Abstract
Human artificial chromosome (HAC)-based vectors offer a promising system for delivery and expression of full-length human genes of any size. HACs avoid the limited cloning capacity, lack of copy number control, and insertional mutagenesis caused by integration into host chromosomes that plague viral vectors. We previously described a synthetic HAC that can be easily eliminated from cell populations by inactivation of its conditional kinetochore. Here, we demonstrate the utility of this HAC, which has a unique gene acceptor site, for delivery of full-length genes and correction of genetic deficiencies in human cells. A battery of functional tests was performed to demonstrate expression of NBS1 and VHL genes from the HAC at physiological levels. We also show that phenotypes arising from stable gene expression can be reversed when cells are “cured” of the HAC by inactivating its kinetochore in proliferating cell populations, a feature that provides a control for phenotypic changes attributed to expression of HAC-encoded genes. This generation of human artificial chromosomes should be suitable for studies of gene function and therapeutic applications.
Keywords: gene delivery vector, gene therapy, transformation-associated recombination cloning
Most advanced current gene therapy systems, such as adenovirus-, lentivirus-, and retrovirus-derived vectors, use cDNA or “minigene” constructs (reviewed in refs. 1–5) that cannot recapitulate the physiological regulation of complex endogenous loci. Viral episomal vectors carrying HSV-1 and EBV amplicons can deliver and express full-length genes as large as approximately 150 kb in size (1, 2). However, several concerns limit the use of HSV-1 and EBV viral vectors: absence of strong copy number control, undesired immunological responses, and occasional integration into the host genome, causing insertional mutagenesis and gene silencing (6).
Human artificial chromosomes (HACs) represent another extrachromosomal gene delivery and gene expression vector system (7–11). Although this technology is less advanced than virus-derived vectors, HACs have several potential advantages over currently used episomal viral vectors for gene therapy applications. First, the presence of a functional centromere provides long-term stable maintenance of HACs as single-copy episomes without integration to the host chromosomes. Second, there is no upper size limit to DNA cloned in a HAC: entire genomic loci with all regulatory elements can be used. Finally, HAC vectors minimize adverse host immunogenic responses and the risk of cellular transformation.
Recent advances have produced a variety of HACs via two different approaches. The “top-down” approach involves modification of human chromosomes in living cells to produce chromosome derivatives (11–18). The “bottom-up” approach involves de novo kinetochore assembly from 50- to 100-kb blocks of centromeric alpha-satellite (i.e., alphoid) DNAs. These are multimerized in human cells, forming HACs with sizes of approximately 1 to 3 Mb (7, 8, 10, 19). Several groups have reported successful expression of full-length genes in HACs (9–11, 20, 21). However, in most cases, genes to be expressed were cotransfected with an alphoid DNA array into human cells and were incorporated into the forming HAC in vivo. Therefore, gene copy number and location of the gene on the HAC were not predetermined. These factors greatly limited the application of de novo-generated HACs as gene delivery vectors.
Recently, this limitation was overcome by construction of an alphoidtetO-HAC carrying loxP sites (22, 23). This circular HAC contains approximately 6,000 copies of the tetracycline operator (tetO) sequence embedded in a synthetic alphoid DNA array. Although the HAC is mitotically stable, tethering certain tetracycline repressor fusion proteins (tetR-KAP1 or tTS) (22, 24) inactivates the centromere, resulting in HAC loss. Thus, the alphoidtetO-HAC and any genes it carries can be eliminated from cell populations, allowing for a regulated “hit-and-run” induction of phenotypic changes.
To minimize problems caused by the lack of certain genes from libraries of BAC and YAC clones or the presence of genes fragmented on multiple vectors, we developed a method for direct gene cloning from genomic DNA. Transformation-associated recombination (TAR) cloning allows isolation of any specific allele of a gene of interest as a predetermined DNA fragment (25, 26).
Here, we describe the combination of the TAR gene-cloning technology with the alphoidtetO-HAC vector for gene delivery. We report a complete cycle starting with selective gene isolation, followed by gene loading into the HAC, and eventually leading to complementation of gene deficiencies in a human cell line (Fig. 1). This approach is useful for studies of gene function and potentially for gene therapy. As a proof of principle, genomic copies of two average-size cancer-associated genes—VHL mutated in von Hippel–Lindau syndrome (VHL; MIM 193300) and NBS1 mutated in Nijmegen breakage syndrome (NBS; MIM 251260)—were isolated by TAR cloning and loaded into the unique loxP site of the alphoidtetO-HAC in CHO cells. HAC transfer into human cells deficient in NBS1 and VHL reveals that both genes are expressed normally and complement the defective endogenous alleles.
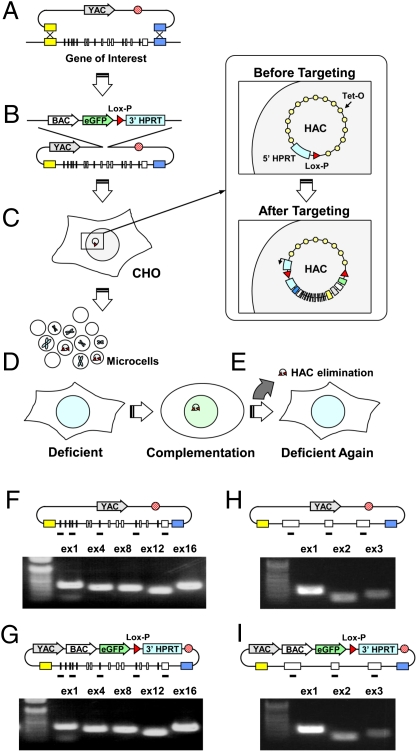
Fig. 1.
