contained 20 μg of RNA. The oligonucleotide used was complementary to positions +5 to −18 with respect to the translational start of nblA. The 5′ ends identified by dRNA-seq are numbered 1–5 (for positions, see Dataset 1, Table S1). (C) Graphical representation of reads mapped to the promoter of alr3808. (D) Northern blot analysis of the alr3808 mRNA in Anabaena 7120 and mutant strain 216 (hetR). Samples contained 10 μg of RNA. The probe used was an internal fragment of alr3808. rnpB (38) was used as a loading control.
TSS in the DIF Category Identify HetR-Regulated Elements and a Sequence Motif Related to Heterocyst-Specific Expression.
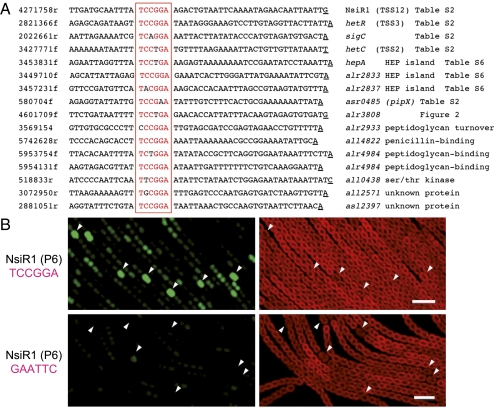
HetR is the earliest known dedicated regulator involved in the differentiation of functional heterocysts. Therefore, transcriptional responses observed in the WT but not in the hetR mutant probably belong to the specific program that leads to the differentiation of these cells. These TSS constitute the DIF+ category (see TSS with at least eightfold change in Dataset S1, Table S5). Their promoters can be analyzed to identify elements that might play a role in the differentiation process. In fact, the TSS for some genes with known HetR-dependent or heterocyst-specific expression (e.g., ntcA, hetR, nsiR1) appear among the TSS exhibiting the highest fold change in this category. A direct search for the 17-base pair palindrome identified as a HetR binding site (12), or for any other conserved element, was unsuccessful. However, we noticed that the promoters for ncRNA NsiR1 (25) were in the DIF+ class. Because NsiR1 is conserved in Nostocales and transcribed from a tandem array of short repeats, we could compare the promoter regions of 51 repeats from five different strains and found a conserved palindrome 5′TCCGGA at or close to the −35 position. Moreover, the same or a very similar motif is present in several other heterocyst-specific promoters (Fig. 3A). A global search identified this motif at similar position in 58 of the 209 DIF+ promoters when a single mismatch was allowed (Dataset S1, Table S7 and selected examples in Fig. 3A). From all remaining 13,496 TSS, only 572 also share this motif (and some of those are DIF+ too, but with a fold change <8). Hence, the enrichment for this motif within the DIF+ category of promoters is nonrandom (P < 2.2e−16 in a χ2 test). We therefore named this sequence the DIF+ motif.
Fig. 3.
Occurrence of a palindrome, 5′ TCCGGA, in promoters of the DIF+ category. (A) Alignment of the heterocyst-specific promoters for NsiR1, the hetR TSS3, sigC, and hetC with selected promoters in the DIF+ category (fold change ≥ 8) containing TCCGGA around position −35 (one mismatch allowed). (B) Cell-specific transcription from the wild-type promoter of NsiR1 (P6; Upper) or a mutated version of P6 carrying GAATTC instead of TCCGGA (Lower). Images corresponding to red autofluorescence (Right) and GFP fluorescence (Left) are shown. White triangles point to proheterocysts. (Scale bars: 10 μm.)
To directly test the functional relevance of the DIF+ motif, the 70-bp promoter from NsiR1 repeat 6 (P6) was placed upstream of a promoter-less GFP. Constructs bearing the intact DIF+ motif expressed strong green fluorescence in (pro)heterocysts (Fig. 3B). Hence, P6 possesses all of the elements for cell-specific expression. In contrast, only very weak, nonheterocyst-specific fluorescence was obtained when the DIF+ motif was replaced by 5′GAATTC (Fig. 3B). Thus, the DIF+ motif is required for heterocyst-specific expression of the nsiR1 promoter.
NtcA Binding Site Revisited: TSS in the DEF Category Define the NtcA Regulon.
