Abstract
Seeds respond to environmental signals, tuning their dormancy cycles to the seasons and thereby determining the optimum time for plant establishment. The molecular regulation of dormancy cycling is unknown, but an extensive range of mechanisms have been identified in laboratory experiments. Using a targeted investigation of gene expression over the dormancy cycle of Arabidopsis seeds in the field, we investigated how these mechanisms are seasonally coordinated. Depth of dormancy and gene expression patterns were correlated with seasonal changes in soil temperature. The results were consistent with abscisic acid (ABA) signaling linked to deep dormancy in winter being repressed in spring concurrent with enhanced DELLA repression of germination as depth of dormancy decreased. Dormancy increased during winter as soil temperature declined and expression of ABA synthesis (NCED6) and gibberellic acid (GA) catabolism (GA2ox2) genes increased. This was linked to an increase in endogenous ABA that plateaus, but dormancy and DOG1 and MFT expression continued to increase. The expression of SNF1-related protein kinases, SnrK 2.1 and 2.4, also increased consistent with enhanced ABA signaling and sensitivity being modulated by seasonal soil temperature. Dormancy then declined in spring and summer. Endogenous ABA decreased along with positive ABA signaling as expression of ABI2, ABI4, and ABA catabolism (CYP707A2) and GA synthesis (GA3ox1) genes increased. However, during the low-dormancy phase in the summer, expression of transcripts for the germination repressors RGA and RGL2 increased. Unlike deep winter dormancy, this represson can be removed on exposure to light, enabling the completion of germination at the correct time of year.
Keywords: environmental signaling, environmental sensing, seed ecology, soil seed bank
Seeds are the mobile phase of the plant's life cycle; vegetative development is suspended as seeds transport the plant's genetic complement through space and time. This is achieved by a seed remaining dormant, potentially for many years/decades in the soil, until conditions occur that are suitable for the resulting plant to survive, be competitive, and reproduce. Field observations by seed ecologists show that seeds act as environmental sensors and adjust their depth of dormancy in response to a range of signals (1). Some signals (e.g., soil temperature and moisture) are related to slow seasonal change that indicates when a suitable time of year and climate space exists (temporal window). These signals are integrated over time to alter the depth of dormancy and therefore the sensitivity to a second set of signals (e.g., light, nitrate, alternating temperatures). This second set of signals indicates in a more immediate way that conditions are suitable to terminate dormancy and induce the completion of germination (spatial window: appropriate soil depth, temperature, and moisture and lack of competing plants). If the correct spatial window does not occur, the temporal window will close for another year.
Despite the obvious importance of dormancy cycling in the natural environment (2), very little is known about its regulation at a molecular level. In contrast, a great deal is known about mechanisms that influence dormancy loss in short-term laboratory experiments (1, 3, 4) that inform us about dormancy in the context of crop seeds. In crops, for obvious reasons, persistent dormancy has been selected against and dormancy must be minimal at harvest and relieved rapidly following sowing in the soil. This laboratory-based work, largely using the model species Arabidopsis, has identified a range of regulatory mechanisms and signaling networks with apparent duplication of function and redundancy. How this complex system is used by the seed in a coordinated way to regulate dormancy cycling in variable field environments is unknown and unstudied.
Recent omics studies [of the transcriptome (5–7) and proteome (8, 9)] show that seeds change and remain responsive at a molecular level in both the imbibed and the dry state. A dynamic balance of the hormones abscisic acid (ABA) and gibberellic acid (GA), resulting from both synthesis and catabolism, is thought to be central to dormancy and the control of germination completion (radical emergence through the seed coat) (1). Regulation of dormancy status results from the response to this balance through hormone-signaling networks that influence sensitivity to ABA and GA (Fig. S1). The influence of other hormones, such as ethylene (10), can be significant, but in general their influence operates through the ABA/GA balance. Recently, the seed transcriptome, and in particular those genes involved in the ABA/GA balance, were shown to be sensitive to maturation temperature, which is important in setting dormancy status before shedding from the mother plant (11).
Here we have adopted a targeted investigation of gene expression combined with a detailed physiological characterization of dormancy-cycling behavior in Arabidopsis seeds in the field. We show that, in addition to the impact of temperature during maturation (11), gene sets related to dormancy and germination potential are highly sensitive to seasonal changes in soil temperature in seeds within the soil seed bank, resulting in continual and dramatic adjustments to the depth of dormancy within the soil seed bank. Gene expression patterns were consistent with the ABA-signaling pathway linked to the establishment of deep dormancy over winter and its repression linked to relief of dormancy in spring/summer and therefore to the sensing of slow seasonal changes in the depth of dormancy (temporal sensing). ABA-signaling repression was concurrent with enhanced DELLA repression of germination, which is consistent with the potential for a rapid response to suitable germination conditions during shallow dormancy (spatial sensing). We interpret these results in the context of the ecological observations reported above to better understand the coordination of molecular mechanisms identified in the laboratory.
