Abstract
Although transcriptional programs associated with T-cell specification and commitment have been described, the functional hierarchy and the roles of key regulators in structuring/orchestrating these programs remain unclear. Activation of Notch signaling in uncommitted precursors by the thymic stroma initiates the T-cell differentiation program. One regulator first induced in these precursors is the DNA-binding protein T-cell factor 1 (Tcf-1), a T-cell–specific mediator of Wnt signaling. However, the specific contribution of Tcf-1 to early T-cell development and the signals inducing it in these cells remain unclear. Here we assign functional significance to Tcf-1 as a gatekeeper of T-cell fate and show that Tcf-1 is directly activated by Notch signals. Tcf-1 is required at the earliest phase of T-cell determination for progression beyond the early thymic progenitor stage. The global expression profile of Tcf-1–deficient progenitors indicates that basic processes of DNA metabolism are down-regulated in its absence, and the blocked T-cell progenitors become abortive and die by apoptosis. Our data thus add an important functional relationship to the roadmap of T-cell development.
T cells are unique among hematopoietic cells because they need a specialized organ, the thymus, to develop. Multipotent progenitors (MPPs) migrating from the bone marrow (BM) through the blood stream seed the thymus (1, 2), whose microenvironment instructs them to lose stem cell properties and alternative developmental potentials and to differentiate along the T-cell lineage (3). One important trigger for T-cell fate is the activation of Notch signaling by Delta-like Notch ligands expressed on thymic stroma. Notch signaling induces the T-cell specification and commitment transcription programs (4). Commitment to the T-cell fate is endorsed as the cells progress from the early thymic progenitor (ETP; Lin−CD44+c-kit+CD25−), the most immature recognizable thymocyte, to the double-negative 3 (DN3; Lin−CD25+CD44−) stage. As part of this process, coordinated changes in the transcription profiles of these progenitors gradually restrict their ability to become B, myeloid, dendritic, and natural killer cells. Sustained Notch signaling is required during T-cell specification and commitment; however, Notch needs to collaborate with numerous other regulators in these processes (5). Final commitment to the T-cell fate at the late DN2 stage is marked by Bcl11b-mediated down-regulation of stem and progenitor cell transcriptional regulators (6–8). Thymus-like organs are present in all animals with adaptive immune systems, further emphasizing the specialized need for this organ (9, 10).
Tcf-1 (product of the Tcf7 gene, referred to as Tcf-1 throughout this article), is a T-lineage–specific, high-mobility group (HMG) box-containing, DNA-binding protein. Tcf-1, like the other T-cell factor/lymphoid enhancer-binding factor (Tcf/Lef) factors, mediates transcriptional activation when bound by β-catenin in response to Wnt signals or transcriptional repression when bound by Groucho (11). It has been well established that Tcf-1 is required in multiple stages of thymic and peripheral T-cell development (12–15). However, Tcf/β-catenin gain of function has not been linked with the up-regulation of T-lineage–specific genes (16) or enhancement of T-cell development (17–19).
In this article, we establish that Tcf-1 is required in the earliest stages of T-cell specification. In the absence of Tcf-1, BM progenitors entering the thymus fail to progress any further, and this effect is cell-intrinsic. The earliest defect detected in Tcf-1–deficient thymocytes is the reduced expression of c-kit at the DN1 stage of development. Tcf-1–deficient cells at this stage show increased apoptosis and have significantly reduced expression of genes involved in DNA metabolic processes, chromatin modification, and response to damage compared with their WT counterparts. We further show that Notch binds the Tcf7 gene locus at a conserved element ∼31.5 kb upstream of the transcription start site, and ectopic expression of Notch1 leads to transcriptional up-regulation of Tcf-1. Thus, our data identify Tcf-1 as one of the earliest direct responders to Notch signaling in T-cell development. Tcf-1 is essential for thymic progenitors to proceed through T-cell determination.
Results
Tcf-1 Is Required for T-Cell Development Starting at the ETP Stage.