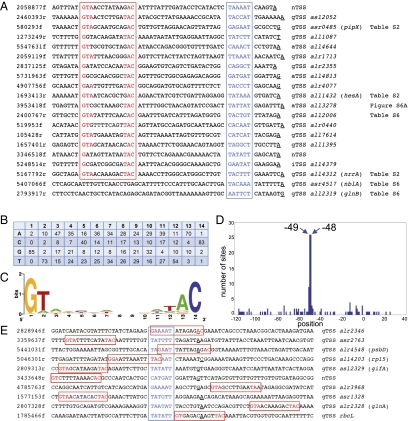
We hypothesized that most nitrogen step-down–induced responses occurring both in the hetR strain and the WT (DEF categories) would not be related to heterocyst differentiation but are likely NtcA-regulated. NtcA binding sites in NtcA-activated promoters overlap in most cases the −35 region and are centered close to position −41.5 (i.e., the first nucleotide is located at −48) with regard to the TSS (3). Fig. 4A shows the promoters of the 20 TSS exhibiting the highest fold change in the DEF+ category. Eighteen of them contain sequences matching the consensus NtcA binding site at the expected position. NtcA-dependent activation was previously described for two of them, 580293f (26) and 5167792r (27), but not for the remaining 16 strongly regulated TSS. Thus, the DEF category defines the NtcA regulon at an unprecedented resolution. A position-specific scoring matrix (PSSM) was defined based on an alignment of all promoter regions for TSS in the DEF+ category with at least eightfold change (SI Appendix, Fig. S5A and Fig. 4 B and C). When all TSS in the DEF+ category were scanned in a sliding window approach for possible NtcA binding sites with a score ≥5, a peak was identified at positions −48 to −49 (first nucleotide of the motif), thereby matching the expected location precisely not only for this dataset with at least eightfold change (Fig. 4D), but also when a larger dataset of 965 TSS with at least twofold change (SI Appendix, Fig. S5) was analyzed. This observation indicated that a significant proportion of the DEF+ TSS (even with relatively low fold changes) contain NtcA binding sites at positions that are compatible with transcriptional activation and that the PSSM as defined here (Fig. 4 and SI Appendix, Fig. S5) is useful for global searches.
Fig. 4.
NtcA-activated and -repressed promoters. (A) Promoter regions of 20 TSS (underlined nucleotide) in the DEF+ category with the highest fold change. Possible −10 elements are highlighted in blue, nucleotides matching the consensus for NtcA binding sites in red. (B) Nucleotide frequencies derived for positions 1–14 of 87 putative NtcA binding sites. (C) Corresponding Weblogo. (D) Position of NtcA binding sites identified in a sliding window approach using the PSSM along the promoters in the DEF+ category (fold change ≥8). The bars indicate the first nucleotide of a putative NtcA binding site and its position with regard to the TSS (E) Putative NtcA binding sites identified at repression-compatible positions around TSS in the DEF− category.
NtcA binding sites incompatible with transcriptional activation (eventually repressing transcription) can be located closer to, and even downstream of, the TSS. To find such elements, we scanned sequences surrounding all TSS in the DEF category in two windows: position −120 to −44 (first nt of motif; activation-compatible sites), and −44 to +41 (repression-compatible sites) and show sites with score ≥5 in Dataset S1, Tables S8 and S9. Fig. 4E shows 10 examples for putative NtcA binding sites at repression-compatible positions around TSS in the DEF− category. Two of these TSS, 2809313r (21) and 2807328f (28), were described as NtcA-repressed. TSS 1785466f (rbcL), also described as containing an NtcA binding site in a repressor-compatible position (29), is included for comparison.
Noncoding RNAs Potentially Involved in the Response to Nitrogen Stress and Heterocyst Differentiation.
Several of the TSS exhibiting the highest fold change correspond to antisense RNA (asRNA) or noncoding RNA (ncRNA) transcripts. The aTSS at position 3953418f, a strongly regulated DEF+ promoter, gives rise to an asRNA for gene all3278, whose mutation leads to the inability to fix N2 in the presence of oxygen (30). Primer extension analysis confirmed the dRNA-seq results SI Appendix, Fig. S6A. The initiation of transcription at this position is strongly induced by nitrogen step-down, independently of HetR, but depending on NtcA, consistent with the identification of a putative NtcA binding site upstream (Dataset S1, Table S8 and Fig. 4A). We also confirmed two strongly regulated nTSS that produce small ncRNAs SI Appendix, Fig. S6 B and C. The coregulation of these TSS with well-studied protein-coding genes in these categories suggests that some of the ncRNAs identified here might be involved in the adaptation to nitrogen stress or the differentiation of heterocysts.