Results
Environmental Regulation of Dormancy Cycling.
To extend our understanding of the molecular physiology of dormancy cycling obtained in laboratory studies (5, 7), we observed the response of Arabidopsis seeds [ecotype Cape Verdi Isle (Cvi)] buried in the field to simulate seed behavior in the soil seed bank. Seeds were exhumed at intervals following burial, and the depth of dormancy (AR50, which is the time required in dry storage to remove dormancy in 50% of the population) (Fig. S2) was determined. The ecotype Cvi requires light to remove the final layer of dormancy to allow germination completion, and consequently all physiological tests were carried out in the light. This absolute requirement for light to germinate is important experimentally to observe changes in dormancy level separately from downstream changes resulting from the germination process.
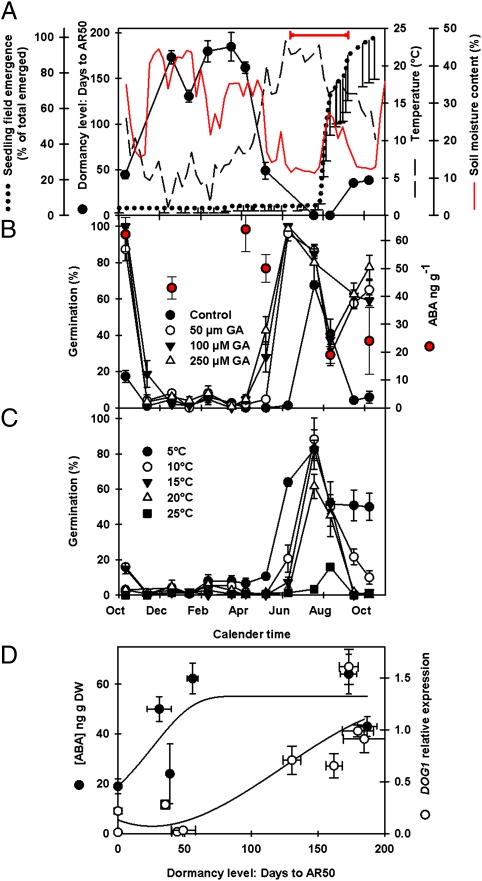
Soil temperature and moisture content recorded alongside the buried seeds were negatively correlated (r = −0.73, P < 0.01) (Dataset S1) and illustrate the large seasonal changes that seeds were subjected to in the soil (Fig. 1A). In response, the depth of seed dormancy (AR50) followed a distinct pattern that was significantly correlated with soil temperature and moisture content (r = −0.92, P < 0.001 and r = 0.77, P < 0.01, respectively) (Dataset S1). Although the linear regression between AR50 and temperature was highly significant, closer inspection suggests a more complex curvilinear relationship between these parameters (R2 = 0.95) (Fig. S3A). Surprisingly, AR50 dramatically increased from a mean of 44 d in primary dormant seeds before burial to 173 d, 2 mo after burial (Fig. 1A). Seeds also rapidly became GA insensitive (range 0–250 μM) as AR50 increased (Fig. 1B). After April, this trend reversed and dormancy was minimal in the summer months as thermodormancy declıned wıth seeds requiring only light for dormancy termination and the completion of germination (Fig. 1C). This is consistent with previous observations (12, 13). The thermal germination window indicates the period where germination is possible (i.e., ambient temperature exceeds the seed's minimum permissive temperatures) if soil moisture is suitable and soil disturbance moves seeds to the surface, exposing them to light. Emergence in the field coincides with this window (Fig. 1A).
Fig. 1.
Seasonal changes in dormancy cycling. (A) Changes in dormancy level (AR50) and soil temperature and moisture content measured at seed depth (5 cm) over 12 mo from October 2007. Mean seedling emergence following monthly soil disturbance (n = 4) is also shown. The red horizontal bar is the thermal germination window (Materials and Methods). (B) ABA concentration and changing sensitivity to gibberellic acid (GA) in Cvi seeds recovered from the field. Following recovery, seeds where incubated in the light at 20 °C in the presence of 50–250 μM GA at pH 5.0. (C) Changing thermodormancy in Cvi seeds recovered from the field. Following recovery, seeds where incubated in the light at 5–25 °C. (D) [ABA] and DOG1 expression in relation to dormancy level in the field (AR50 in A). The ABA regression was fitted using the Weibull function (R2 = 0.524). The DOG1 regression was fitted using a cubic polynomial function (R2 = 0.784). Error bars indicate SEM; n = 3.