Earlier reports demonstrated that ablation of Tcf-1 causes an age-dependent degeneration of T-cell development (20). These studies showed that the DN1 population, thought at the time to represent the earliest thymocyte subset, was abundantly present in Tcf-1−/− mice, whereas the subsequent DN2, DN3, double-positive, and single-positive populations were dramatically reduced. It is now understood that DN1 cells are highly heterogeneous and can be subdivided into at least five subsets (DN1a–DN1e) (21). The c-kit–expressing DN1a and DN1b subsets contain the most potent T-cell progenitors and are otherwise known as ETPs (22). However, T cells can also be derived from the atypical DN1c–DN1e subsets, although inefficiently (21). In light of these recent findings, we reanalyzed the earliest thymic subpopulations in Tcf-1−/− mice, including the classical DN1 (Lin−CD25−CD44+) (Fig. 1A), the ETP (Lin−CD44+c-kit+CD25−) (Fig. 1B), and the DN1a–DN1e subsets (Fig. 1C). Surprisingly, these analyses showed that Tcf-1−/− thymi had a previously unappreciated 100-fold reduction in absolute DN1 numbers and a dramatic 300-fold reduction in ETPs compared with Tcf-1+/− or wt controls (Fig. 1D). Tcf-1−/− DN1 cells had reduced levels of surface c-kit (Fig. 1E), and an increased fraction of cells expressed intermediate levels of CD25 (Fig. 1 A and C). In agreement with earlier reports, DN2 cells and all subsequent thymic subsets were dramatically reduced. Thus, Tcf-1 is required earlier than previously described: at, or before, thymic entry of uncommitted thymus-seeding progenitors. Although the profile of early thymocytes in Tcf-1−/− mice resembled that of Notch1-deficient, Notch2-haploinsufficient (MxCreN1fl/flN2fl/+) thymocytes (Fig. S1), no accumulation of B cells or macrophages was observed (Fig. S2), indicating that Tcf-1 may not be involved in the suppression of B-cell fate.
Fig. 1.
Early thymocyte development in Tcf-1−/− mice. Profiles of gated Tcf-1−/− and control Lin− DN thymocytes are shown. (A) CD44 versus CD25 profiles. The DN1–DN4 subsets are indicated. Numbers indicate the frequencies of each subset. (B Left) CD44 versus c-kit profiles show the electronic gating for pro-T cells. (Right) c-kit versus CD25 profiles of gated pro-T cells define the ETP, DN2a, and DN2b subsets as indicated. (C) Analysis of the DN1 subsets in Tcf-1−/− and control mice. (Left) CD44 versus CD25 profiles show the gating of DN1 prothymocytes. (Right) CD24 versus c-kit profiles of gated DN1 cells define the DN1a (c-kit+CD24−), DN1b (c-kit+CD24+), DN1c (c-kitloCD24+), DN1d (c-kit−CD24+), and DN1e (c-kit−CD24−) subsets. (D) Bar histograms of cell numbers in the indicated subsets of Tcf-1−/− and Tcf-1+/− mice (n = 4–6). (E) Histogram overlay of c-kit levels in gated DN1 thymocytes from the indicated mice. MFI, mean fluorescence intensity of c-kit.
Tcf-1 Is Dispensable for Progenitor Migration Through the Bloodstream.
To establish the onset of Tcf-1 requirement, we evaluated prethymic hematopoietic development in Tcf-1−/− mice. Subset distribution, cellularity, and proliferation properties of BM hematopoietic progenitors, including the hematopoietic stem cell (HSC; Lin−Sca-1+c-kit+Flt3+), MPP (Lin−Sca-1+c-kit+Flt3lo), lymphoid-primed MPP (LMPP; Lin−Sca-1+c-kit+Flt3hi), and common lymphoid progenitor (CLP; Lin−IL7Rα+Flt3hiSca-1loc-kitlo), were not detectably impacted by the Tcf-1 deficiency (Fig. 2 A and B and Fig. S3A). This finding is in agreement with our observation that, among BM progenitors, only the HSCs express Tcf-1 (Fig. S3B). Thus, the reduction of ETPs in Tcf-1−/− mice could reflect either a defect in migrating from the BM to the thymus or a cell-intrinsic inability of mutant thymus-seeding progenitors to develop toward the T-cell lineage.
Fig. 2.
BM and circulating blood progenitors in Tcf-1−/− mice. (A) Analysis of gated Lin− BM cells is shown. c-kit versus Sca-1 profiles show the gating for LSK progenitors. c-kit versus Flt3 profiles show the HSC, MPP, and LMPP subsets as indicated. Flt3 versus IL7Rα profiles show the gating for Lin−Flt3+IL7Rα+ cells, and c-kit versus Sca-1 plots depict the CLP. (B) Bar histograms show absolute numbers of HSCs, MPPs, LMPPs, and CLPs in the indicated mice (n = 3–6). Error bars are SD. Data are representative of three experiments. (C–E) Lymphocytes (0.6–1 × 106) isolated from the blood of six mice for each group were stained for Lin as well as the indicated markers. (C) Gating strategy for blood LSK cells in the indicated mice. (D) Gating strategy for blood CLPs. (E) CTPs were gated as Lin−, HuCD25+. Histogram overlays show the surface profile for markers that define the CTP in the indicated mice. Unshaded histograms depict the CTPs, and shaded histograms depict an unstained negative control. Data are representative of two experiments.
