Abstract
Activated transcription of the bacteriophage T4 late genes, which is coupled to concurrent DNA replication, is accomplished by an initiation complex containing the host RNA polymerase associated with two phage-encoded proteins, gp55 (the basal promoter specificity factor) and gp33 (the coactivator), as well as the DNA-mounted sliding-clamp processivity factor of the phage T4 replisome (gp45, the activator). We have determined the 3.0 Å-resolution X-ray crystal structure of gp33 complexed with its RNA polymerase binding determinant, the β-flap domain. Like domain 4 of the promoter specificity σ factor (σ4), gp33 interacts with RNA polymerase primarily by clamping onto the helix at the tip of the β-flap domain. Nevertheless, gp33 and σ4 are not structurally related. The gp33/β-flap structure, combined with biochemical, biophysical, and structural information, allows us to generate a structural model of the T4 late promoter initiation complex. The model predicts protein/protein interactions within the complex that explain the presence of conserved patches of surface-exposed residues on gp33, and provides a structural framework for interpreting and designing future experiments to functionally characterize the complex.
Keywords: , Gp33, replication-coupled gene expression, X-ray crystallography
Transcription initiation in bacteria depends on the RNA polymerase (RNAP) catalytic core (subunit composition α2ββ′ω) and the promoter specificity subunit σ, which combine to create the RNAP holoenzyme. The σ subunit recruits RNAP to promoters through sequence-specific interactions with two conserved hexameric DNA sequence motifs, the -10 and -35 promoter elements. The -10 element is recognized by structural domain 2 of σ (σ2), positioned on the clamp helices of the RNAP β′-subunit, while the -35 element is recognized by σ4 positioned on the flap-tip-helix (FTH) of the RNAP β-subunit flap domain (1, 2). Transcription from weak promoters may be modulated by activators that typically bind to specific DNA sequences, or operators, located upstream of the -10/-35 promoter elements, and stabilize the initiation complex through protein/protein interactions directly with the RNAP holoenzyme, often with the α-subunit C-terminal domain or the σ4/β-flap subcomplex (3).
Activated transcription of the bacteriophage T4 late genes, which couples transcription of more than one-third of the phage genome to concurrent DNA replication, is accomplished by a mechanism that is apparently unique (4). Activation requires the function of two phage-encoded RNAP-binding proteins, gp55 and gp33, as well as DNA-mounted gp45, the sliding-clamp processivity factor for the phage T4 replisome (5, 6). Both gp33 and gp55 interact with the sliding-clamp via a hydrophobic and acidic C-terminal tail, the sliding-clamp binding motif (SCBM) that is attached to the body of each protein through a linker (7, 8).
Gp55 is a highly diverged member of the σ70 family (9, 10) that binds to Escherichia coli (Eco) RNAP core on the β′-subunit clamp helices (11, 12), in competition with 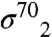 . The Gp55-holoenzyme accurately initiates low level (basal) transcription from T4 late promoters (13), which comprise only a -10-like motif with consensus TATAAATA (14). Gp55 lacks a σ4-equivalent domain. Instead, gp33 (112 amino acids) binds to the β-flap (11), in competition with
. The Gp55-holoenzyme accurately initiates low level (basal) transcription from T4 late promoters (13), which comprise only a -10-like motif with consensus TATAAATA (14). Gp55 lacks a σ4-equivalent domain. Instead, gp33 (112 amino acids) binds to the β-flap (11), in competition with 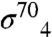 (15). Gp33 has no recognizable homology with other proteins (including σ4) and does not bind DNA; instead it represses gp55-dependent basal transcription (5, 16). However, in the presence of DNA-mounted sliding-clamp gp45, gp55-dependent transcription is activated more than 1,000-fold through a direct protein/protein interaction with the β-flap-bound gp33 (17). This requirement of the sliding-clamp for activation couples T4 late transcription to DNA replication (18).
(15). Gp33 has no recognizable homology with other proteins (including σ4) and does not bind DNA; instead it represses gp55-dependent basal transcription (5, 16). However, in the presence of DNA-mounted sliding-clamp gp45, gp55-dependent transcription is activated more than 1,000-fold through a direct protein/protein interaction with the β-flap-bound gp33 (17). This requirement of the sliding-clamp for activation couples T4 late transcription to DNA replication (18).
Other requirements for activated late transcription include the phage-encoded sliding-clamp loader (gp44/62), ATP or dATP hydrolysis, and a DNA template with a properly placed single-strand break or end, all needed to load the gp45 sliding-clamp onto DNA. The single-strand break, which serves as the sliding-clamp loading site, must be on the nontranscribed strand (so that the sliding-clamp is loaded in the proper orientation), and has the properties of an enhancer, in that it can be placed kilobases away from the promoter. The enhancer acts strictly in cis and requires a continuous, unobstructed DNA path between itself and the promoter (reviewed in ref. 6).