The AR50 increased again as seeds in the soil entered secondary dormancy in the autumn. Changes in AR50 in the autumn and spring occur when soil is moist (>20%) and seeds are therefore likely to be hydrated. This has significant ecological implications as it is widely considered that the release of dormancy via afterripening occurs in the dry state (14). Thus, in damp temperate soils, the rapid loss of dormancy occurring in hydrated seeds indicates that dry afterripening—traditionally considered to be the primary driving force for loss of dormancy (in both laboratory and field) (15–17)—may have a limited role during dormancy cycling in soil seed banks of temperate regions such as the United Kingdom.
Hormonal Regulation of Dormancy Cycling.
To further investigate the role of the hormones ABA and GA in dormancy cycling, the ABA content of seeds from selected field harvests was measured, as were the expression levels of key genes in the synthesis of ABA and GA and their signaling pathways. Gene family members were selected that exhibited distinct seed expression patterns in our previous laboratory-based microarray analysis of dormancy cycling (5, 7) (Dataset S2). Genes chosen were those involved in ABA and GA biosynthesis (NCED6, GA3ox1) and catabolism (CYP707A2, GA2ox2), ABA signaling (PYR1, PYL7, ABI2, SnrK2.1 and -2.4, ABI3, ABI4, and ABI5) (18), the ABA-signaling regulator MFT (19), GA signaling (GID1A, RGL2, RGA2), members of the phytochrome interacting factor (PIF) family (PIL5 and SPT), and the dormancy-associated DOG1 (20). The putative involvement of these genes in the regulation of dormancy is summarized in Fig. S1. FLC, a major flowering-time gene recently linked with germination timing (21), was also included. Of these 19 genes, 13 have expression patterns that are significantly (P < 0.05) correlated with soil temperature, 9 with AR50, but only 4 with soil moisture and 1 with ABA content (Dataset S1). However, it should be noted that there are fewer degrees of freedom in correlations with AR50 (df = 8) and ABA (df = 3) than with soil temperature and moisture (df = 10). Here we show that the expression patterns of these genes fall into two groups: one that is negatively related to temperature (Dataset S1) and associated with slow seasonal change (temporal sensing and deep dormancy) and the second that is positively related to temperature (Dataset S1) and associated with sensitivity to rapid environmental change (spatial sensing, shallow dormancy, and increased germination potential).
Slow Seasonal Response and Deep Dormancy.
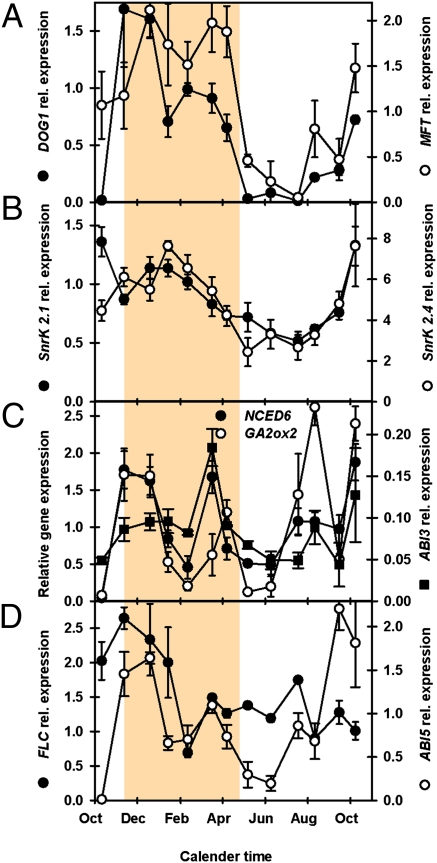
Of the genes investigated, two (DOG1 and MFT) have a pattern of expression that is highly negatively correlated, such as AR50 with temperature (P < 0.001, Dataset S1; Fig. 2A; and Fig. S4). DOG1 is the gene at the loci with the strongest dormancy association in QTL analyses (20), and its expression, although not directly associated with ABA, is altered by environmental conditions (22) and may enhance ABA sensitivity (23). MFT, a homolog of the antagonistic flowering-time regulators FLOWERING LOCUS T (FT) and TERMINAL FLOWER 1 (TFL1) is a proposed ABA-induced negative regulator of ABA signaling that promotes embryo growth in germinating seeds (19). However, in wheat, an MFT homolog is more highly expressed under low-temperature seed maturatıon conditions and acts as a repressor of germination potential (24); the data presented here are consistent with this observation. The ABA content of seeds has been linked to differences in dormancy within the Cvi ecotype (25). To investigate whether this was also true in seeds in the soil seed bank, ABA content was measured at six points along the annual dormancy cycle (Fig. 1B) and plotted against the AR50 value for that harvest (Fig. 1D). The ABA concentration in mature seeds was significant, and changes following burial were in line with the depth of dormancy until an AR50 value of ∼50 d was reached, after which ABA reached a plateau (Fig. 1D). In contrast, when DOG1 expression is plotted, expression is low when dormancy is low and increases only above an AR50 of 50 d (Fig. 1D). These results are consistent with [ABA] being important in the control of the dormancy typically seen in primary dormant seeds used in the majority of laboratory studies. However, once seeds enter the deep dormancy observed here in the field, DOG1 expression may be the dominant factor by influencing ABA signaling and therefore sensitivity. Thus, dormancy is enhanced without necessitating an increase in ABA. Dormant seeds at maturity and seeds in deep dormancy are transcriptionally distinct as demonstrated by principal component analysis of whole-transcriptome data (7), which lends weight to the suggestion that DOG1 enhances ABA sensitivity (23).