BM progenitors migrate to the thymus via the bloodstream, which has been shown to contain progenitor subsets with T-lineage potential, including blood LSK (Lin−Sca-1+c-kit+) cells, CLPs (Lin−IL7Rα+Flt3+c-kitloSca-1lo) (2), and the circulating T-cell progenitors (CTPs) that express a human CD25 (HuCD25) reporter of the pre–T-cell receptor α chain (1, 23). To examine whether Tcf-1−/− progenitors efficiently migrate through the bloodstream, we compared the presence of blood LSK cells, CLPs, and CTPs in Tcf-1−/− mice and littermate controls. LSK cells and CLPs were present in the blood of Tcf-1−/− mice at frequencies comparable to those of control littermates (Fig. 2 C and D). The presence of CTPs was surveyed in the blood of Tcf-1−/− mice crossed to the transgenic HuCD25 reporter that was used for their identification (1, 23). As for LSK cells and CLPs, CTPs were also present in the blood of Tcf-1−/− mice in frequencies comparable to those of control littermates (Fig. 2E). Thus, Tcf-1−/− progenitors are able to migrate through the bloodstream.
Tcf-1−/− Progenitors Are Cell-Intrinsically Defective in T-Cell Specification.
The normal distribution of BM and blood progenitors compared with the reduced presence of Tcf-1−/− ETPs suggests a functional inability to progress along the T-cell lineage. To address this possibility, we examined the T-cell potential of hematopoietic progenitors from Tcf-1−/− mice in cocultures on OP9-DL1 stroma cells, which instruct differentiation to the T-cell lineage because they express Delta-1–like ligands to stimulate Notch signaling. HSCs, MPPs, LMPPs, and CLPs were isolated from Tcf-1−/− (CD45.2+) and WT (CD45.1+) mice. Tcf-1−/− and WT sorted subsets were mixed at a 1:1 ratio and cocultured on OP9-DL1 stroma (Fig. 3A). Tcf-1−/− cells failed to up-regulate CD25 and progress toward the T-cell lineage (Fig. 3B). Additional experiments revealed that such nondifferentiating Tcf-1−/− progenitors contained a larger fraction of early apoptotic (annexin V+) cells compared with their control (Tcf-1+/−) counterparts (Fig. 3C).
Fig. 3.
T-cell potential of Tcf-1−/− progenitors on OP9-DL1 cocultures. (A) Experimental scheme. Lin− BM precursors or progenitor subsets, as defined in Fig. 1, were sorted from Tcf-1−/− or WT mice. Equal numbers of Tcf-1−/− or WT progenitors were mixed and cocultured on OP9-DL1 stroma for 10 d. (B) OP9-DL1 cultures of the indicated progenitors. CD45.1 versus forward-scatter (FSC) plots define the gating of Tcf-1−/− (CD45.1−) versus WT (CD45.1+) cells. CD44 versus CD25 plots of the gated cells as indicated by arrows. Similar results were obtained in three independent experiments. (C) Apoptosis of Tcf-1−/− and control progenitors in OP9-DL1 cultures was measured by annexin V staining. (Left) Histograms show annexin V staining of control (Upper) and Tcf-1−/− (Lower) Lin− BM progenitors cultured on OP9-DL1 cocultures for 10 d. (Right) Histogram bars show the average frequency of annexin V+ cells from the indicated mice in three cocultures. Error bars are SD.
To establish that Tcf-1 deficiency did not interfere with thymic entry, we generated competitive BM chimeras by using Lin− BM progenitors from Tcf-1−/− and WT mice. Equal numbers of Tcf-1−/− progenitors were combined with WT progenitors and injected i.v. to reconstitute lethally irradiated syngeneic mice (Fig. 4A). At 10 wk after adoptive transfer, BM, thymi, and spleens were analyzed to compare the relative contribution of Tcf-1−/− and WT progenitors to the various stages and lineages of hematopoietic development. As expected, Tcf-1−/− cells efficiently reconstituted BM progenitor subsets and other mature blood lineages as determined by their frequency among B cells (B220+) and dendritic cells (CD11c+) (Fig. 4B and Fig. S4). Interestingly, within the thymi of the chimeras, Tcf-1−/− progenitors did repopulate the ETP stage (Fig. 4C). Their fraction in this subset was reduced compared with BM progenitors; however, this reduction was not statistically significant, indicating that Tcf-1−/− progenitors can efficiently seed the thymus. Tcf-1−/− progenitors were strikingly outcompeted in all subsequent stages of intrathymic and peripheral T-cell development (Fig. 4 D–E and Fig. S4). Altogether, our findings in vivo and in vitro lead to the conclusion that Tcf-1−/− uncommitted progenitors can efficiently migrate through the bloodstream and enter the thymus, but they are cell-intrinsically defective in T-cell commitment.
Fig. 4.