In this scheme, gp55 acts as the basal promoter specificity factor, gp45 acts as a DNA-tracking transcriptional activator, and the single-strand break acts as an unusual, enhancer-like operator. The properties of gp33 fit perfectly to the definition of a coactivator (19).
Here, we present the X-ray crystal structure of gp33 bound to its binding determinant on Eco RNAP, the β-subunit’s flap domain, allowing us to compare gp33 structural and functional characteristics to those of σ4 and revealing the basis for many functional characteristics of gp33, including its repression of basal transcription in the absence of gp45, as well as details of its interaction with the β-flap. Finally, the new structure, in combination with other information, allows us to generate a model of the T4 late promoter activated transcription complex that provides a structural framework for interpreting previous results, and serves as a guide for the design of further experiments.
Results and Discussion
Crystallization and Structure Determination of the gp33/β-flap Complex.
Gp33 binds the FTH of the Eco RNAP β-flap domain, and this interaction is required both for repression of gp55-mediated basal transcription by gp33 and gp45-mediated transcriptional activation (15). To elucidate the details of the gp33/RNAP interaction, we determined the crystal structure of the gp33/β-flap complex. The coexpressed (20) and purified gp33/β-flap complex (Fig. S1 A and B), comprising full-length T4 gp33 (12.8 kDa), and residues 831–1057 of the Eco RNAP β-subunit (25.2 kDa), was crystallized. The structure was determined using phases obtained from a molecular replacement solution (using the Taq RNAP β-flap, with the flexible flap-tip removed, as a search model) combined with single anomalous dispersion phases collected from crystals containing selenomethionyl-substituted (SeMet) proteins (21) (Fig. S1C). An atomic model was built and refined to an R/Rfree of 0.263/0.290 at 3.0 Å-resolution (Table S1).
Overall Architecture of the gp33/β-flap Complex.
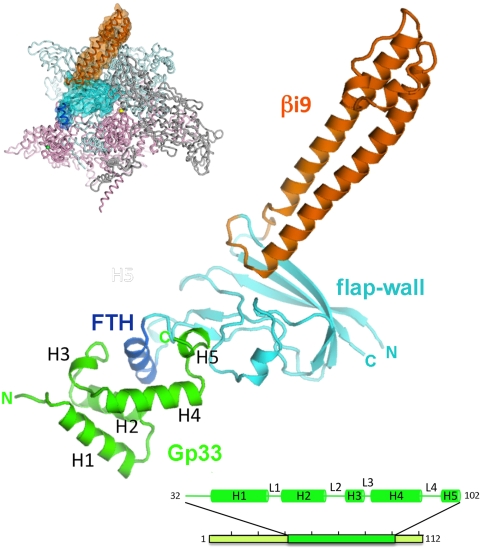
The two proteins form a complex with 1∶1 stoichiometry (Fig. 1, Fig. S1 B and D). The β-flap residues 831–937 and 1,043–1,057 form the conserved flap-wall (FW) and flap-tip (FT), with residues 938–1,042 constituting the lineage-specific insertion βi9 (22, 23) (Fig. 1). The FT (residues 891–912) is composed of the FTH (residues 897–906) that is connected to the FW by two flexible loops. The FW (residues 831–890/913–937/1,043–1,057) aligns well with the corresponding residues in previously solved structures of bacterial RNAP, from Thermus aquaticus (24) and Thermus thermophilus (25), with rmsd over α-carbon positions of 1 Å and 0.9 Å, respectively. The FT forms a flexible appendage, which is conserved in fold but found in different orientations in various structures (26). The structure of βi9 (22), a eubacterial lineage-specific insert of unknown function, has been described previously (23). The βi9 does not interact with gp33 (Fig. 1), nor is it involved in gp33 function (Fig. S2), so it is not discussed further here.
Fig. 1.
Structure of the gp33/Eco RNAP β-flap complex, shown as a ribbon diagram. Gp33 is green, the β-flap cyan, the FTH slate blue, βi9 orange. The small inset (left) illustrates the β-flap structure in the context of Eco RNAP (23), with β colored light cyan, β′ light pink. The β-flap is shown as a transparent surface and color-coded as in the ribbon diagram. The bottom bar illustrates the crystallized full-length gp33, but only the green portion (residues 32–102) was ordered and observed in the electron density. Observed secondary structure features are shown schematically and labeled (helices H1-5, and loops L1-L4).
The entire β-flap was built into the electron density map, but there was no clear density for two terminal segments of gp33, so only residues 32–102 (of the full 112) were modeled in the structure (Fig. 1). Analysis of the crystals indicated that they contained both proteins without proteolytic degradation (Fig. S1C), so the N-terminal 31 and C-terminal 10 residues of gp33 are presumed disordered in the crystals. In an alignment of gp33 homologs found in the bacteriophage T4 group, the N-terminal region is poorly conserved, and many homologs lack this region entirely (Fig. 2A, Fig. S3). Furthermore, a 29-residue N-terminal truncation of gp33 was shown to bind RNAP as effectively as full-length gp33 (15). The C-terminal tail of gp33 contains the SCBM, which would not be expected to interact with RNAP, but instead to be available for interaction with gp45. Thus, gp33[32–102], visualized in the crystal structure, represents the entire, functionally relevant RNAP-interaction determinant. Throughout the remainder of this manuscript, gp33[32–102] is referred to as gp33.