Fig. 2.
Gene expression in the deep dormancy phase. (A) Expression of DOG1 (dormancy) and MFT (ABA-induced germination repressor). (B) Expression of two members (SnrK2.1 and -2.4) of the SNF1-related protein kinase subfamily (positive regulators of ABA signaling). (C) Changes in ABI3 (dormancy), NCED6 (ABA biosynthesis), and GA2ox2 (GA catabolism) expression. (D) Expression of FLC (a flowering-time regulator) and ABI5 (ABRE-regulated transcription factor). Shaded area represents the region of deep dormancy. Error bars indicate SEM; n = 3.
The SNF1-related protein kinases, SnrK 2.1 and 2.4, are positive regulators of ABA signaling acting downstream of ABA receptors to activate transcription regulators (Fig. S1). Expression of these genes follows a pattern similar to depth of dormancy, which is negatively correlated (P < 0.05) to temperature and positively correlated (P < 0.01) to DOG1 and MFT expression (Fig. 2B, Dataset S1, and Fig. S3B). These patterns are consistent with ABA sensitivity being modulated by seasonal soil temperature. Genes representative of ABA biosynthesis (NCED6) and GA catabolism (GA2ox2) are positively correlated (P < 0.05) with ABI3 and ABI5 expression (Fig. 2 C and D and Dataset S1). Interestingly, none of these genes are directly correlated with the environmental variables soil temperature and soil moisture, or the variables AR50 and ABA, but NCED6 is correlated with DOG1 (P < 0.05) (Dataset S1), again indicating a key role for DOG1 in deep dormancy. ABI3, like DOG1, has been strongly associated with dormancy. Both have similar high levels of expression in dormant seed transcriptomes (Dataset S2) (4, 5, 20). However, ABI3 expression shows a relatively small increase/decrease as dormancy increases and decreases in the field (Fig. 2C). The exception to this occurs with changes in soil moisture content in late winter/early spring and late summer/early autumn periods immediately before dynamic changes in dormancy level. This is followed by an increase in expression as seeds enter secondary dormancy. DOG1 and the SnrK genes (SnrK2.1 and -2.4) (Fig. 2 A and B) do not show this increase during shallow dormancy, indicating that they respond only to slow seasonal change in the maintenance of deep dormancy.
FLC was recently linked to the regulation of temperature-dependent germination in Arabidopsis; seeds with high-temperature thermodormancy exhibited high FLC expression during germination at low temperature (21). In the field, FLC expression increases with falling temperature and increasing dormancy (Fig. 2D) and then subsequently declines with increasing exposure to low temperature. This pattern of expression was positively correlated (P < 0.05) with DOG1 expression, but with no other gene or environmental variable (Dataset S1). Further work is required to determine whether FLC has a role in the regulation of dormancy in buried seeds.
Seeds have developed a checkpoint before testa rupture that is controlled by ABI5 and leads to accumulation of polygalacturonase inhibitor proteins that block cell degradation in the testa (Dataset S2) (26, 27). In the field, ABI5 expression increased in line with increasing dormancy before increasing with the onset of secondary dormancy (Fig. 2D), but its expression was correlated only with that of NCED6.
Slow Seasonal Response and Shallow Dormancy.