Tcf-1−/− progenitors are selectively defective in T-cell development. (A) Experimental scheme. Lin− BM progenitors (5 × 105) CD45.2+ sorted from Tcf-1−/− were mixed with an equal number of WT CD45.1+ Lin− BM progenitors and injected i.v. into lethally irradiated mice. After 10 wk, cells were harvested from BM, spleens, and thymi and analyzed. Progenitor subsets and mature lineages were gated as indicated to determine the fraction of CD45.2+ Tcf-1−/− cells versus CD45.2− WT cells in each population. (B) Histogram bars depict the relative contribution of Tcf-1−/− versus WT progenitors in reconstituting the indicated progenitor subsets and mature lineages. Values represent the average of five independent mice. (C–E) Gating strategy for ETP (C), DN1–DN4 (D), and double-positive (DP) and single-positive subsets (E). Similar results were obtained with more than 10 reconstituted mice in three independent experiments.
Tcf-1 Is Specifically Required at the c-kit+ DN1 Cell Subset.
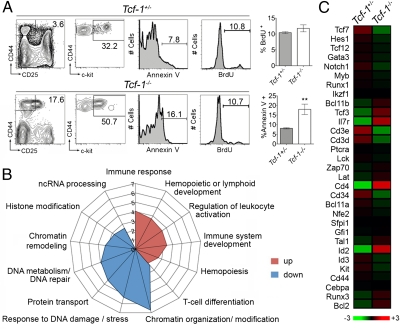
The earliest defect observed in Tcf-1−/− thymic development was the reduced level of c-kit expression at the DN1 stage and the presence of an increased fraction of CD25int CD44+ DN thymocytes (Fig. 1 A and E). Similar thymic phenotypes have been observed in mice deficient for transcription factors implicated in early T-cell differentiation, including Gfi1 (24). To gain insights into the properties of the c-kitlo DN1 Tcf-1−/− thymocytes, we compared them to the c-kit–expressing DN1 thymocytes in control mice. Tcf-1−/− thymi had a 30-fold reduction in c-kitlo DN1 cells compared with controls. Although these cells proliferated at similar rates to their control counterparts, they were significantly more apoptotic (Fig. 5A). We further determined the molecular impact of Tcf-1 ablation in these cells by comparing their global expression profiles to that of c-kit+ DN1 cells from Tcf-1+/− controls. To this aim, RNA was prepared from Lin−CD44+CD25−kit+ cells (Fig. 5A) and profiled on microarrays. Gene ontology analysis revealed an overall enrichment in the expression of genes involved in hematopoiesis and immune system development (Fig. 5B). Although genes involved in T-cell specification, including Notch1, Hes1, Gata3, Bcl11b, Runx1, and Ikzf1 (Ikaros), were largely unchanged in Tcf-1−/− cells (Fig. 5C), expression of some genes, such as Id2, Tcf3 (E2a), and Il7r, was elevated. Most T-cell signature genes showed unchanged expression (Ptcra, Lck, Zap70, and Lat), but Cd3d and Cd3e were down- regulated, and Cd4 was up-regulated. Interestingly, these analyses indicate a dramatic overall down-regulation of genes associated with response to DNA damage stimuli, chromatin remodeling, and DNA metabolic processes in the Tcf-1–deficient progenitors. This finding is consistent with our observation that these cells also show increased cell death. Our findings therefore indicate that Tcf-1 is required to maintain basic metabolic processes in early thymic immigrants that, in its absence, become abortive and die by apoptosis.
Fig. 5.
Comparison of the c-kit+ DN1 thymocytes subset in Tcf-1−/− versus WT mice. (A Left) CD44 versus CD25 plots depict the gating of DN1 cells in Tcf-1+/− versus Tcf-1−/− mice. CD44 versus c-kit plots are DN1 cells from Tcf-1+/− and Tcf-1−/− mice as indicated. The gate in these plots depicts Lin−CD44+c-kit+CD25− (ETP) cells analyzed for apoptosis proliferation and global gene expression. (Right) Annexin+ and BrdU histograms depict the fraction of apoptotic and proliferating ETPs, respectively, in the indicated mice (Materials and Methods). (B) Gene ontology analysis of up- and down-regulated genes in the indicated processes in Tcf-1−/− versus Tcf-1+/− ETPs. Scale shows fold enrichment. (C) Heat map showing the mean expression of genes representative of the ETP/T-cell signature from the indicated mice. Means are the average of three biological replicates.
Direct Activation of Tcf-1 by Notch1.
Transcriptional up-regulation of Tcf-1 starts at the ETP stage (Fig. S3B and ref. 25) and coincides with the developmental block in Tcf-1−/− progenitors. Given the key role that Notch-1 plays in triggering the T-cell differentiation process and considering the profound and early defects resulting from Tcf-1 ablation, we hypothesized that Notch-1 could directly activate Tcf-1. Intriguingly, chromatin immunoprecipitation followed by next-generation sequencing (ChIP-Seq) of activated Notch-1 and CSL in the mouse T-cell lymphoma cell line T6E (26) revealed that both bind to an evolutionarily conserved CSL consensus site ∼31.5 kb upstream of the Tcf-1 transcription start site (Fig. 6A). ChIP-Seq for Tcf-1 was performed in primary thymocytes of WT mice and also revealed strong Tcf-1 binding to an adjacent Tcf consensus site. The evolutionary conservation of this region, and the active binding of Notch-1, CSL, and Tcf-1 to it, suggest that it may represent an enhancer element.