Fig. 2.
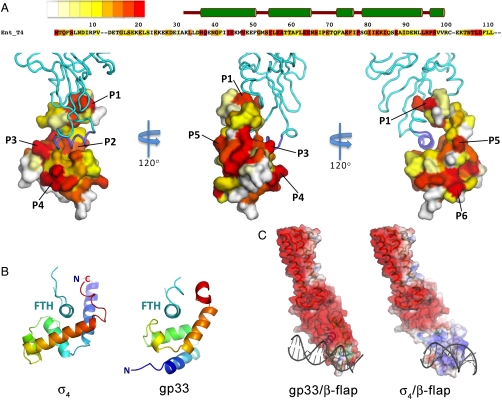
Sequence and structural properties of gp33. (A) Top: The sequence of T4 gp33 is shown, color-coded according to conservation score [red, highest identity; (31)] from an alignment of gp33 homologs from the T4 bacteriophage group (the full alignment is shown in Fig. S3). Below: three views of the gp33/β-flap structure, with gp33 shown as a molecular surface, color-coded according to the conservation shown in the alignment, and the β-flap shown as an α-carbon backbone worm, colored cyan or slate blue (FTH). Six conserved, surface-exposed patches are labeled (P1-P6). (B) Structural comparison of β-flap-bound σ4 (left) and gp33 (right). The view is down the axis of the FTH (cyan), which is shown as a thin α-carbon backbone worm; σ4 and gp33 are shown as ribbon diagrams and color-coded in a ramp from N (blue) to C terminus (red). (C) Comparison of the electrostatic surface distributions (42) of Eco β-flap bound to gp33 (left) and 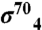 (right, generated from a homology model). Blue represents positively charged surfaces (+5 kT) and red, negatively charged surfaces (-5 kT).
(right, generated from a homology model). Blue represents positively charged surfaces (+5 kT) and red, negatively charged surfaces (-5 kT).
Gp33 is composed of five helices (H1-H5) connected by short loops (L1-L4; Fig. 1). The overall shape resembles the letter “C” with a bulbous base. The opening of the C accomodates the FTH (Fig. 1, 2 A and B).
Comparison of Gp33 and σ4.
Because both gp33 and σ4 interact with RNAP via the FT (1, 15, 27), it was proposed that gp33 might act as a functional analog of σ4 despite having no discernible sequence homology. Both gp33 and σ4 are helical proteins that clamp onto the FTH, but their overall folds are not related (Fig. 2B). In addition, gp33 and σ4 have very different surface properties (Fig. 2C). RNAP-bound σ4 presents a positive electrostatic surface to, and interacts extensively with, the DNA (28) (Fig. 2C). When bound to the β-flap, gp33 presents an overall negatively charged surface except for one small slightly basic patch (Fig. 2C), explaining how gp33 might inhibit nonspecific interactions with upstream DNA in the absence of the gp45 sliding-clamp activator (29).
Conservation Among gp33 Homologs.
An alignment (30) of selected gp33 homologs from the large family of T4-related bacteriophages allowed us to map conserved residues onto the surface of the structure (31) (Fig. 2A, Fig. S3). There are six distinct surface patches of conserved residues (P1–P6; Fig. 2A, Table S2). Two of these patches, P1 (L95, R96, P97, S98) and P2 (L56, I82, I85), are involved in interactions with the FW and FTH, respectively (Figs. 2A, 3). These interactions are presumably critical for gp33 function across species, explaining their conservation. P5 (E45, V48, Y55, K84, E88) comprises residues that are involved in interhelical contacts between H1, H2, and H4 and are thus likely critical to maintain the tertiary structure of gp33. The other surface patches (P3–E57; P4–E64, E65, N66, S67; P6–Q38, R37, K75), as well as an additional possible function for residues of P5, are discussed below in the context of a model of the T4 late promoter activation complex.
Fig. 3.