A scheme of consecutive experimental steps from selective gene isolation to its expression in gene-deficient human cells. (A) Direct TAR isolation of the NBS1 and VHL genes from human genomic DNA. TAR vectors contain two gene targeting hooks (yellow and blue boxes). (B) Retrofitting of the circular YAC containing the full-length gene by the pJBRV1 vector containing a 3′ HPRT-loxP-eGFP cassette. Recombination of the BamHI-linearized pJBRV1 vector with a YAC in yeast leads to replacement of the ColE1 origin of replication by the F′ factor origin of replication, allowing subsequent propagation in BAC form. (C) Gene loading into a unique loxP site of the alphoidtetO-HAC by Cre-loxP recombination in CHO cells. (D) MMCT of alphoidtetO-HAC/NBS1 or VHL from CHO into gene-deficient cells for complementation analysis (E) Elimination of the alphoidtetO-HAC/NBS1 or VHL from cells by expression of the tTS fusion construct. (F and G) Analysis of TAR clones containing the full-length NBS1 gene for the presence of exons before (F) and after (G) retrofitting. (H and I) Analysis of TAR clones containing the full-length VHL gene for the presence of exons before (H) and after (I) retrofitting.
Results
Isolation of Genomic Regions Containing NBS1 and VHL Genes from Human Genome by TAR Cloning.
The TAR cloning scheme for isolating the NBS1 and VHL genes is shown in Fig. 1A. For cloning purposes, TAR vectors were designed containing short targeting hooks specific to 5′ and 3′ sequence ends of the genes (SI Materials and Methods). Human genomic DNA was transformed into yeast spheroplasts along with linearized TAR vectors. Recombination between the hooks and genomic DNA fragments results in isolation of NBS1 and VHL genes as circular YACs with insert sizes of approximately 55 kb and 25 kb, respectively. Five independent TAR YAC isolates were obtained for each gene. The cloned NBS1 locus contains approximately 5 kb sequence upstream of the ATG codon and 1.5 kb sequence downstream of the stop codon. The cloned VHL locus contains approximately 10 kb sequence upstream of the ATG codon and 7.3 kb sequence downstream of the stop codon. PCR analysis and physical characterization of the TAR isolates for each gene confirmed that the clones contain all exons (Fig. 1 F and H) and did not reveal any structural rearrangements. Analysis of the NBS1 genomic segments is shown in Fig. S1.
Conversion of NBS1- and VHL-YACs into BACs with a LoxP Cassette for Gene Loading into AlphoidtetO-HAC.
A yeast-bacteria-mammalian cell shuttle vector, pJBRV1, was constructed to retrofit YAC gene isolates into YAC/BACs (Figs. S2 and S3). The vector contains a 3′ HPRT-loxP-eGFP cassette, allowing gene loading into a unique loxP site of the alphoidtetO-HAC in CHO cells. An F′ factor origin of replication allows YAC propagation as a BAC molecule. Conversion of the YAC into a BAC is advantageous because purification of circular DNA molecules is much easier from Escherichia coli than from yeast cells. The protocol for retrofitting is shown in Fig. 1B and Fig. S2. Integrity of NBS1- and VHL-containing BACs was confirmed by PCR amplification of all exons (Fig. 1 G and I) and analysis of their inserts by pulse-gel electrophoresis (Fig. S1D).
Loading of NBS1 and VHL Genes into AlphoidtetO-HAC by Cre-loxP–Mediated Recombination in CHO Cells.
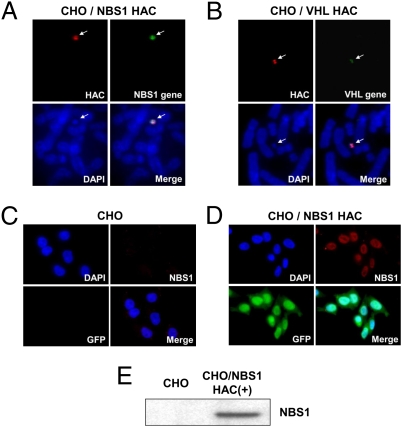
The alphoidtetO-HAC with a unique gene loading site was used for this purpose. This HAC was identified among several HAC clones carrying a loxP cassette(s) (23) by Southern blot hybridization. To insert genomic copies of the NBS1 and VHL genes into the alphoidtetO-HAC, the appropriate BAC constructs and a Cre-recombinase expression vector were cotransfected into hprt-minus CHO cells carrying the HAC and HPRT-plus colonies were selected on HAT medium (Fig. 1C). Insertion of genes into the HAC was confirmed by PCR by using specific primers (Table S1) to detect reconstitution of the HPRT gene, which accompanies Cre/Lox targeting. FISH images of alphoidtetO-HAC/NBS1 and alphoidtetO-HAC/VHL are shown in Fig. 2 A and B. The HACs are maintained autonomously and the HAC signal colocalizes with NBS1 and VHL gene signals on metaphase chromosome spreads. Immunocytochemistry detected homogeneous pNBS1 expression in the nucleus of all cells (Fig. 2 C and D). Western blot analysis of the alphoidtetO-HAC/NBS1 clone with human-specific antibodies against pNBS1 revealed that the NBS1 gene inserted into the alphoidtetO-HAC expresses a protein of the predicted size (Fig. 2E). Because antibodies against human pVHL produce a band with the mobility of hVHL on Western blots in naive CHO cell lysates, we used RT-PCR analysis to prove expression of the VHL gene. RT-PCR products of the predicted size were obtained (Fig. S4A), and their identity was confirmed by sequencing.