Discussion
In this study, we have defined a set of >10,000 putative TSS for Anabaena 7120. We did not require these 5′ ends to be linked to a classical −10 element because the differentiation process could involve alternative sigma factors recognizing different promoter elements, because the quality of data appeared high (3,401 TSS were identified on the basis of >300 reads) and because our dataset was experimentally validated in several ways. This dataset confirms most of the previously defined TSS for this organism (SI Appendix, Table S2), while identifying unique nitrogen-regulated TSS for the majority of genes previously reported as involved in heterocyst differentiation or adaptation to nitrogen stress (SI Appendix, Table S6). Additionally, using primer extension and Northern blot analysis, we have confirmed several TSS in complex promoter regions or corresponding to asRNAs or ncRNAs. When considering potential −10 elements, 9,885 TSS remain in the dataset at a threshold of +3.0 (Dataset S1, Table S1). Approximately one-third of all TSS were on the reverse complementary strand of 2,412 genes, suggesting antisense transcription to 39% of all genes. This number seems high but is consistent with observations for several other bacteria (31). A total of 1,414 TSS located in the intergenic regions >200 nt away from any annotated gene indicated a high number of ncRNAs, although some of these nTSS drive the transcription of mRNAs with very long leaders and, therefore, are functional gTSS (see comments in Dataset S1, Table S1).
Our data provide insight into the complexity of the primary transcriptome of Anabaena 7120 under standard growth conditions and at a relatively early step of heterocyst differentiation. The use of the hetR strain, unable to start the transcriptional program leading to heterocyst differentiation, allowed us to separate transcriptional changes specifically related to this developmental process (DIF) from other nitrogen-stress responses that are still observed in the hetR mutant (DEF), thus likely unrelated to heterocyst differentiation but rather involved in other aspects of the adaptation to nitrogen stress. We thus defined sets of specifically regulated promoters belonging to the DEF and DIF categories. TSS included in these two categories defined both the NtcA and the HetR regulons. As exemplified by the cases of hetR (Fig. 1A), nblA, or alr3808 (Fig. 2), the use of TEX-treated samples allowed the identification of multiple TSS in a given promoter region. Complex promoter regions with several TSS are commonly found in genes involved in heterocyst differentiation and patterning, probably due to differential TSS use in the two cell types of the filament (e.g., TSS for ntcA, hetR, devB, and hetC in SI Appendix, Table S2).
Over the last decades, genes involved in heterocyst differentiation and N2 fixation were primarily identified by mutagenesis and screening of strains unable to grow in the absence of combined nitrogen (2, 30). Here, comparison of the wild type to the hetR transcriptome upon nitrogen step-down yielded the DIF category, i.e., the HetR regulon, which now can be searched for HetR-dependent TSS of genes potentially involved in the differentiation process. This regulon includes, for instance, genes related to cell wall synthesis and/or remodeling (Fig. 3), a key aspect of heterocyst differentiation. The DIF category also provides a valuable dataset to identify sequence motifs potentially involved in heterocyst-specific expression. Indeed, we identified the DIF+ motif common to many heterocyst-specifically expressed promoters. It consists of a short palindrome 5′ TCCGGA, centered at or close to position −35, suggesting it might be recognized by a specific sigma factor.