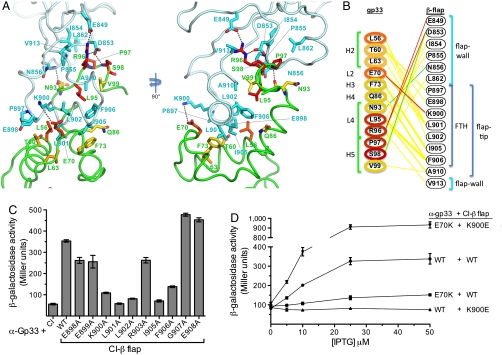
Gp33/β-flap interactions. (A). Residues that interact are drawn in stick format on gp33 (green worm) and the β-flap (cyan). Gp33 side chain carbons are color coded by conservation as in Fig. 2A. The right-hand view looks down the axis of the FTH and illustrates hydrophobic interactions between it and gp33. (B). Interaction schematic. Gp33 residues (left column) are colored according to conservation as in Fig. 2A. The nature of the interactions between each residue pair is color coded: nonpolar interactions, yellow; hydrogen-bonds, green; hydrogen-bonds/salt-bridges, red. Structural elements of gp33 (left) and the β-flap (right) are denoted. (C). Use of the bacterial two-hybrid assay (schematic in Fig. S3A); (33) to test the effects of alanine substitutions at β-flap residues 898-908 (except A904) on the gp33/β-flap interaction. Results of β-galactosidase assays performed with reporter strain cells containing one plasmid that encoded the α-gp33 fusion protein and a compatible plasmid that encoded either λCI or the indicated λCI-β-flap variant. The plasmids directed the synthesis of the fusion proteins under the control of IPTG-inducible promoters and the cells were grown in the presence of 50 μM IPTG. The bar graph shows the averages of three independent measurements and standard deviations. (D). Bacterial two-hybrid screen for substitutions in α-gp33 that restored the interaction of the gp33 moiety with the β-flap moiety of the λCI-β-flap-K900E fusion protein suggests that oppositely charged side chains of gp33-E70 and β-K900 interact at the gp33/β-flap interface. Results of β-galactosidase assays performed with reporter strain cells containing compatible plasmids that directed the synthesis of the indicated fusion proteins under the control of IPTG-inducible promoters. Cells were grown in the absence of IPTG or in the presence of 5, 10, 25, or 50 μM IPTG. The line graph shows the averages of three independent measurements for each IPTG concentration and standard deviations.
Gp33/β-Flap Interactions.
Gp33 establishes two distinct interaction surfaces with the β-flap, one at the FT (consisting mostly of the FTH), and one within the FW (Fig. 3
A and B). The protein/protein complex buries a substantial surface area of 1,900 Å2. Interactions between gp33 and the FT are mediated largely by hydrophobic van der Waals contacts (Fig. 3B). One face of the FTH presents a hydrophobic surface to a concave hydrophobic groove of gp33. The hydrophobic interaction surface of the FTH includes several residues that are also important for 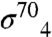 binding [L901, L902, I905, and F906 (32)]. The nonpolar alkyl side chain of β-FTH-K900 contributes to the hydrophobic interface, and this residue is also important for
binding [L901, L902, I905, and F906 (32)]. The nonpolar alkyl side chain of β-FTH-K900 contributes to the hydrophobic interface, and this residue is also important for 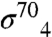 binding (Fig. S4). The hydrophobic surface of gp33 that interacts with the FT includes residues of conserved patches P1 and P2 (Figs. 2A, 3
A and B). In addition to contributing to the mostly hydrophobic interface between the FT and gp33, FTH-K900 is poised to form a salt-bridge with conserved gp33-E70 (Fig. 3).
binding (Fig. S4). The hydrophobic surface of gp33 that interacts with the FT includes residues of conserved patches P1 and P2 (Figs. 2A, 3
A and B). In addition to contributing to the mostly hydrophobic interface between the FT and gp33, FTH-K900 is poised to form a salt-bridge with conserved gp33-E70 (Fig. 3).
Interactions between gp33 and the FW are mediated mostly by four conserved residues comprising P1 (Figs. 2A, 3 A and B) located at the C-terminal end of the resolved portion of gp33. Conserved R96 of gp33 forms salt bridges with β-E849 and D853 as well as hydrophobic interactions with D853 and V913 (Fig. 3 A and B). The negatively charged β-flap residues that are key to this interaction, E849 and D853, are conserved throughout proteobacteria but are substituted by positively charged residues in more distantly related phyla (22). In addition, absolutely conserved S98 of gp33 engages β-D853 in polar and nonpolar contacts (Fig. 3 A and B).
A previous study identified gp33 mutants bearing substitutions F62A, K84E, E88K, and double substitution E64A/E65A as being defective both in binding to RNAP and in suppressing basal transcription (15). The structure shows that F62, K84, and E88 do not interact with the β-flap, but instead participate in intramolecular contacts that likely stabilize the gp33 structure. E64 and E65 also do not contact the β-flap, nor are they involved in intramolecular interactions (we suggest other interactions below).
Genetic Data Correlate with the Observed Structure.