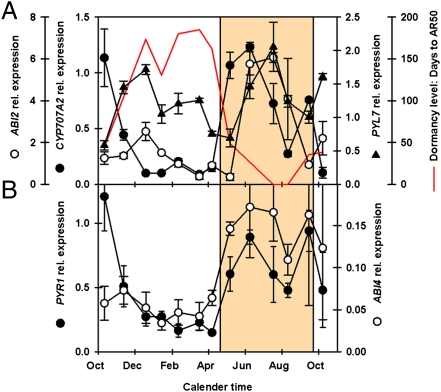
From April to May a rapid decrease in dormancy level occurred in the field. A period of shallow dormancy then persisted until August after which dormancy levels increased (Fig. 1A). During this period of shallow dormancy, genes expressed for ABA catabolism (CYP707A2), the ABA receptor (PYR1), the repressor of ABA signaling (ABI2), and the ABA-induced transcription factor (ABI4) all showed increased expression (Fig. 3) that was positively correlated with soil temperature (Dataset S1). This pattern is generally the inverse of that described for the genes linked to deep dormancy and consistent with the repression of ABA signaling. Increased transcript numbers in microarray data (Dataset S2) suggest that the ABA receptor PYR1 (possibly in conjunction with PYL4) functions in shallow dormancy, whereas PYL7 functions in deep dormancy. However, PYL7 expression (Fig. 3A) was positively correlated (P < 0.05) (Dataset S1) with that of ABI2 and no other hormone-signaling genes in the present study.
Fig. 3.
Gene expression in the shallow dormancy phase. (A) Expression of ABI2 (repressor of ABA signaling), PYL7 (ABA receptor), and CYP707A2 (ABA catabolism) in relation to the changing dormancy level (AR50). (B) Expression of PYR1 (ABA receptor) and ABI4 (control of energy utilization). Shaded area represents the region of shallow dormancy and increasing germination potential. Error bars indicate SEM; n = 3.
During the transition to shallow dormancy ABI2 expression increases (Fig. 3A), consistent with the repression of ABA signaling linked to the inhibition of germination, Although germination potential increases (Fig. 1C), seeds remain dormant (light requiring) throughout, and thus conservation of energy remains essential until the final environmental signal required to remove dormancy (light) is received. ABI4, a repressor of lipid mobilization in the embryo (28), is highly positively correlated with soil temperature and negatively correlated with AR50 (P < 0.001) (Dataset S1). The high expression of ABI4 (Fig. 3B) during shallow dormancy is consistent with this requirement for energy conservation.
Rapid Response for Spatial Sensing.
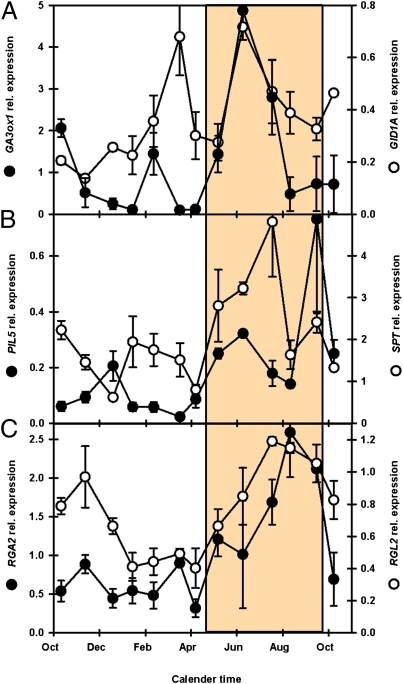
On transition from deep to shallow dormancy, the expression of several key genes was positively correlated (P < 0.05) (Dataset S1) with temperature increased. These include genes known to enhance germination potential such as the GA biosynthesis gene GA3ox1 and the GA receptor gene GID1A (Fig. 4A). Countering this, the DELLA family are major repressors of germination potential and GA signaling, and the high expression of RGA2 and RGL2 (Fig. 4C) suggests that they prevent germination during shallow dormancy in the absence of light. However, in contrast to the ABA repression during deep dormancy, this DELLA repression can be rapidly removed on exposure to light. For example, the increase in GA3ox1 transcripts in the dark is greatly enhanced (∼600-fold) if these shallow dormant seeds are exposed to light (5); deeply dormant seeds are unresponsive to light.
Fig. 4.
Gene expression related to germination potential during spatial sensing. (A) Expression of GA3ox1 (GA biosynthesis) and GID1A (GA receptor). (B) Expression of PIL5 and SPT (bHLH transcription factors of the PIF family: germination repressors). (C) Expression of RGA2 and RGL2 (DELLAs: germination repressors). Shaded panel represents the region of increasing germination potential. Error bars indicate SEM; n = 3.
While repressing germination DELLAs maintain low metabolic activity in seeds by repressing a range of genes related to carbohydrate, lipid, and protein metabolism, potentially acting in concert with ABI4. They also repress genes involved in cell-wall loosening and organization of the cytoskeleton, events crucial for endosperm weakening and radical extension during germination. DELLAs also up-regulate a range of ABA and stress-related genes (29). The PIF family members PIL5 and SPT are similarly expressed (Fig. 4B) and also repress expression of genes related to increased germination potential (30, 31). PIL5 and SPT in turn are inactivated by RGL2 and RGA, which form an inactive complex with them (32). PIF proteins are released when the GID protein–GA complex binds DELLA proteins to target their degradation by the proteosome (33).