NtcA-mediated regulation is operated by binding to a consensus sequence, GTAN8TAC, first described for strongly regulated promoters in Synechococcus (5). Promoters that are directly activated by NtcA contain an NtcA binding site that, in most cases, is centered close to position −41.5 with respect to the TSS (although some NtcA binding sites further upstream are also described). In such promoters, NtcA activates transcription in a manner that resembles CAP-mediated regulation at class II promoters. On the other hand, NtcA-mediated repression is operated by interaction with NtcA binding sites at a position that makes binding incompatible with the normal operation of the promoter. In the case of gifA from Anabaena, one NtcA binding site is centered at position −28.5 (21). The DEF category defined here identifies transcriptional responses that in many cases were directly regulated by NtcA as deduced from the identification of NtcA binding sites located in positions compatible with transcriptional activation (DEF+) or repression (DEF−) (Dataset S1, Tables S8 and S9 and Fig. 4). Comparison with a computational prediction (32) of the NtcA regulon (SI Appendix, Table S10) revealed that many of the TSS in the DEF category correspond to previously unknown NtcA-regulated promoters, thereby expanding the known NtcA regulon. The absence of NtcA binding sites in the promoters for several other TSS in the DEF categories (such as TSS1 for nblA; Figs. 2B and 4A) suggests their expression either is not directly regulated by NtcA or operated by binding to different positions.
Finally, as observed in other cyanobacteria (15, 33, 34), our dataset indicates the abundant transcription of antisense and ncRNAs (e.g., ncRNA T1 in Fig. 1B, associated with >600,000 reads). As previously described for NsiR1, a short ncRNA, whose expression is induced specifically in proheterocysts upon nitrogen step-down (25), expression of some of these transcripts is regulated by nitrogen availability (SI Appendix, Fig. S6), suggesting that antisense and noncoding transcripts might be involved in the regulation of nitrogen assimilation and heterocyst differentiation. The annotated primary transcriptome of Anabaena 7120 during the transition from ammonium utilization to N2 fixation will greatly facilitate the use of this organism as a model for prokaryotic cell differentiation and N2 fixation in an oxygenic phototroph.
Methods
Full protocols are available in SI Appendix, SI Methods.
Growth Conditions.
Cultures of Anabaena 7120 WT, hetR mutant 216 (8), and ntcA mutant CSE2 (6) were bubbled with an air/CO2 mixture (1% vol/vol) and grown photoautotrophically at 30 °C in BG110C medium lacking NaNO3 but containing 6 mM NH4Cl, 10 mM NaHCO3, and 12 mM N-Tris (hydroxymethyl) methyl-2-aminoethanesulfonic acid-NaOH buffer (pH 7.5). Four RNA samples were isolated for dRNA-seq analysis from cells taken at t = 0 h (WT-0 and hetR-0) and t = 8 h (WT-8 and hetR-8) after removing all combined nitrogen from the media.
Preparation and Analysis of RNA.
Total RNA was isolated by using hot phenol (35) with modifications. Northern blot hybridization and primer extension analysis of 5′ ends was performed as described (13, 34, 36). The cDNA libraries were prepared by vertis Biotechnologie, Germany (www.vertis-biotech.com) after enrichment for primary transcripts by treatment with TEX (Epicentre) and analyzed on an Illumina sequencer as described (16). Based on tetranucleotide tags (Dataset S1, Table S11), 5.153.094, 4.690.212, 6.398.708, and 5.497.219 sequence reads were assigned to the WT-0, the WT-8, the hetR-0, and hetR-8 populations, respectively, and matched against the sequences of the chromosome or plasmids of Anabaena 7120.
Computational Methods.
Reads <18 nt and those corresponding to the ribosomal clusters were filtered out. Remaining reads were mapped to the genome by using the segemehl algorithm (37), with default parameters. Reads were pooled from the four samples and their 5′ ends were binned within a 5-nt section. The position within the window where the most reads began was considered to be the initial TSS. Because we noticed a few cases of initiation of transcription from a broader window, this dataset was clustered to allow the combination of initial TSS, which were not further than 5 nt apart. The position within this window where the greatest number of reads began was considered a TSS when a minimum of 50 reads was associated with it. For ratio calculation, the number of reads for the four samples were normalized (19), and single pseudocounts were added. The resulting ratios were classified and filtered into DEF and DIF categories of regulated promoters. Possible −10 elements were searched 6–8 nt upstream of all putative TSS and scored according to a PSSM derived from this dataset (Dataset S1, Table S1 and SI Appendix, Fig. S7). To construct a PSSM for the NtcA binding site, all promoter regions for the 129 TSS in the DEF+ category with at least eightfold change were aligned. From these, 81 possessed an element matching at least 4 nt of the GTAN8TAC motif at position 22/23 upstream of the −10 element. These sites were used together with six additional experimentally defined sites (SI Appendix, Table S2) in the construction of the matrix. For the DIF+ motif, the regions −44 to −25 of all mapped TSS were searched in two distinct datasets: One consisted of all 209 TSS in the DIF+ class and the second of the remaining 13,496 putative TSS. Statistical significance of an enrichment for the DIF+ motif in the DIF+ class was tested in a Pearson's χ2 test.