To obtain a functional picture of the gp33/β-flap interface, we employed an in vivo bacterial two-hybrid assay (33) (Fig. S4). In this assay, contact between a protein domain fused to the RNAP α-subunit and a partner domain fused to the bacteriophage λ CI protein activates transcription from a promoter bearing a λ operator. We began by introducing random mutations into the β-flap module of either a λCI-β-flap or an α-β-flap fusion protein and screening for amino acid substitutions that reduced expression of the two-hybrid reporter gene in the presence of the partner fusion protein (either α-gp33 or λCI-gp33). To exclude mutations resulting in general effects on the structure of the β-flap moiety, we performed two secondary screens by taking advantage of our ability to detect interactions between the β-flap and 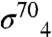 or
or 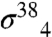 (the stationary phase-specific σ factor). We thus screened for amino acid substitutions in the β-flap that disrupted its interaction with gp33, but did not disrupt its interaction with at least one of the σ4 moieties. We identified six amino acid substitutions (K900E, K900I, K900N, I905V, I905T, and F906S) at three positions in the FTH that met these criteria, consistent with previous data (15) and our structure, showing that the FTH is an important determinant for gp33 binding to the β-flap. We also used the two-hybrid assay to test the effect of individual alanine substitutions at β-flap positions 898-909. Substitutions K900A, L901A, L902A, I905A, and F906A each decreased reporter gene transcription by a factor of at least two (Fig. 3C). These results closely paralleled those obtained with
(the stationary phase-specific σ factor). We thus screened for amino acid substitutions in the β-flap that disrupted its interaction with gp33, but did not disrupt its interaction with at least one of the σ4 moieties. We identified six amino acid substitutions (K900E, K900I, K900N, I905V, I905T, and F906S) at three positions in the FTH that met these criteria, consistent with previous data (15) and our structure, showing that the FTH is an important determinant for gp33 binding to the β-flap. We also used the two-hybrid assay to test the effect of individual alanine substitutions at β-flap positions 898-909. Substitutions K900A, L901A, L902A, I905A, and F906A each decreased reporter gene transcription by a factor of at least two (Fig. 3C). These results closely paralleled those obtained with 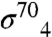 with one exception—substitution E898A was strongly disruptive for the
with one exception—substitution E898A was strongly disruptive for the 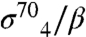 -flap interaction (Fig. S4), but not for the gp33/β-flap interaction (Fig. 3C). These data, along with previous observations (15), specify that gp33 and
-flap interaction (Fig. S4), but not for the gp33/β-flap interaction (Fig. 3C). These data, along with previous observations (15), specify that gp33 and 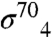 share overlapping but not identical molecular contacts on the β-flap. It should be noted that gp33 interacts much more extensively with the FW than does σ4 (2, 25) and that these interactions would be critical for reorienting the FTH relative to the FW.
share overlapping but not identical molecular contacts on the β-flap. It should be noted that gp33 interacts much more extensively with the FW than does σ4 (2, 25) and that these interactions would be critical for reorienting the FTH relative to the FW.
Focusing on charge reversal substitution β-K900E, we used mutant-suppressor analysis to identify a functional partner residue in gp33. Thus, we introduced random mutations into the gp33 moiety of the α-gp33 fusion protein and screened for amino acid substitutions that restored stimulated expression of the two-hybrid reporter gene in the presence of the λCI-β-flap-K900E fusion protein. One α-gp33 mutant, gp33-E70K, specifically suppressed the effect of β-flap substitution K900E (Fig. 3D), suggesting that these oppositely charged side chains form a salt bridge when gp33 binds the RNAP β-flap.
To determine whether substitutions gp33-E70K and β-K900E were also mutually suppressive in the context of gp33-bound RNAP, we analyzed the repressive effect of gp33 on basal transcription by the gp55-holoenzyme (E•gp55) in vitro. Gp33-E70K was less efficient than wild-type gp33 in repressing transcription by E•gp55 reconstituted with wild-type β, but significantly more efficient than wild-type gp33 in repressing transcription by E•gp55 reconstituted with β-K900E (Fig. S5). These results indicate that the molecular details of the interaction between gp33 and E•gp55 are recapitulated in our two-hybrid assay and provide further support for the interaction between gp33-E70 and β-K900 at the gp33-β-flap interface.
Structure-Based Model of a T4 Late Promoter Transcription Initiation Complex.
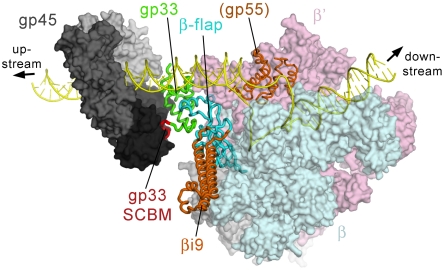
A recently described complete molecular model of Eco core RNAP (23), along with a previously determined structure of the Taq RNAP promoter initiation complex and derived open complex models (1), provide the basis for a model of gp33 bound to a T4 late promoter initiation complex (Fig. 4; Materials and Methods). The model allows us to visualize the overall architecture of the T4 late gene transcription initiation complex. The positioning of the sliding-clamp and the associated SCBM brings the N terminus of the SCBM [corresponding to residue 104 of gp33 according to Table 1 of (34)] within 3.94 Å of the C terminus of the gp33 structure (residue 102; α-carbon to α-carbon distance). The positioning of gp45 and gp33 correlates accurately with biochemical data: (i) when added to a preformed open T4 late promoter complex, gp33 changes the DNase I protection pattern from about -23 to -32 (29), (ii) protection is extended upstream to about -41 in the presence of gp45, and (iii) both gp33 and gp45 cross link to DNA at -34, -35 and -39 (35, 36). In our model, gp33 overlaps in space with 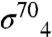 and is located near the DNA from about -29 to -35, whereas gp45 covers the DNA from about -33 to -45, consistent with footprinting and cross-linking analysis.
and is located near the DNA from about -29 to -35, whereas gp45 covers the DNA from about -33 to -45, consistent with footprinting and cross-linking analysis.