Fig. 6.
Tcf-1 is the target of Notch1. (A) Visualization of ChIP-Seq data in the Tcf-1 (Tcf7) locus. Notch-1 and CSL ChIP-Seq data are from the T6E cell line, and Tcf-1 ChIP-Seq data are from WT thymocytes. Mammalian conservation (conservation) is shown under the data tracks. Magnification of the indicated area strongly bound by Notch-1, CSL, and Tcf-1 is shown (∼31.5 kb upstream of the Tcf-1 transcription start site). This region contains conserved CSL and Tcf binding sites as indicated. (B) EMSA with nuclear extracts from the 1F9 T-cell acute lymphoblastic leukemia (T-ALL) cell line (lanes 2–4) or purified CSL (lanes 5–7). Probes are from the conserved CSL binding site in the putative Tcf-1 enhancer region, starting at position 52127891 on chromosome 11 (mm9). (C) Heat map depicting the expression of the indicated genes in LSK BM progenitors sorted before or after MxCre-mediated induction of a dormant oncogenic Notch1-IC (N1-IC+). Fold change of expression between N1-IC+ and WT LSK samples is shown.
To further confirm Notch binding to this sequence, we performed electrophoretic mobility-shift assay (EMSA) with oligonucleotides containing the conserved CSL motif. Nuclear extracts from the E2A-deficient 1F9 lymphoma cell line that has active Notch signaling (27, 28) and purified CSL were probed for their ability to bind to this site. Mutant oligonucleotides in which the central 4 nt of the CSL consensus-binding site have been converted to adenines (A) were designed as negative controls (Materials and Methods). These analyses revealed specific binding of purified CSL to the oligonucleotide probe but not to the negative control (Fig. 6B, lanes 5–7). When incubated with nuclear extracts, the oligonucleotide probe migrated more slowly than the complex with only purified CSL did (Fig. 6B, compare lanes 2 and 5). This slower migration of the protein–DNA complex observed in 1F9 extracts suggests the presence of a multiprotein complex comprising factors such as Notch in addition to CSL at this site.
In a third approach to establish that Notch directly regulates Tcf-1, we examined the levels of Tcf-1 expression in LSK BM progenitors ectopically expressing an activated, intracellular Notch (Notch1-IC). To this aim, we interrogated our microarray data [Gene Expression Omnibus database accession no. GSE27799 (29)] and compared the expression profiles of LSK BM progenitors sorted before or after MxCre-mediated induction of a dormant oncogenic Notch1-IC (30). Indeed, Tcf-1 was sharply up-regulated by more than 10-fold in response to Notch1-IC induction, paralleling the behavior of a panel of known Notch targets (Fig. 6C). In conclusion, the temporal regulation of Tcf-1 expression, the ChIP-Seq and EMSA data, as well as the up-regulation of Tcf-1 in response to ectopic Notch1-IC activation show that Tcf-1 is directly regulated by Notch.
Discussion
The present study defines Tcf-1 as a gatekeeper of the earliest phase of T-cell specification when Notch signals initiate the T-lineage program in uncommitted progenitors. Tcf-1 is up-regulated upon entry of seeding progenitors to the thymus. We show here that transcriptional up-regulation of Tcf-1 is essential to enable progenitors to progress beyond the ETP stage. We provide evidence that the expression of factors involved in basic processes of DNA metabolism and chromatin organization is compromised, offering an explanation for why Tcf-1−/− ETPs are more apoptotic than control counterparts are. Importantly, the expression of most T-cell specification factors remains unaltered. Finally, we show that Notch1 directly mediates Tcf-1 transcription.
Wnt signaling has been previously implicated in the self-renewal of HSCs (31). BM development is normal in Tcf-1−/− mice, indicating that other members of the Tcf/Lef family may mediate Wnt signaling in HSCs. The dispensability of Tcf-1 for BM development is also in line with our detection of only low or no Tcf-1 transcripts in most BM progenitor subsets. Moreover, both control and Tcf-1−/− mice had comparable frequencies of circulating progenitors with T-cell potential, such as the LSK cells, CLPs, (2), and CTPs (1), suggesting that Tcf-1 is not required for progenitor emigration from the BM. Finally, the presence of ETP-stage Tcf-1−/− progenitors in the thymi of the competitive chimeras indicates that the Tcf-1 deficiency does not prevent T-cell progenitors from successfully entering the thymus. It is currently unclear why, despite the presence of ETPs in competitive chimeras, only a few ETPs were detected in thymi of constitutive Tcf-1−/− mice. This finding may indicate defects in the Tcf-1−/− stroma cells that compromise the maintenance of the thymus-seeding progenitors in these mice. Alternatively, the presence of WT thymocytes in the competitive chimeras may provide an environment that improves the survival of Tcf-1−/− thymus-seeding progenitors. The complete block at the ETP stage seen in BM chimeras also contrasts with the development of small numbers of T cells in Tcf-1−/− mice (12, 20). The presence of more mature developmental stages in Tcf-1−/− thymi may reflect compensatory expansion of these intrathymic populations and has been previously noted in other mouse models (32, 33).