Fig. 2.
Expression of NBS1 and VHL genes loaded into the HAC in CHO cells. (A) FISH analysis of the alphoidtetO-HAC/NBS1 in CHO cells with specific probes for the BAC vector (in red) and for gene sequences (green). (B) FISH analysis of alphoidtetO-HAC/VHL in CHO cells. (C) Immunocytochemical staining of CHO cells and (D) isogenic cells with the alphoidtetO-HAC/NBS1 using Abs against pNBS1 (red) and GFP (green). (E) Western blot analysis of a CHO clone containing alphoidtetO-HAC/NBS1 with human-specific Abs against pNBS1. NBS1 inserted into the alphoidtetO-HAC produces a protein of the predicted size.
Functional Complementation of Genetic Deficiency by AlphoidtetO-HAC/NBS1.
To prove its use as a genetic complementation vector, the NBS1-containing HAC was transferred to NBS1-deficient GM07166 (657del5) human cells via microcell-mediated chromosome transfer (MMCT; Fig. 1D). FISH analysis confirmed that alphoidtetO-HAC/NBS1 propagates autonomously without detectable integration into chromosomes (Fig. 3A). Western blot analysis confirmed that this clone expresses pNBS1 (Fig. 3B). To prove the functionality of the conditional kinetochore and confirm that pNBS1 is expressed from the HAC, we targeted the tTS transcriptional repressor to the HAC centromere in the alphoidtetO-HAC/NBS1–containing GM07166 cells (Fig. 1E). This targeting results in a high frequency of HAC loss, as described previously (23, 24) (Fig. S5). Based on FISH and Western blot analysis, cells that lost the HAC no longer expressed pNBS1 (Fig. 3B).
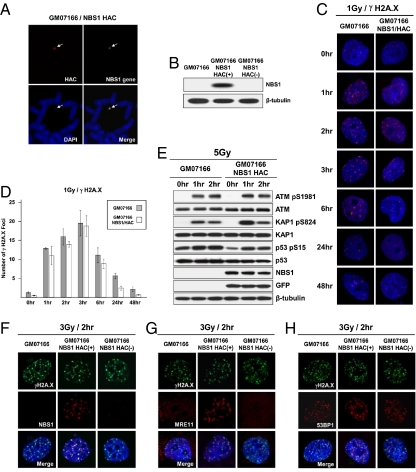
Fig. 3.
pNBS1 expression in NBS1-deficient cells. (A) FISH analysis of the alphoidtetO-HAC/NBS1 in GM07166 cells with specific probes for the BAC vector (red) and NBS1 gene sequences (green). (B) Western blot analysis of NBS1-deficient GM07166 cells, alphoidtetO-HAC/NBS1-containing GM07166 cells before and after HAC elimination from the cells by expression of the tTS fusion construct. (C and D) Kinetics of disassembly of γ-H2A.X foci in NBS1-deficient GM07166 cells and the same cells with alphoidtetO-HAC/NBS1. (C) Immunostaining of irradiated cells. Cells were stained with anti–γ-H2A.X antibodies (red) and with DAPI (blue) after exposition to 1 Gy IR and then collected after the times indicated. (D) Quantitative analysis of disassembly of γ-H2A.X foci. Cells were collected after the time indicated, and the average number of γ-H2A.X foci was quantified over time. (E) Western analysis of GM07166 cells and cells with alphoidtetO-HAC/NBS1. As predicted, p53, ATM, and KAP1 are phosphorylated in response to irradiation. (F–H) Analysis of colocalization of γ-H2A.X, MRE11, 53BP1, and NBS1 proteins after irradiation. NBS1-deficient cells (GM07166) and the same cells with alphoidtetO-HAC/NBS1 before and after HAC loss were fixed 2 h after irradiation with 3 Gy and were double-stained with anti–γ-H2A.X, anti-hMRE11, anti-53BP, and anti-hNBS1 antibodies. (F) NBS1 is colocalized with γ-H2A.X in cells carrying the alphoidtetO-HAC/NBS1. (G) γ-H2A.X and MRE11 are colocalized in foci in a pNBS1-dependent manner. (H) 53BP1 foci formation is not affected by absence of pNBS1.