Fig. 4.
Structural Model of a T4 late gene Transcription Initiation Complex. A composite model of the complete transcription initiation complex of T4 late gene promoters (see Materials and Methods). Eco RNAP is shown as a molecuar surface (α, ω, gray; β, light cyan; β′, light pink) except the β-flap (from the gp33/β-flap structure), which is shown as a backbone worm and colored as in Fig. 1. Other components of the model include gp55 (thin orange backbone worm, modeled as conserved regions 1.2 and 2 of Taq σA), gp33 (green) but with the SCBM colored red, the gp45 heterotrimer (shown as a molecular surface, with each monomer a different shade of gray). DNA is shown as a yellow backbone worm.
The model allows us to propose functions for conserved gp33 surface patches P3, P4, P5, and P6, that are otherwise difficult to assign, all involving putative protein/protein interactions within the complex. First, side chains of conserved patch P4 are positioned to interact with several basic side chains of the β′-Zinc-Binding-Domain [ZBD (11)], specifically Eco β′-R77 and K79 (Fig. S6A), explaining previous data suggesting that E64 and E65 are involved in contacting RNAP (15). Second, gp33 conserved patch P5 faces the approaching sliding-clamp, suggesting a possible gp33/sliding-clamp interaction in addition to tethering by the gp33-SCBM. The third putative interaction is between gp33 conserved patch P3 (E57) plus additional gp33 residues D91/E92/N93, and a basic helix of β′ on the opposite side of the RNA exit channel (β′-K395-A-A-K-M-V-R403). The side chains of these residues approach within 10 Å, and the β′-helix is connected by loops that may be flexible enough to accommodate electrostatic interactions between the two proteins. Gp33-E57 is important for RNAP binding (15) and is absolutely conserved within the group of gp33 homologs listed in Fig. 2A.
The model further allows us to explain biochemical observations that gp33 reduces the nonspecific DNA binding of core RNAP and represses basal transcription in the absence of the gp45 sliding-clamp (5). Gp33 does not bind DNA sequence-specifically, and the gp33 surface is largely negatively charged (Fig. 2C) so would likely repel DNA. Interestingly, in our transcription complex model, the point of closest approach of the overall negatively charged gp33 with the upstream DNA is the small “basic spot” (Fig. 2C) created by gp33 conserved patch P6 residues R37 and K75.
The model contains ambiguities in several details that prevent us from specifying the rotational setting of the sliding-clamp in the promoter complex (see SI Text) and does not specify the attachment site of the gp55-SCBD; the evidence at hand (37) suggests that, in contrast to gp33, the gp55-SCBD is flexibly linked to the body of this protein.
In the RNAP holoenzyme, σ3.2 in the RNA exit channel and σ4 bound to the β-flap block the exit path for the nascent transcript and must be displaced to allow further extension of the RNA and transition into the elongation complex (38, 39). Modeling of gp33 onto the RNAP ternary elongation complex (40) suggests that gp33 would not occlude the RNA exit channel and could remain bound to the β-flap during elongation (Fig. S6B). This suggestion is consistent with previous observations indicating that gp33 may remain associated with elongation complexes (29). A system wherein gp33 does not need to cycle on and off RNAP for successive rounds of activated transcription might help to optimize late gene transcription.
Conclusion
The T4 late gene promoter complex has been characterized by biochemical and genetic analyses extending over the last four decades, providing a detailed paradigm for how a pathogen manipulates the host transcriptional machinery, and revealing a unique mode of transcription regulation (6). Studies of this system led to the first identification of a master regulator of a complex gene expression pathway (41). In this pathway, gp33 functions as a simple coactivator (19). Structures of most components of the T4 late promoter complex have either been determined or could be accurately modeled based on homologous structures, but gp33 has been a structural “black-box” because it has no homologs in the databases, preventing structural modeling of the entire complex. Using the gp33/β-flap structure reported here, we generated a structural model of the T4 late promoter initiation complex, including Eco RNAP, DNA, the T4-encoded promoter specificity factor gp55, the coactivator gp33, and the activator gp45. The model is consistent with previously determined biochemical and biophysical parameters (29, 35, 36) and predicts additional protein/protein interactions that explain conserved patches of surface-exposed residues on gp33 (Fig. 2A). Some of these predicted protein/protein interactions, between gp33-P4 and the RNAP β′-ZBD (Fig. S6A) and between gp33-P3 and β′[394–403], help to rationalize previously mysterious properties of gp33 point mutants (15). An additional protein/protein interaction, between gp33-P5 and gp45, has not been addressed with biochemical or genetic studies, suggesting future experiments to further characterize this complex.