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft program “Sensory and regulatory RNAs in Prokaryotes” SPP1258 Grant HE 2544 4-2; German Federal Ministry of Education and Research Grant 0313921 (to W.R.H.); and by the Ministerio de Ciencia e Innovación Grants BFU2007-60651 (to A.V.) and BFU2010-14821 (to A.M.P.) cofinanced by European Regional Development Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Supplementary data files 1-8 are available at http://www.cyanolab.de/Supplementary.html.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112724108/-/DCSupplemental.
References
- 1.Flores E, Herrero A. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat Rev Microbiol. 2010;8:39–50. doi: 10.1038/nrmicro2242. [DOI] [PubMed] [Google Scholar]
- 2.Kumar K, Mella-Herrera RA, Golden JW. Cyanobacterial heterocysts. Cold Spring Harb Perspect Biol. 2010;2:a000315. doi: 10.1101/cshperspect.a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herrero A, Muro-Pastor AM, Flores E. Nitrogen control in cyanobacteria. J Bacteriol. 2001;183:411–425. doi: 10.1128/JB.183.2.411-425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herrero A, Muro-Pastor AM, Valladares A, Flores E. Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol Rev. 2004;28:469–487. doi: 10.1016/j.femsre.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Luque I, Forchhammer K. Nitrogen assimilation and C/N balance sensing. In: Herrero A, Flores E, editors. The Cyanobacteria: Molecular Biology, Genomics and Evolution. Norfolk, UK: Caister Academic; 2008. pp. 335–382. [Google Scholar]
- 6.Frías JE, Flores E, Herrero A. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol Microbiol. 1994;14:823–832. doi: 10.1111/j.1365-2958.1994.tb01318.x. [DOI] [PubMed] [Google Scholar]
- 7.Wei TF, Ramasubramanian TS, Golden JW. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J Bacteriol. 1994;176:4473–4482. doi: 10.1128/jb.176.15.4473-4482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buikema WJ, Haselkorn R. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev. 1991;5:321–330. doi: 10.1101/gad.5.2.321. [DOI] [PubMed] [Google Scholar]
- 9.Black TA, Cai Y, Wolk CP. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol Microbiol. 1993;9:77–84. doi: 10.1111/j.1365-2958.1993.tb01670.x. [DOI] [PubMed] [Google Scholar]
- 10.Huang X, Dong Y, Zhao J. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc Natl Acad Sci USA. 2004;101:4848–4853. doi: 10.1073/pnas.0400429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim YC, et al. Structure of transcription factor HetR required for heterocyst differentiation in cyanobacteria. Proc Natl Acad Sci USA. 2011;108:10109–10114. doi: 10.1073/pnas.1106840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higa KC, Callahan SM. Ectopic expression of hetP can partially bypass the need for hetR in heterocyst differentiation by Anabaena sp. strain PCC 7120. Mol Microbiol. 2010;77:562–574. doi: 10.1111/j.1365-2958.2010.07257.x. [DOI] [PubMed] [Google Scholar]
- 13.Muro-Pastor AM, Valladares A, Flores E, Herrero A. Mutual dependence of the expression of the cell differentiation regulatory protein HetR and the global nitrogen regulator NtcA during heterocyst development. Mol Microbiol. 2002;44:1377–1385. doi: 10.1046/j.1365-2958.2002.02970.x. [DOI] [PubMed] [Google Scholar]
- 14.Flaherty BL, Van Nieuwerburgh F, Head SR, Golden JW. Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combined-nitrogen deprivation. BMC Genomics. 2011;12:332. doi: 10.1186/1471-2164-12-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitschke J, et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci USA. 2011;108:2124–2129. doi: 10.1073/pnas.1015154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma CM, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 17.Buikema WJ, Haselkorn R. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc Natl Acad Sci USA. 2001;98:2729–2734. doi: 10.1073/pnas.051624898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajagopalan R, Callahan SM. Temporal and spatial regulation of the four transcription start sites of hetR from Anabaena sp. strain PCC 7120. J Bacteriol. 