We used physiological tests to prove that alphoidtetO-HAC/NBS1 produces functional pNBS1. pNBS1 plays a critical role in damage responses to DNA double-strand breaks (DSBs). Within seconds of generation of DSBs, histone H2A.X molecules are phosphorylated by ATM and ATR on serine residues. The resulting γ-H2A.X spreads along megabase chromatin domains flanking the DSB site (reviewed in refs. 27, 28). pNBS1 then recruits the Mre11/Rad50/NBS1 complex to the break site. Although γ-H2A.X foci formed in NBS1-deficient GM07166 cells after irradiation, as expected (ref. 29 and references therein), MRE11 protein did not colocalize with γ-H2A.X (Fig. 3 F–H). In contrast, in GM07166 cells bearing alphoidtetO-HAC/NBS1, localization of NBS1 and MRE11 to damage-induced γ-H2A.X foci was observed (Fig. 3 F–H). This colocalization was lost in GM07166 cells that had been “cured” of the HAC (Fig. 3 F–H). As a control, we examined colocalization of γ-H2A.X and 53BP1 in the same cells. As shown by others, 53BP1 localization to damage foci is not affected by absence of pNBS1 (ref. 29 and references therein). Indeed, γ-H2A.X and 53BP1 irradiation-induced foci colocalized independently of the presence of functional pNBS1 in GM07166 cells (Fig. 3 F–H and Fig. S6).
Analysis of three proteins—ATM, KAP1, and p53—that should be modified in response to irradiation revealed phosphorylation of the predicted amino acid residues in the analyzed cells (Fig. 3E). We then analyzed kinetics of the disassembly of γ-H2A.X foci, which closely parallels the rate of DSB repair. The kinetics slows down in NBS1-deficient cells similar to that in cells deficient for ATM and 53BP1 (30). GM07166 cells bearing alphoidtetO-HAC/NBS1 showed a more efficient disassembly of foci compared with cells without HAC after 24 h (Fig. 3 C and D). The dynamics of foci disassembly in the cells without and with the HAC was identical to that observed in cultured lymphocytes from patients with NBS and normal individuals, correspondingly (31). Thus, these results indicate a higher rate of rejoining of DSBs in GM07166 cells bearing alphoidtetO-HAC/NBS1. Altogether, our observations demonstrate that the alphoidtetO-HAC/NBS1 vector expresses a functional protein.
Functional Complementation of Genetic Deficiency by AlphoidtetO-HAC/VHL.
When transferred to 786-0 renal carcinoma cells (RCCs) via MMCT, alphoidtetO-HAC/VHL propagated autonomously without integrating into host chromosomes (Fig. 4A). These cells are VHL-negative as a result of a frameshift mutation in the coding region of the gene (311delG). Expression of VHL in 786-0 cells bearing alphoidtetO-HAC/VHL was confirmed by RT-PCR by using primers that specifically amplify the WT but not the mutant allele of VHL (Fig. S4B). To prove that VHL is expressed from the HAC, a vector expressing a tTS fusion was introduced into cells to induce HAC loss (23, 24). Several GFP-negative clones were selected. Based on FISH and RT-PCR analyses, these clones lost the VHL along with the HAC.
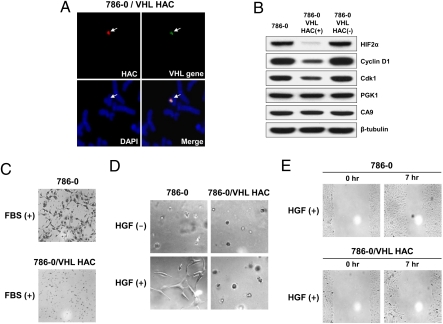
Fig. 4.
pVHL expression in VHL-deficient cells. (A) FISH analysis of the alphoidtetO-HAC/VHL in VHL-deficient 786-0 RCC4 cells. (B) Effect of pVHL on the level of HIF2-α, CyclinD1, and Cdk1/CDC2 proteins. Immunoblots of whole cell extracts isolated from the 786-0 cell line, the same cell line containing alphoidtetO-HAC/VHL before and after induction of HAC loss. CA9 and PGK1 proteins, which are not affected by pVHL status in 786-0 cells, were used as controls. (C) The level of invasiveness of VHL-deficient 786-0 cells and the same cell line containing alphoidtetO-HAC/VHL in the presence of FBS. (D) pVHL regulation of HGF-mediated (HGF+) 786-0 cell branching morphogenesis in the absence and presence of functional pVHL (with or without alphoidtetO-HAC/VHL). (E) pVHL regulation of 786-0 cell migration. pVHL expression in 786-0 cells results in reduction of cellular migration. A clear difference was observed after 7 h when representative fields were photographed. Each experiment was performed in triplicate.
Several specific tests based on the known functions of pVHL were carried out to prove functional stable expression of pVHL from the HAC in recipient cells. A critical role of pVHL in down-regulation of HIF, CyclinD1, and Cdk1/CDC2 is well documented (32–35). pVHL has E3 ubiquitin ligase activity and targets α-subunits of HIF for rapid degradation by the proteasome under normoxia. Those subunits are stabilized by hypoxia (reviewed in ref. 36). We compared the levels of HIF2α, CyclinD1, and CDC2 expression in VHL-deficient and isogenic cells containing alphoidtetO-HAC/VHL by Western blot analysis. As predicted, levels of HIF2α, CyclinD1, and CDC2 were significantly lower in cells carrying alphoidtetO-HAC/VHL (Fig. 4B), suggesting that pVHL expressed from the HAC restores their normal down-regulation. Indeed, 786-0 cells cured of the alphoidtetO-HAC/VHL vector by centromere inactivation expressed the VHL-regulated proteins at their original elevated levels (Fig. 4B). PGK1 and CA9 expression is regulated by hypoxia and more specifically by the HIF1-α isoform (PubMed ID no. 15964822). Because the 786-0 cell line lacks expression of HIF1-α, no reduction of CA9 and PGK1 was observed after induction of pVHL in 786-0 cells (Fig. 4B).