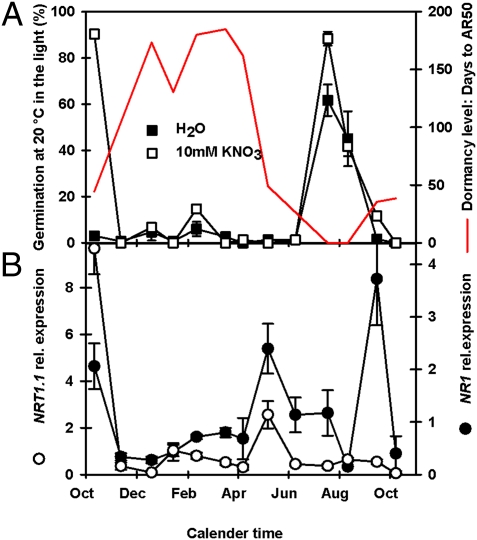
Nitrate sensitivity is related to the enhancement of germination in the light when dormancy levels are low (34). Dormant seeds were nitrate sensitive before burial when expression of the nitrate transporter 1.1 (NRT1.1) and nitrate reductase 1 (NR1) genes was high (Fig. 5). Expression rapidly dropped on burial, increasing in May immediately before the decline in thermodormancy (Fig. 5B). Nitrate also induces expression of the primary gene of ABA catabolism, CYP707A2 (35). In seeds recovered from the field, the expression of CYP707A2 (Fig. 3A) was positively correlated (P < 0.05) (Dataset S1) with NR1 with expression decreasing following burial and increasing in May and again in October (Figs. 3A and 5B). The single peak in NRT1.1 suggests a possible adaptation to gap sensing where, in addition to light intensity, soil nitrate increases in vegetation gaps; thus nitrate sensitivity could complement light sensitivity in spatial sensing (36). It should also be noted that light and possibly nitrate can act as temporal signals because both can decrease as competing plants progressively grow. The final peak in NR1 expression (Fig. 5B) may be linked to removing nitrate from the system potentially by channeling nitrates into amino acid biosynthesis to sequester nitrate in storage proteins, resulting in the down-regulation of CYP707A2 as seeds in the field enter secondary dormancy.
Fig. 5.
Nitrate sensitivity during dormancy cycling in the field. (A) Germination in the light at 20 °C ± 10 mM KNO3 in relation to changing dormancy level (AR50). (B) Expression of NRT1.1 (nitrate transporter) and NR1 (nitrate reductase). Error bars indicate SEM; n = 3.
Discussion
We present insight into the necessarily complex nature of dormancy regulation by interpreting mechanisms identified in the laboratory using field observations (summarized in Fig. 6). Our analysis indicates that the depth of dormancy (AR50) is largely determined by soil temperature with some influence of soil moisture content. These environmental signals similarly affected ABA synthesis and related signaling. However, following an initial rise in [ABA] with dormancy, [ABA] reached a plateau while depth of dormancy continued to increase, showing that the final depth of dormancy is not set during seed maturation. This indicates that ABA signaling and sensitivity are more likely regulators of dormancy than the absolute level of ABA. The expression of SnrK 2.1 and 2.4 follows a pattern similar to depth of dormancy, consistent with changes in ABA signaling and sensitivity, which is negatively correlated with soil temperature. DOG1 expression is highly negatively correlated to soil temperature and appears sensitive to thermal time and could therefore act as part of a thermal-sensing mechanism to influence dormancy level by altering sensitivity to ABA. The observation that the DOG1 mutant is dormant under cool maturation conditions (11) suggests that the ABA-induced germination repressor MFT (24), which is also negatively correlated with soil temperature, may also function in dormancy regulation.
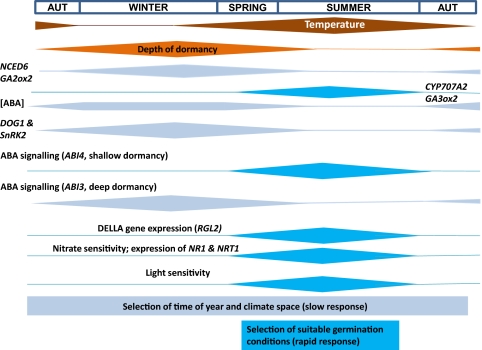
Fig. 6.