Materials and Methods
Full details of the methods used are presented in the SI Text.
Protein Expression and Purification.
T4 gp33 and an Eco rpoB fragment encoding the β-flap were coexpressed (Fig. S1A) as described (21). The complex was purified by: Ni2+-affinity chromatography, removal of the His6-tag and uncleaved complex by subtractive Ni2+-affinity and size-exclusion chromatographies (Fig. S1A).
Crystallization and Structure Determination.
Crystals were grown by hanging-drop vapor diffusion at 22 °C. The structure was solved by a combination of molecular replacement and single-wavelength anomalous dispersion with data collected from selenomethionyl-substituted proteins.
Supplementary Material
Acknowledgments.
We thank Andy Yuan for assistance in the initial stages of this project, and Deena Oren of The Rockefeller University Structural biology Resource Center (RU-SBRC) for expert assistance. We thank Wuxian Shi at the National Synchrotron Light Source beamline X3A for assistance with data collection. The use of the Rigaku/MSC microMax 07HF in the RU-SBRC was made possible by grant number 1S10RR022321-01 from the National Center for Research Resources of the National Institutes of Health (NIH). This work was supported by NIH Grants GM44025 (A.H.) and GM053759 (S.A.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3TBI).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113328108/-/DCSupplemental.
References
- 1.Murakami K, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: An RNA polymerase holoenzyme/DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 2.Murakami K, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 3.Rhodius VA, Busby SJ. Positive activation of gene expression. Curr Opin Microbiol. 1998;1:152–159. doi: 10.1016/s1369-5274(98)80005-2. [DOI] [PubMed] [Google Scholar]
- 4.Brody EN, et al. Old phage, new insights: Two recently recognized mechanisms of transcriptional regulation in bacteriophage T4 development. FEMS MIcrobiol Lett. 1995;128:1–8. doi: 10.1111/j.1574-6968.1995.tb07491.x. [DOI] [PubMed] [Google Scholar]
- 5.Herendeen DR, Williams KP, Kassavetis GA, Geiduschek EP. An RNA polymerase-binding protein that is required for communication between an enhancer and a promoter. Science. 1990;278:573–578. doi: 10.1126/science.2185541. [DOI] [PubMed] [Google Scholar]
- 6.Geiduschek EP, Kassavetis GA. Transcription of the T4 late genes. Virol J. 2010;7:288–300. doi: 10.1186/1743-422X-7-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanders GM, Kassavetis GA, Geiduschek EP. Dual targets of a transriptional activator that tracks on DNA. EMBO J. 1997;16:3124–3132. doi: 10.1093/emboj/16.11.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong K, Geiduschek EP. Activator-sigma interaction: a hydrophobic segment mediates the interaction of a sigma family promoter recognition protein with a sliding clamp transcription activator. J Mol Biol. 1998;284:195–203. doi: 10.1006/jmbi.1998.2166. [DOI] [PubMed] [Google Scholar]
- 9.Gribskov M, Burgess RR. Sigma factors from E coli, B. subtilis, phage SPO1, and phage T4 are homologous proteins. Nucleic Acids Res. 1986;14:6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lonetto M, Gribskov M, Gross CA. The σ70 family: Sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lane WJ, Darst SA. Molecular evolution of multi-subunit RNA polymerases: structural analysis. J Mol Biol. 2010;395:686–704. doi: 10.1016/j.jmb.2009.10.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong K, Kassavetis GA, Leonetti J-P, Geiduschek EP. Mutational and functional analysis of a segment of the sigma family bacteriophage T4 late promoter recognition protein gp55. J Biol Chem. 2003;278:7073–7080. doi: 10.1074/jbc.M211447200. [DOI] [PubMed] [Google Scholar]
- 13.Kassavetis GA, Geiduschek EP. Defining a bacteriophage T4 late promoter: bacteriophage T4 gene 55 protein suffices for directing late promoter recognition. Proc Natl Acad Sci USA. 1984;81:5101–5105. doi: 10.1073/pnas.81.16.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen AC, Young ET. T4 late transcripts are initiated near a conserved DNA sequence. Nature. 1982;299:369–371. doi: 10.1038/299369a0. [DOI] [PubMed] [Google Scholar]
- 15.Nechaev S, Kamali-Moghaddam M, Andre E, Leonetti J-P, Geiduschek EP. The bacteriophage T4 late-transcription coactivator gp33 binds the flap domain of Escherichia coli RNA polymerase. Proc Natl Acad Sci USA. 2004;101:17365–17370. doi: 10.1073/pnas.0408028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams KP, Muller R, Ruger W, Geiduschek EP. Overproduced bacteriophage T4 gene 33 protein binds RNA polymerase. J Bacteriol. 1989;171:3579–3582. doi: 10.1128/jb.171.6.3579-3582.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolesky SE, Ouhammouch M, Geiduschek EP. The mechanism of transcriptional activation by the topologically DNA-linked sliding clamp of bacteriophage T4. J Mol Biol. 2002;321:767–784. doi: 10.1016/s0022-2836(02)00732-5. [DOI] [PubMed] [Google Scholar]