Three additional tests confirmed the functional expression of pVHL in recipient alphoidtetO-HAC/VHL cells. In RCCs, hepatocyte growth factor (HGF) and FBS stimulate branching morphogenesis, cell invasiveness, and cell migration via a VHL-dependent pathway (37, 38). We therefore assessed those phenotypes in the VHL-deficient 786-0 RCC parental cell line and its isogenic derivative containing the alphoidtetO-HAC/VHL vector (Fig. 4 C–E). Addition of exogenous HGF resulted in branching of VHL-deficient 786-0 cells in a growth factor-reduced Matrigel assay. In contrast, stable expression of pVHL from the alphoidtetO-HAC/VHL vector abrogated this morphologic response of 786-0 cells to HGF (Fig. 4D). Similarly, the relatively high basal level of invasiveness of VHL-deficient 786-0 cells was markedly stimulated by addition of FBS. This response was dramatically suppressed in the isogenic 786-0 cell line containing the alphoidtetO-HAC/VHL vector (Fig. 4C). Finally, migration of 786-0 cells with alphoidtetO-HAC/VHL was also reduced relative to the parental cell line (Fig. 4E).
This battery of functional tests shows that the VHL gene expressed from the alphoidtetO-HAC/VHL produces a functional product that complements the VHL-deficiency in 786-0 cells.
Proximity of CENP-A Chromatin Is Compatible with Gene Expression.
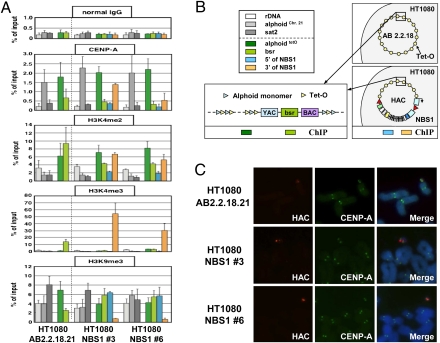
During de novo HAC formation, input DNA is multimerized and then assembled into different types of chromatin, including arrays of CENP-A nucleosomes that are interspersed with those carrying different modifications of histone H3 (39). To better understand the chromatin structure of regions flanking the NBS1 gene in the alphoidtetO HAC, alphoidtetO-HAC/NBS1 was transferred back to human HT1080 cells (Fig. S7) via MMCT. The alphoidtetO-HAC was generated in this host and its chromatin structure in those cells has been previously characterized (22). After MMCT, two clones (NBS1 no. 3 and NBS1 no. 6) were selected (Fig. 5). Immuno-FISH analysis confirmed the assembly of CENP-A centromeric chromatin on the HAC sequences (Fig. 5C). ChIP analysis of these clones showed a chromatin pattern similar to that in the original alphoidtetO-HAC clone (AB2.2.18.21) (22) for CENP-A chromatin as well as H3K4me3 (a marker for transcriptionally active chromatin), H3K4me2 (a marker for open chromatin) and H3K9me3 (a marker for heterochromatin) on alphoidtetO DNA (Fig. 5 A and B). These results confirm that the alphoidtetO-HAC/NBS1 retained its kinetochore chromatin structure after several rounds of MMCT and gene insertion. ChIP analysis of regions flanking the NBS1 gene revealed a robust level of CENP-A chromatin enrichment at the 3′ end of NBS1 in clone NBS1 no. 3 and a lower, but still significant, enrichment in clone NBS1 no. 6 (Fig. 5A). This indicates that the CENP-A chromatin domain extends to beyond the gene insertion site. CENP-A enrichment at the 3′ NBS1 sequence may be explained by the ability of CENP-A chromatin to spread onto noncentromeric DNA (16, 39, 40). The observed stable gene expression in the HAC suggests that close proximity of the CENP-A domain does not prevent expression of Pol II-transcribed genes.
Fig. 5.
Immuno-FISH and ChIP analyses of the alphoidtetO-HAC/NBS1. (A) ChIP analysis of centromeric chromatin in alphoidtetO-HAC/NBS1 clones 3 and 6. Normal mouse IgG (Top), antibodies against CENP-A, H3K4me2, H3K4me3, and H3K9me3 were used for analysis. The assemblies of these proteins on the alphoid DNA of the original alphoidtetO-HAC in AB2.2.18.21 cell line (Left), the alphoidtetO-HAC carrying the human NBS1 gene in NBS1 no. 3 and NBS1 no. 6 cell lines (Right), are shown. The bars show the percentage recovery of the various target DNA loci by immunoprecipitation with each antibody to input DNA. Error bars indicate SD (n = 2 or 3). Analyzed loci are alphoidtetO (alphoid DNA with tetO motif on the alphoidtetO-HAC), bsr (the marker gene in the HAC vector sequence), and 3′ and 5′ ends of NBS1. rDNA (5S ribosomal DNA), alphoidchr.21 (centromeric alphoid DNA of chromosome 21), and sat2 (pericentromeric satellite 2) were used as controls. Immunoprecipitated DNAs were quantified by real-time PCR. (B) Positions of probes for ChIP analysis in the original HAC and in the HAC carrying the NBS1 gene are shown by colored boxes. (C) Immuno-FISH analysis of metaphase chromosome spreads containing the alphoidtetO-HACs. Cells with the original alphoidtetO-HAC (AB2.2.18.21) and with alphoidtetO-HAC carrying the NBS1 gene (clones 3 and 6) were used for analysis. Immunolocalization of the centromeric protein CENP-A on metaphases was performed by indirect immunofluorescence with anti–CENP-A antibody and Alexa 488-conjugated secondary antibody (green). HAC-specific DNA sequence (BAC) was used as a FISH probe to detect the HAC (red). CENP-A and BAC signals on the HACs overlap one another.