Seasonal timing of molecular physiological events during dormancy cycling in the field. The height of each bar indicates the amplitude of the response measured across four seasons. Temperature represents the annual fluctuation in soil temperature at seed depth. Depth of dormancy is related to the AR50 of Cvi seeds in the field. Other bars represent seasonal changes in [ABA], gene expression and environmental responses. Temporal sensing represents the slow seasonal change in dormancy for the selection of time of year and climate space (light blue bars). Spatial sensing represents the period of potential rapid change during shallow dormancy if suitable germination conditions are detected (dark blue bars).
The transition to shallow dormancy appears linked to repression of ABA signaling and is marked by increased nitrate sensitivity and an associated ABA catabolism (35). At this time, the GA3ox repressors SPT and PIL5 are highly expressed as are the DELLA genes RGA2 and RGL2. PIL5 is also known to repress CYP707A2 (ABA catabolism) and induce DELLAs, GA2ox2 (GA catabolism), and NCED6 (ABA biosynthesis) (31), which would reduce germination potential in the absence of light.
Crucially for the regulation of dormancy cycling (Fig. 6), seeds in deep dormancy, which appears to be determined by enhanced sensitivity to ABA, change slowly (temporal sensing) and do not respond to signals that inform about germination conditions (e.g., light), whereas seeds in shallow dormancy (light requiring only), resulting largely from germination repression by DELLAs, can respond rapidly to favorable germination conditions (spatial sensing). For example, exposure to light dramatically enhances GA3ox expression to remove DELLA repression (5). Thus, the purpose and coordination of molecular mechanisms that regulate dormancy identified in the laboratory is revealed by field observations. Furthermore, the framework that we have identified will allow existing data sets to be placed in an ecological context and enable fresh approaches to be developed to further elucidate the operation of these pathways in response to the environment.
Materials and Methods
Seeds were produced (February–May 2007) in a temperature-controlled glasshouse. Mature seeds were harvested by hand threshing and equilibrated at 15% relative humidity/5 °C for 7 d to produce an equilibrium moisture content of 5–7% on a dry-weight basis. Seeds were stored at −80 °C in sealed tubes.
Seeds were dispersed in soda lime Ballotini balls, placed into nylon-mesh bags, and buried in the field at a depth of 5 cm. Soil temperature and moisture were monitored throughout the experimental period. Seeds were recovered from the field at monthly intervals. Seeds for molecular analysis were recovered in the dark and processed under a green safe light before freezing at −80 °C. Seeds for physiological analysis were recovered and processed in the light. Germination testing was carried out immediately following recovery (details of seed burial, ABA analysis, seedling emergence, germination tests, and subsequent analysis are described in SI Material and Methods).
RNA was extracted from seeds using the RNAqueous kit (Ambion) in conjunction with a plant RNA isolation aid (Ambion). Quantitative PCR was performed in triplicate on each of three independent biological samples. Gene expression levels were determined using a cDNA dilution series of the primer pairs of each gene (Table S1) of interest with normalization against the housekeeping gene At4g34270 (Tip 4-like). For more details, see SI Material and Methods.
Supplementary Material
Acknowledgments
We thank Linda Brown and Katherine Dent for assistance; Dr. Karl Morris for assistance and review of the manuscript; the Plant Biotechnology Institute of the National Research Council of Saskatoon, Canada, for the analysis of seed ABA and metabolites; and the Warwick University Hortservices staff for assistance in the field. This project was funded by the UK Department of Environment, Food and Rural Affairs.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116325108/-/DCSupplemental.