Initiation of the T-cell program is marked by activation of Notch signaling by Delta-like ligands expressed on thymic stroma cells. The data presented here suggest that the next critical step in the signaling hierarchy of early T-cell specification is the induction of Tcf-1. We showed transcriptional up-regulation of Tcf-1 upon normal as well as ectopic Notch1 activation. Moreover, we have identified a conserved element upstream of the Tcf-1 gene that is occupied by Notch, CSL, and Tcf-1 itself. Tcf-1 has been previously reported to promote its own expression (34, 35). These findings suggest that the identified region has enhancer properties and that Tcf-1 is a direct target of Notch. Notch1 ablation has been shown to redirect thymocytes to the B-cell lineage (36). Because Tcf-1–deficient thymi show no accumulation of B cells, we conclude that Tcf-1 is not required for the suppression of the B-cell differentiation program. Tcf-1 may be required in progenitors that have already lost B-cell potential.
Overall, the expression profile of the c-kitlo Tcf1−/− ETPs is similar to that of control ETPs. However, a significant down-regulation of pathways involved in DNA metabolism and response to damage may explain increased apoptosis observed in Tcf-1–deficient thymocytes. This apoptosis cannot be explained by pro- or anti-survival factors because their expression was not significantly altered in Tcf-1−/− ETPs (Fig. 6C). Although it has previously been reported that Il7r is induced by β-catenin/Tcf signals (37), here we show in Tcf-1−/− ETPs that IL7r is up-regulated. Our findings are consistent with the increased surface IL7Rα expression previously noted in Tcf1−/− DN1 cells (38). Their IL7Rα+ c-kitlo surface profile renders the Tcf1−/− ETPs phenotypically similar to CLPs, which have not previously been detected in the thymus. Further analyses will be required to address this possibility.
Similar findings with respect to the Tcf-1–dependent block in early thymocyte development and the regulation of Tcf-1 expression by Notch were reported while our article was being considered for publication (39). These authors also propose that Tcf-1 may directly up-regulate the expression of T-cell essential genes. However, our profiling of ex vivo kitlo Tcf-1−/− and control ETPs argues that Tcf-1 does not generally affect the expression of T-cell specification factors, at least at this early stage. Further studies will be needed to determine the physiological significance of a few expression changes we did observe, such as the up-regulation of Id2 and Tcf3 (E2a).
In conclusion, our findings establish Tcf-1 as an essential early responder to Notch signals that is critically required for further progression to the T-cell lineage. We show that, in the absence of Tcf-1, progenitor thymocytes down-regulate DNA metabolic processes and become abortive.
Materials and Methods
Animals.
All mice were kept in the animal facilities of the University of Chicago according to protocol no. 71880 approved by the Institutional Animal Care and Use Committee. Tcf-1−/− mice on the C57BL/6 background were obtained from Hans Clevers (Hubrecht Institute, Utrecht, The Netherlands) and Frank Staal (Erasmus University, Rotterdam, The Netherlands) (12). We have previously reported the generation of the pre–T-cell receptor α HuCD25 reporter mice (40). The generation of Notch1fl/fl, Notch2fl/fl, and MxCre mice is described in refs. 41–43.
Monoclonal Antibodies and Flow Cytometry.
Multicolor FACS staining was performed for analysis and cell sorting of primary thymocytes on LSR II, FACSCanto, or FACSAria instruments (BD Biosciences). Antibodies were from BD Biosciences or eBioscience: CD4 (L3T4), CD8 (53-6.7), CD11b (M1/70), CD11c (N418), CD19 (6D5), CD25 (PC61.5), CD44 (1M7), B220 (RA3-6B2), CD45 (30-F11), CD45.1 (A20), CD45.2 (104), (53-2.1), CD122 (5H4), c-kit (2B8), Gr1 (RB6-8C5), Sca-1 (D7), TCRβ (H57-597), TCRγδ (eBioGL3), NK1.1 (PK136), DX5 (DX5), and Ter119 (Ter119). Biotinylated antibodies were detected with streptavidin–phycoerythrin/Cy5.5 or eFluor780. An annexin V–phycoerythrin labeling kit (BD Biosciences) was used according to the manufacturer's instructions. Data were analyzed in FlowJo software (Tree Star).
BM Chimeras.