Discussion
HACs represent a unique system for delivery and expression of full-length genes that has several advantages over currently used episomal virus-based vectors. Several groups have reported successful expression of full-length genes in HACs (9–11, 20, 21). The most advanced top-down HAC vectors are truncated derivatives of chromosome 21. Determination of the structure of one such derivative, 5-Mb 21ΔpqHAC, opened the way for its use in clinical applications (18). Recently, this HAC was successfully used for delivery and expression of several full-length human genes, including the 2-Mb Duchenne Muscular Dystrophy (DYS) gene (41). However, a common limitation of all HACs constructed so far is the inability to induce HAC loss to enable transient gene expression or to have a negative control for phenotypic changes attributed to expression of the gene loaded into the HAC.
In our previous work, we developed a synthetic alphoidtetO-HAC with a conditional centromere that can be inactivated (22). This HAC contains a unique loxP site that allows selection for gene loading (23).
In the present study, we analyzed the capacity of the alphoidtetO-HAC to deliver genomic copies of average-size genes into human cells and complement genetic deficiencies. We found that the VHL and NBS1 genes obtained by TAR cloning can be efficiently and accurately introduced into the alphoidtetO-HAC, resulting in artificial chromosomes that can be propagated indefinitely in CHO cells. Because CHO cells exhibit a high efficiency of microcell formation, this enables transfer of the HAC to other cell types, including mouse ES cells, via MMCT (23, 42).
The HACs were transferred into cell lines derived from patients with deficiencies in VHL or NBS1. Functional expression of pVHL and pNBS1 in recipient cells was demonstrated by a set of specific tests based on the known functions of proteins. Importantly, corresponding controls could be conducted following specific elimination (i.e., curing) of the HAC from the cells following targeted inactivation of the kinetochore.
No significant changes in the level of expression of pNBS1 were detected for more than 3 mo following introduction of the alphoidtetO-HAC into patient-derived cell lines (Fig. S8). Although additional studies are needed to determine whether genes inserted into the alphoidtetO-HAC may eventually be subject to silencing, our results and those obtained with other HACs (21, 41) support the view that proximity to a functional kinetochore does not negatively affect gene expression. It is possible that, in the human genome, centromeric chromatin may be a privileged region for Pol II-transcribed genes, as previously shown for rice centromeres (43) and suggested by the resemblance of the centromeric chromatin profile to the downstream region of transcribed genes in human cells (39, 44). Indirectly, this hypothesis is supported by our data on spreading of CENP-A chromatin to the 3′ end of NBS1 and transcriptional activity of genes in neocentromeres (reviewed in ref. 40).
The alphoidtetO-HAC, with its conditional centromere, has significant advantages over other expression/cloning systems when transient expression of a gene of interest is needed. For example, generation of iPS cells requires only transient expression of specific cellular factors such as OCT4, SOX2, KLF4, cMYC, and LIN28 (45). Therefore, the ability to eliminate the HAC along with any stem cell-inducing genes carried on it could provide a strategy to avoid insertional mutagenesis and cell transformation, complications that are frequently observed during cell reprogramming (46). To adopt the alphoidtetO-HAC vector for such purpose, a HAC inactivation module consisting of the doxycycline-regulated tTS fusion is being incorporated into the HAC allowing induction of its loss by a simple addition of a ligand. For gene therapy, the HAC may be inserted into stem cells in the laboratory and then these stem cells may be introduced into the patient. It is worth noting that efficient formation and stable maintenance of HACs has been demonstrated in stem cells (47).
To summarize, in this work, we demonstrate the utility of an alphoidtetO-HAC vector for delivery of full-length genes and correction of genetic deficiencies in human cells. We also demonstrate the benefit of coupling the TAR gene cloning technology, which provides an effectively unlimited resource of full-length human genes, with the tetO-loxP-HAC gene delivery and expression system. Because the structure of the alphoidtetO-HAC is determined, this system may be potentially useful for gene therapy.
Materials and Methods
Construction of TAR Cloning Vectors.
The TAR circularizing vectors, pVC-NBS1 and pVC-VHL, containing 5′ and 3′ sequences of the NBS1 and VHL genes, were constructed by using the basic vector pVC604 (26). Details of the construction are described in SI Materials and Methods.
Construction of pJBRV1 Retrofitting Vector and Conversion of NBS1- and VHL-YACs into BACs Containing a loxP Site.
A diagram of the pJBRV1 vector and retrofitting of a circular YAC into a BAC is shown in Figs. S2 and S3. Details are described in SI Materials and Methods.