- 18.Riva S, Cascino A, Geiduschek EP. Coupling of late transcription to viral replication in bacteriophage T4 development. J Mol Biol. 1970;54:85–102. doi: 10.1016/0022-2836(70)90447-x. [DOI] [PubMed] [Google Scholar]
- 19.Lewin B. Commitment and activation at pol II promoters: a tail of protein-protein interactions. Cell. 1990;61:1161–1164. doi: 10.1016/0092-8674(90)90675-5. [DOI] [PubMed] [Google Scholar]
- 20.Campbell EA, Darst SA. The anti-σ factor SpoIIAB forms a 2∶1 complex with σF, contacting multiple conserved regions of the σ factor. J Mol Biol. 2000;300:17–28. doi: 10.1006/jmbi.2000.3838. [DOI] [PubMed] [Google Scholar]
- 21.Hendrickson W, Norton JR, LeMaster DM. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD) EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane WJ, Darst SA. Molecular evolution of multi-subunit RNA polymerases: sequence analysis. J Mol Biol. 2010;395:671–685. doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Opalka N, et al. Complete structural model of Escherichia coli RNA polymerase from a hybrid approach. PLoS Biol. 2010;8:e1000483. doi: 10.1371/journal.pbio.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang G, et al. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 25.Vassylyev DG, et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 26.Kuznedelov K, et al. Recombinant Thermus aquaticus RNA polymerase for structural studies. J Mol Biol. 2006;359:110–121. doi: 10.1016/j.jmb.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Kuznedelov K, et al. A role for interaction of the RNA polymerase flap domain with the sigma subunit in promoter recognition. Science. 2002;295:855–857. doi: 10.1126/science.1066303. [DOI] [PubMed] [Google Scholar]
- 28.Campbell EA, et al. Structure of the bacterial RNA polymerase promoter specificity sigma factor. Mol Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 29.Nechaev S, Geiduschek EP. The role of an upstream promoter interaction in initiation of bacterial transcription. EMBO J. 2006;25:1700–1709. doi: 10.1038/sj.emboj.7601069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landau M, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geszvain K, Gruber TM, Mooney RA, Gross CA, Landick R. A hydrophobic patch on the flap-tip helix of E. coli RNA polymerase mediates σ70 region 4 function. J Mol Biol. 2004;343:569–587. doi: 10.1016/j.jmb.2004.08.063. [DOI] [PubMed] [Google Scholar]
- 33.Dove SL, Hochschild A. A bacterial two-hybrid system based on transcription activation. Methods in Molecular Biology. 2004;261:231–246. doi: 10.1385/1-59259-762-9:231. [DOI] [PubMed] [Google Scholar]
- 34.Shamoo Y, Steitz TA. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- 35.Tinker RL, Sanders GM, Severinov K, Kassavetis GA, Geiduschek EP. The COOH-terminal domain of the RNA polymerase a subunit in transcriptional enhancement and deactivation at the bacteriophage T4 late promoter. J Biol Chem. 1995;270:15899–15907. doi: 10.1074/jbc.270.26.15899. [DOI] [PubMed] [Google Scholar]
- 36.Tinker RL, Williams KP, Kassavetis GA, Geiduschek EP. Transcriptional activation by a DNA-tracking protein: structural consequences of enhancement at the T4 late promoter. Cell. 1994;77:225–237. doi: 10.1016/0092-8674(94)90315-8. [DOI] [PubMed] [Google Scholar]
- 37.Nechaev S, Geiduschek EP. Dissection of the bacteriophage T4 late promoter complex. J Mol Biol. 2008;379:402–413. doi: 10.1016/j.jmb.2008.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nickels BE, et al. The interaction between σ70 and the σ-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc Natl Acad Sci USA. 2005;102:4488–4493. doi: 10.1073/pnas.0409850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nickels BE, Roberts CW, Roberts JW, Hochschild A. RNA-mediated destabilization of the σ70 region 4/β flap interaction facilitates engagement of RNA polymerase by the Q antiterminator. Mol Cell. 2006;24:457–468. doi: 10.1016/j.molcel.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 41.Epstein RH, et al. Physiological studies of conditional lethal mutants of bacteriophage T4D. Cold Spring Harbor Symp Quant Biol. 1963;28:375–394. [Google Scholar]
- 42.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.