References
- 1.Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytol. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- 2.Baskin CC, Baskin JM. Seeds: Ecology, Biogeography, and Evolution of Dormancy and Germination. London: Academic Press; 1998. [Google Scholar]
- 3.Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annu Rev Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- 4.Holdsworth MJ, Bentsink L, Soppe WJJ. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008;179:33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- 5.Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 2006;46:805–822. doi: 10.1111/j.1365-313X.2006.02738.x. [DOI] [PubMed] [Google Scholar]
- 6.Carrera E, et al. Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. Plant J. 2008;53:214–224. doi: 10.1111/j.1365-313X.2007.03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finch-Savage WE, Cadman CSC, Toorop PE, Lynn JR, Hilhorst HWM. Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 2007;51:60–78. doi: 10.1111/j.1365-313X.2007.03118.x. [DOI] [PubMed] [Google Scholar]
- 8.Gallardo K, et al. Proteomics of Arabidopsis seed germination: A comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol. 2002;129:823–837. doi: 10.1104/pp.002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajjou L, et al. The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 2004;134:1598–1613. doi: 10.1104/pp.103.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linkies A, et al. Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: A comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell. 2009;21:3803–3822. doi: 10.1105/tpc.109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kendall SL, et al. Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell. 2011;23:2568–2580. doi: 10.1105/tpc.111.087643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derkx MPM, Karssen CM. Are seasonal dormancy patterns in Arabidopsis thaliana regulated by changes in seed sensitivity to light, nitrate and gibberellin? Ann Bot (Lond) 1994;73:129–136. [Google Scholar]
- 13.Baskin JM, Baskin CC. Ecological life-cycle and physiological ecology of seed germination of Arabidopsis thaliana. Can J Bot. 1972;50:353–360. [Google Scholar]
- 14.Leopold AC, Glenister R, Cohn MA. Relationship between water-content and afterripening in red rice. Physiol Plant. 1988;74:659–662. [Google Scholar]
- 15.Bair NB, Meyer SE, Allen PS. A hydrothermal after-ripening time model for seed dormancy loss in Bromus tectorum L. Seed Sci Res. 2006;16:17–28. [Google Scholar]
- 16.Gianinetti A, Cohn MA. Seed dormancy in red rice. XII: Population-based analysis of dry-afterripening with a hydrotime model. Seed Sci Res. 2007;17:253–271. [Google Scholar]
- 17.Probert RJ. The role of temperature in the regulation of seed dormancy and germination. In: Fenner M, editor. Seeds: The Ecology of Regeneration in Plant Communities. 2nd Ed. Wallingford: CABI; 2000. pp. 261–292. [Google Scholar]
- 18.Nambara E, et al. Abscisic acid and the control of seed dormancy and germination. Seed Sci Res. 2010;20:55–67. [Google Scholar]
- 19.Xi WY, Liu C, Hou XL, Yu H. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell. 2010;22:1733–1748. doi: 10.1105/tpc.109.073072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang GCK, Barua D, Kramer EM, Amasino RM, Donohue K. Major flowering time gene, flowering locus C, regulates seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2009;106:11661–11666. doi: 10.1073/pnas.0901367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiang GCK, et al. DOG1 expression is predicted by the seed-maturation environment and contributes to geographical variation in germination in Arabidopsis thaliana. Mol Ecol. 2011;20:3336–3349. doi: 10.1111/j.1365-294X.2011.05181.x. [DOI] [PubMed] [Google Scholar]
- 23.Teng S, Rognoni S, Bentsink L, Smeekens S. The Arabidopsis GSQ5/DOG1 Cvi allele is induced by the ABA-mediated sugar signalling pathway, and enhances sugar sensitivity by stimulating ABI4 expression. Plant J. 2008;55:372–381. doi: 10.1111/j.1365-313X.2008.03515.x. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura S, et al. A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell. 2011;23:3215–3229. doi: 10.1105/tpc.111.088492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ali-Rachedi S, et al. Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: Studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta. 2004;219:479–488. doi: 10.1007/s00425-004-1251-4. [DOI] [PubMed] [Google Scholar]
- 26.Kanai M, Nishimura M, Hayashi M. A peroxisomal ABC transporter promotes seed germination by inducing pectin degradation under the control of ABI5. Plant J. 2010;62:936–947. doi: 10.1111/j.1365-313X.2010.04205.x. [DOI] [PubMed] [Google Scholar]
- 27.Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002;32:317–328. doi: 10.1046/j.1365-313x.2002.01430.x. [DOI] [PubMed] [Google Scholar]
- 28.Penfield S, Li Y, Gilday AD, Graham S, Graham IA. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell. 2006;18:1887–1899. doi: 10.1105/tpc.106.041277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao DN, Cheng H, Wu W, Soo HM, Peng JR. Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 2006;142:509–525. doi: 10.1104/pp.106.082289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penfield S, et al. Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr Biol. 2005;15:1998–2006. doi: 10.1016/j.cub.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 31.Oh E, et al. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell. 2009;21:403–419. doi: 10.1105/tpc.108.064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallego-Bartolomé J, et al. Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Mol Biol Evol. 2010;27:1247–1256. doi: 10.1093/molbev/msq012. [DOI] [PubMed] [Google Scholar]
- 33.Davière JM, de Lucas M, Prat S. Transcriptional factor interaction: A central step in DELLA function. Curr Opin Genet Dev. 2008;18:295–303. doi: 10.1016/j.gde.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Hilhorst HWM, Karssen CM. Dual effect of light on the gibberellin-stimulated and nitrate-stimulated seed-germination of Sisymbrium officinale and Arabidopsis thaliana. Plant Physiol. 1988;86:591–597. doi: 10.1104/pp.86.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matakiadis T, et al. The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol. 2009;149:949–960. doi: 10.1104/pp.108.126938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pons TL. Breaking of seed dormancy by nitrate as a gap detection mechanism. Ann Bot (Lond) 1989;63:139–143. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.