Loading of Genomic Fragments into Unique loxP Site of AlphoidtetO-HAC.
Details of gene loading are described in SI Materials and Methods.
MMCT.
AlphoidtetO-HAC/NBS1 and alphoidtetO-HAC/VHL were transferred from CHO cells to GM07166 and RCC 786-0 cell lines deficient for NBS1 and VHL genes, respectively, by using a standard MMCT protocol (23, 41, 42). Details of MMCT are described in SI Materials and Methods.
AlphoidtetO-HAC Elimination by Its Targeting with Chromatin Modifiers.
Induction of HAC loss by inactivation of its kinetochore was performed as previously described (23, 24). The kinetics of HAC loss in response to tTS expression are shown in Fig. S5.
Antibodies.
The antibodies used for immunoblotting and immunostaining are described in SI Materials and Methods.
In Vitro Invasion, Cell Migration, and Branching-Morphogenesis Assays.
Cell invasion, cell migration, and branching-morphogenesis assays of 786-0 cells and 786-0 cells containing alphoidtetO-HAC/VHL were carried out as described previously (37, 38).
ChIP Analysis.
ChIP analysis was performed as described previously (22, 24, 44). Details of ChIP are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Hee-Sheung Lee for technical advice and help with microcell-mediated chromosome transfer experiments. We also thank Olga Aprelikova and Bill Bonner [National Cancer Institute (NCI), National Institutes of Health (NIH)] for helpful discussion. This study was supported by the Intramural Research Program of the NIH NCI Center for Cancer Research (V.L.); grant-in-aid from the Ministry of Education, Science, Sports and Culture of Japan (H.M.); and the 21st Century Center of Excellence program from the Ministry of Education, Culture, Sports, Science and Technology of Japan (M.O.). Work in the laboratory of W.C.E. is funded by the Wellcome Trust, of which W.C.E. is a principal research fellow.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114483108/-/DCSupplemental.
References
- 1.Lufino MM, Edser PA, Wade-Martins R. Advances in high-capacity extrachromosomal vector technology: Episomal maintenance, vector delivery, and transgene expression. Mol Ther. 2008;16:1525–1538. doi: 10.1038/mt.2008.156. [DOI] [PubMed] [Google Scholar]
- 2.Epstein AL. Progress and prospects: Biological properties and technological advances of herpes simplex virus type 1-based amplicon vectors. Gene Ther. 2009;16:709–715. doi: 10.1038/gt.2009.42. [DOI] [PubMed] [Google Scholar]
- 3.Chailertvanitkul VA, Pouton CW. Adenovirus: A blueprint for non-viral gene delivery. Curr Opin Biotechnol. 2010;21:627–632. doi: 10.1016/j.copbio.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Maier P, von Kalle C, Laufs S. Retroviral vectors for gene therapy. Future Microbiol. 2010;5:1507–1523. doi: 10.2217/fmb.10.100. [DOI] [PubMed] [Google Scholar]
- 5.Mátrai J, Chuah MK, VandenDriessche T. Recent advances in lentiviral vector development and applications. Mol Ther. 2010;18:477–490. doi: 10.1038/mt.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li CM, et al. Insertional mutagenesis of the mouse acid ceramidase gene leads to early embryonic lethality in homozygotes and progressive lipid storage disease in heterozygotes. Genomics. 2002;79:218–224. doi: 10.1006/geno.2002.6686. [DOI] [PubMed] [Google Scholar]
- 7.Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 8.Ikeno M, et al. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 9.Grimes BR, Monaco ZL. Artificial and engineered chromosomes: Developments and prospects for gene therapy. Chromosoma. 2005;114:230–241. doi: 10.1007/s00412-005-0017-5. [DOI] [PubMed] [Google Scholar]
- 10.Basu J, Willard HF. Artificial and engineered chromosomes: Non-integrating vectors for gene therapy. Trends Mol Med. 2005;11:251–258. doi: 10.1016/j.molmed.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Monaco ZL, Moralli D. Progress in artificial chromosome technology. Biochem Soc Trans. 2006;34:324–327. doi: 10.1042/BST20060324. [DOI] [PubMed] [Google Scholar]
- 12.Mills W, Critcher R, Lee C, Farr CJ. Generation of an approximately 2.4 Mb human X centromere-based minichromosome by targeted telomere-associated chromosome fragmentation in DT40. Hum Mol Genet. 1999;8:751–761. doi: 10.1093/hmg/8.5.751. [DOI] [PubMed] [Google Scholar]
- 13.Shen MH, et al. A structurally defined mini-chromosome vector for the mouse germ line. Curr Biol. 2000;10:31–34. doi: 10.1016/s0960-9822(99)00261-4. [DOI] [PubMed] [Google Scholar]
- 14.Yang JW, et al. Human mini-chromosomes with minimal centromeres. Hum Mol Genet. 2000;9:1891–1902. doi: 10.1093/hmg/9.12.1891. [DOI] [PubMed] [Google Scholar]
- 15.Spence JM, et al. Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha-satellite array. EMBO J. 2002;21:5269–5280. doi: 10.1093/emboj/cdf511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong LH, Saffery R, Choo KH. Construction of neocentromere-based human minichromosomes for gene delivery and centromere studies. Gene Ther. 2002;9:724–726. doi: 10.1038/sj.gt.3301756. [DOI] [PubMed] [Google Scholar]
- 17.Katoh M, et al. Construction of a novel human artificial chromosome vector for gene delivery. Biochem Biophys Res Commun. 2004;321:280–290. doi: 10.1016/j.bbrc.2004.06.145. [DOI] [PubMed] [Google Scholar]