Lethally irradiated (950 rad; Gammacell 40) CD45.1+ C57BL/6 mice (host) were injected with a 1:1 mixture of FACS-sorted host and Tcf-1−/− donor lineage (B220, CD3, CD8, CD4, CD11b, CD11c, CD19, NK1.1,or Ter119) negative BM (1 × 106 cells per mouse). Bactrim was added to the drinking water for the time of observation (4–10 wk).
OP9-DL1 Cell-Culture Conditions.
A 1:1 mixture of 104 FACS-sorted CD45.1+ WT and CD45.2+ Tcf-1−/− cells from corresponding progenitor subsets were transferred to a 24-well plate seeded with OP9-DL1 at 24 h before coculture to be ∼60% confluent. Progenitors were cocultured as described previously (44). Cultures were carried out for 10 d before analysis.
In Vivo BrdU-Incorporation Assay.
Mice were injected retroorbitally with 0.5 mg of BrdU (Sigma) per 5 g of body weight at 2 h before analysis of BM and thymic subpopulations by flow cytometry. BrdU staining was performed with the FITC BrdU Flow Kit (BD Biosciences) according to the manufacturer's instructions.
RNA Extraction and Quantitative Real-Time RT-PCR.
Cells were lysed, and RNA was extracted with the RNeasy Micro kit (Qiagen). cDNA was prepared with the SuperScript III RT kit (Invitrogen). Quantitative PCR was performed on an ABI7300 machine (Life Technologies) relative to Gapdh expression with TaqMan Gene Expression Assays from Applied Biosystems (Life Technologies). Data were analyzed according to the relative ΔΔCT method. Experiments were done in triplicate.
Gene-Expression Microarrays.
Approximately 1,000 ETPs were sorted directly into 100 μL of lysis buffer, and RNA was extracted as recommended (Arcturus PicoPure RNA Isolation kit; Life Technologies). RNA quality and concentration were estimated by using the Bioanalyzer Pico Chip and RNA 6000 Pico Assay reagents (Agilent). Average yield from 1,000 cells was 3 ng of total RNA. All material was amplified with Ovation Pico WTA System (NuGen). Labeling, fragmentation, and hybridization to Mouse Genome 430 2.0 Arrays were done according to the manufacturer's instructions (Affymetrix). Comparative analysis of gene-expression profiles from WT and mutant progenitors was performed with the Bioconductor limma package (45). Gene ontology analysis of target genes was conducted with the Functional Annotation Tool of DAVID software.
Supplementary Material
Acknowledgments
We thank J. Y. Park for excellent technical support for microarray analysis and B. Kee for helpful suggestions. This work was supported in part by National Institutes of Health Grants R01AI059676 (to F.G.) and F31 AI830542 (to K.G.); R01CA133379, R01CA105129, R21CA141399, and R01CA149655 (to I.A.); a Leukemia and Lymphoma Society Translational Research Program grant (to I.A.); American Cancer Society Grant RSG0806801 (to I.A.); the Irma T. Hirschl Charitable Trust; the Dana Foundation; the Mallinckrodt Foundation; the Gabrielle's Angels Foundation; and the Alex's Lemonade Stand Foundation (I.A.). C.L. is supported by the Helen L. and Martin S. Kimmel Center for Stem Cell Biology fellowship.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE33513).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110230108/-/DCSupplemental.
References
- 1.Krueger A, von Boehmer H. Identification of a T lineage-committed progenitor in adult blood. Immunity. 2007;26:105–116. doi: 10.1016/j.immuni.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwarz BA, Bhandoola A. Circulating hematopoietic progenitors with T lineage potential. Nat Immunol. 2004;5:953–960. doi: 10.1038/ni1101. [DOI] [PubMed] [Google Scholar]
- 3.Rothenberg EV, Zhang J, Li L. Multilayered specification of the T-cell lineage fate. Immunol Rev. 2010;238:150–168. doi: 10.1111/j.1600-065X.2010.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sultana DA, Bell JJ, Zlotoff DA, De Obaldia ME, Bhandoola A. Eliciting the T cell fate with Notch. Semin Immunol. 2010;22:254–260. doi: 10.1016/j.smim.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li P, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikawa T, et al. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–96. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- 9.Bajoghli B, et al. A thymus candidate in lampreys. Nature. 2011;470:90–94. doi: 10.1038/nature09655. [DOI] [PubMed] [Google Scholar]
- 10.Boehm T. Design principles of adaptive immune systems. Nat Rev Immunol. 2011;11:307–317. doi: 10.1038/nri2944. [DOI] [PubMed] [Google Scholar]
- 11.Willert K, Jones KA. Wnt signaling: Is the party in the nucleus? Genes Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- 12.Verbeek S, et al. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 13.Zhou X, et al. Differentiation and persistence of memory CD8+ T cells depend on T cell factor 1. Immunity. 2010;33:229–240. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeannet G, et al. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc Natl Acad Sci USA. 2010;107:9777–9782. doi: 10.1073/pnas.0914127107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Q, et al. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-γ. Nat Immunol. 2009;10:992–999. doi: 10.1038/ni.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staal FJ, et al. Wnt target genes identified by DNA microarrays in immature CD34+ thymocytes regulate proliferation and cell adhesion. J Immunol. 2004;172:1099–1108. doi: 10.4049/jimmunol.172.2.1099. [DOI] [PubMed] [Google Scholar]