2010;192:1088–1096. doi: 10.1128/JB.01297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frías JE, Flores E, Herrero A. Nitrate assimilation gene cluster from the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1997;179:477–486. doi: 10.1128/jb.179.2.477-486.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galmozzi CV, Saelices L, Florencio FJ, Muro-Pastor MI. Posttranscriptional regulation of glutamine synthetase in the filamentous cyanobacterium Anabaena sp. PCC 7120: Differential expression between vegetative cells and heterocysts. J Bacteriol. 2010;192:4701–4711. doi: 10.1128/JB.00222-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baier K, Lehmann H, Stephan DP, Lockau W. NblA is essential for phycobilisome degradation in Anabaena sp. strain PCC 7120 but not for development of functional heterocysts. Microbiology. 2004;150:2739–2749. doi: 10.1099/mic.0.27153-0. [DOI] [PubMed] [Google Scholar]
- 23.Ehira S, Ohmori M. NrrA, a nitrogen-responsive response regulator facilitates heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Mol Microbiol. 2006;59:1692–1703. doi: 10.1111/j.1365-2958.2006.05049.x. [DOI] [PubMed] [Google Scholar]
- 24.Ow SY, et al. Quantitative shotgun proteomics of enriched heterocysts from Nostoc sp. PCC 7120 using 8-plex isobaric peptide tags. J Proteome Res. 2008;7:1615–1628. doi: 10.1021/pr700604v. [DOI] [PubMed] [Google Scholar]
- 25.Ionescu D, Voss B, Oren A, Hess WR, Muro-Pastor AM. Heterocyst-specific transcription of NsiR1, a non-coding RNA encoded in a tandem array of direct repeats in cyanobacteria. J Mol Biol. 2010;398:177–188. doi: 10.1016/j.jmb.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Valladares A, et al. Specific role of the cyanobacterial PipX factor in the heterocysts of Anabaena sp. strain PCC 7120. J Bacteriol. 2011;193:1172–1182. doi: 10.1128/JB.01202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muro-Pastor AM, Olmedo-Verd E, Flores E. All4312, an NtcA-regulated two-component response regulator in Anabaena sp. strain PCC 7120. FEMS Microbiol Lett. 2006;256:171–177. doi: 10.1111/j.1574-6968.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- 28.Valladares A, Muro-Pastor AM, Herrero A, Flores E. The NtcA-dependent P1 promoter is utilized for glnA expression in N2-fixing heterocysts of Anabaena sp. strain PCC 7120. J Bacteriol. 2004;186:7337–7343. doi: 10.1128/JB.186.21.7337-7343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramasubramanian TS, Wei TF, Golden JW. Two Anabaena sp. strain PCC 7120 DNA-binding factors interact with vegetative cell- and heterocyst-specific genes. J Bacteriol. 1994;176:1214–1223. doi: 10.1128/jb.176.5.1214-1223.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lechno-Yossef S, Fan Q, Wojciuch E, Wolk CP. Identification of ten Anabaena sp. genes that under aerobic conditions are required for growth on dinitrogen but not for growth on fixed nitrogen. J Bacteriol. 2011;193:3482–3489. doi: 10.1128/JB.05010-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev. 2011;75:286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novichkov PS, et al. RegPrecise: A database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 2010;38(Database issue):D111–D118. doi: 10.1093/nar/gkp894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Georg J, et al. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Syst Biol. 2009;5:305. doi: 10.1038/msb.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steglich C, et al. The challenge of regulation in a minimal photoautotroph: Non-coding RNAs in Prochlorococcus. PLoS Genet. 2008;4:e1000173. doi: 10.1371/journal.pgen.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohamed A, Jansson C. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol Biol. 1989;13:693–700. doi: 10.1007/BF00016024. [DOI] [PubMed] [Google Scholar]
- 36.Muro-Pastor AM, Valladares A, Flores E, Herrero A. The hetC gene is a direct target of the NtcA transcriptional regulator in cyanobacterial heterocyst development. J Bacteriol. 1999;181:6664–6669. doi: 10.1128/jb.181.21.6664-6669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffmann S, et al. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLOS Comput Biol. 2009;5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vioque A. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 1992;20:6331–6337. doi: 10.1093/nar/20.23.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.