- 18.Kazuki Y, et al. Refined human artificial chromosome vectors for gene therapy and animal transgenesis. Gene Ther. 2010;10:1–10. doi: 10.1038/gt.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebersole T, et al. Rapid generation of long synthetic tandem repeats and its application for analysis in human artificial chromosome formation. Nucleic Acids Res. 2005;33:e130. doi: 10.1093/nar/gni129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moralli D, Simpson KM, Wade-Martins R, Monaco ZL. A novel human artificial chromosome gene expression system using herpes simplex virus type 1 vectors. EMBO Rep. 2006;7:911–918. doi: 10.1038/sj.embor.7400768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breman AM, Steiner CM, Slee RB, Grimes BR. Input DNA ratio determines copy number of the 33 kb Factor IX gene on de novo human artificial chromosomes. Mol Ther. 2008;16:315–323. doi: 10.1038/sj.mt.6300361. [DOI] [PubMed] [Google Scholar]
- 22.Nakano M, et al. Inactivation of a human kinetochore by a specific targeting of chromatin modifiers. Dev Cell. 2008;14:1–16. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iida Y, et al. Human artificial chromosome with a conditional centromere for gene delivery and gene expression. DNA Res. 2010;17:293–301. doi: 10.1093/dnares/dsq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardinale S, et al. Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol Biol Cell. 2009;20:4194–4204. doi: 10.1091/mbc.E09-06-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kouprina N, Larionov V. TAR cloning: Insights into gene function, long-range haplotypes and genome structure and evolution. Nat Rev Genet. 2006;7:805–812. doi: 10.1038/nrg1943. [DOI] [PubMed] [Google Scholar]
- 26.Kouprina N, Larionov V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat Protoc. 2008;3:371–377. doi: 10.1038/nprot.2008.5. [DOI] [PubMed] [Google Scholar]
- 27.Tauchi H, et al. Nijmegen breakage syndrome gene, NBS1, and molecular links to factors for genome stability. Oncogene. 2002;21:8967–8980. doi: 10.1038/sj.onc.1206136. [DOI] [PubMed] [Google Scholar]
- 28.Lamarche BJ, Orazio NI, Weitzman MD. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010;584:3682–3695. doi: 10.1016/j.febslet.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura K, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Noon AT, et al. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- 31.Porcedda P, et al. Impaired elimination of DNA double-strand break-containing lymphocytes in ataxia telangiectasia and Nijmegen breakage syndrome. DNA Repair (Amst) 2006;5:904–913. doi: 10.1016/j.dnarep.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 33.Sherr CJ. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 34.Kim M, et al. Recombinant adenovirus expressing von Hippel-Lindau-mediated cell cycle arrest is associated with the induction of cyclin-dependent kinase inhibitor p27Kip1. Biochem Biophys Res Commun. 1998;253:672–677. doi: 10.1006/bbrc.1998.9839. [DOI] [PubMed] [Google Scholar]
- 35.Baba M, et al. Loss of von Hippel-Lindau protein causes cell density dependent deregulation of CyclinD1 expression through hypoxia-inducible factor. Oncogene. 2003;22:2728–2738. doi: 10.1038/sj.onc.1206373. [DOI] [PubMed] [Google Scholar]
- 36.Baldewijns MM, et al. VHL and HIF signalling in renal cell carcinogenesis. J Pathol. 2010;221:125–138. doi: 10.1002/path.2689. [DOI] [PubMed] [Google Scholar]
- 37.Jeffers M, Rong S, Woude GF. Hepatocyte growth factor/scatter factor-Met signaling in tumorigenicity and invasion/metastasis. J Mol Med (Berl) 1996;74:505–513. doi: 10.1007/BF00204976. [DOI] [PubMed] [Google Scholar]
- 38.Koochekpour S, et al. The von Hippel-Lindau tumor suppressor gene inhibits hepatocyte growth factor/scatter factor-induced invasion and branching morphogenesis in renal carcinoma cells. Mol Cell Biol. 1999;19:5902–5912. doi: 10.1128/mcb.19.9.5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci USA. 2006;103:4186–4191. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alonso A, Hasson D, Cheung F, Warburton PE. A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin. 2010;3:6. doi: 10.1186/1756-8935-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kazuki Y, et al. Complete genetic correction of ips cells from Duchenne muscular dystrophy. Mol Ther. 2010;18:386–393. doi: 10.1038/mt.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fournier RE, Ruddle FH. Microcell-mediated transfer of murine chromosomes into mouse, Chinese hamster, and human somatic cells. Proc Natl Acad Sci USA. 1977;74:319–323. doi: 10.1073/pnas.74.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagaki K, et al. Sequencing of a rice centromere uncovers active genes. Nat Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- 44.Bergmann JH, et al. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 47.Mandegar MA, et al. Functional human artificial chromosomes are generated and stably maintained in human embryonic stem cells. Hum Mol Genet. 2011;20:2905–2913. doi: 10.1093/hmg/ddr144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.