- 17.Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat Immunol. 2006;7:1048–1056. doi: 10.1038/ni1381. [DOI] [PubMed] [Google Scholar]
- 18.Scheller M, et al. Hematopoietic stem cell and multilineage defects generated by constitutive β-catenin activation. Nat Immunol. 2006;7:1037–1047. doi: 10.1038/ni1387. [DOI] [PubMed] [Google Scholar]
- 19.Baba Y, Garrett KP, Kincade PW. Constitutively active β-catenin confers multilineage differentiation potential on lymphoid and myeloid progenitors. Immunity. 2005;23:599–609. doi: 10.1016/j.immuni.2005.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schilham MW, et al. Critical involvement of Tcf-1 in expansion of thymocytes. J Immunol. 1998;161:3984–3991. [PubMed] [Google Scholar]
- 21.Porritt HE, et al. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2004;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Allman D, et al. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 23.Gounari F, et al. Tracing lymphopoiesis with the aid of a pTα-controlled reporter gene. Nat Immunol. 2002;3:489–496. doi: 10.1038/ni778. [DOI] [PubMed] [Google Scholar]
- 24.Yücel R, Karsunky H, Klein-Hitpass L, Möröy T. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J Exp Med. 2003;197:831–844. doi: 10.1084/jem.20021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.David-Fung ES, et al. Transcription factor expression dynamics of early T-lymphocyte specification and commitment. Dev Biol. 2009;325:444–467. doi: 10.1016/j.ydbio.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci USA. 2011;108:14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engel I, Murre C. Disruption of pre-TCR expression accelerates lymphomagenesis in E2A-deficient mice. Proc Natl Acad Sci USA. 2002;99:11322–11327. doi: 10.1073/pnas.162373999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spaulding C, et al. Notch1 co-opts lymphoid enhancer factor 1 for survival of murine T-cell lymphomas. Blood. 2007;110:2650–2658. doi: 10.1182/blood-2007-04-084202. [DOI] [PubMed] [Google Scholar]
- 29.Klinakis A, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buonamici S, et al. CCR7 signalling as an essential regulator of CNS infiltration in T-cell leukaemia. Nature. 2009;459:1000–1004. doi: 10.1038/nature08020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reya T, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 32.Zlotoff DA, et al. CCR7 and CCR9 together recruit hematopoietic progenitors to the adult thymus. Blood. 2010;115:1897–1905. doi: 10.1182/blood-2009-08-237784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krueger A, Willenzon S, Lyszkiewicz M, Kremmer E, Förster R. CC chemokine receptor 7 and 9 double-deficient hematopoietic progenitors are severely impaired in seeding the adult thymus. Blood. 2010;115:1906–1912. doi: 10.1182/blood-2009-07-235721. [DOI] [PubMed] [Google Scholar]
- 34.Roose J, et al. Synergy between tumor suppressor APC and the β-catenin-Tcf4 target Tcf1. Science. 1999;285:1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- 35.Hovanes K, et al. β-Catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat Genet. 2001;28:53–57. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- 36.Wilson A, MacDonald HR, Radtke F. Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J Exp Med. 2001;194:1003–1012. doi: 10.1084/jem.194.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu Q, Xu M, Sen JM. β-Catenin expression enhances IL-7 receptor signaling in thymocytes during positive selection. J Immunol. 2007;179:126–131. doi: 10.4049/jimmunol.179.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goux D, et al. Cooperating pre-T-cell receptor and TCF-1-dependent signals ensure thymocyte survival. Blood. 2005;106:1726–1733. doi: 10.1182/blood-2005-01-0337. [DOI] [PubMed] [Google Scholar]
- 39.Weber BN, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gounari F, et al. Somatic activation of β-catenin bypasses pre-TCR signaling and TCR selection in thymocyte development. Nat Immunol. 2001;2:863–869. doi: 10.1038/ni0901-863. [DOI] [PubMed] [Google Scholar]
- 41.Radtke F, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 42.McCright B, Lozier J, Gridley T. Generation of new Notch2 mutant alleles. Genesis. 2006;44:29–33. doi: 10.1002/gene.20181. [DOI] [PubMed] [Google Scholar]
- 43.de Boer J, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 44.Garbe AI, Krueger A, Gounari F, Zúñiga-Pflücker JC, von Boehmer H. Differential synergy of Notch and T cell receptor signaling determines αβ versus γδ lineage fate. J Exp Med. 2006;203:1579–1590. doi: 10.1084/jem.20060474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.