Abstract
Arenaviruses are important agents of zoonotic disease worldwide. The virions expose a tripartite envelope glycoprotein complex at their surface, formed by the glycoprotein subunits GP1, GP2 and the stable signal peptide. This complex is responsible for binding to target cells and for the subsequent fusion of viral and host-cell membranes for entry. During this process, the acidic environment of the endosome triggers a fusogenic conformational change in the transmembrane GP2 subunit of the complex. We report here the crystal structure of the recombinant GP2 ectodomain of the lymphocytic choriomeningitis virus, the arenavirus type species, at 1.8-Å resolution. The structure shows the characteristic trimeric coiled coil present in class I viral fusion proteins, with a central stutter that allows a close structural alignment with most of the available structures of class I and III viral fusion proteins. The structure further shows a number of intrachain salt bridges stabilizing the postfusion hairpin conformation, one of which involves an aspartic acid that appears released from a critical interaction with the stable signal peptide upon low pH activation.
Keywords: enveloped viruses, membrane fusion, RNA viruses, emerging viruses
Arenaviruses are agents of emerging zoonotic diseases such as severe hemorrhagic fever. They cause chronic infections in rodents, the natural hosts, and transmission to humans often occurs as a result of man-driven changes in the ecosystem that alter the rodent population. Examples of recently emerged arenavirus infections in humans are those of Chapare and Lujo hemorrhagic fever viruses in Bolivia and South Africa, respectively (1, 2).
The Arenaviridae family contains a single genus subdivided in two separate serological groups, the Old World and the New World arenaviruses. The lymphocytic choriomeningitis virus (LCMV), Lujo virus, and the highly pathogenic Lassa fever virus, which is endemic of West Africa, are members of the first group. The second one includes several South American hemorrhagic fever viruses—among them Machupo, Junin, Sabia, Guanarito, and Chapare viruses. LCMV is the arenavirus type species and is widely used as experimental model for the study of viral persistence and pathogenesis (3). LCMV infections in humans occur worldwide, and, although often asymptomatic, they can cause a spectrum of illnesses ranging from isolated fever to meningitis and encephalitis (4, 5). In particular, a number of fatal cases of transplant-associated LCMV infections have been recently reported, illustrating the threat to patients undergoing immunosuppressive treatment (6–8).
Arenavirus virions are pleomorphic enveloped particles containing a bisegmented, ambisense RNA genome with four open reading frames. The long (L) RNA segment encodes the viral RNA-dependent RNA polymerase (protein L, 200 kDa) and a small RING-finger protein (Z, 11 kDa) involved viral replication. The short (S) segment encodes the nucleoprotein (NP, 63 kDa) and the viral glycoprotein complex precursor (GPC, 75 kDa). Proteolytic maturation of the 498-amino acid GPC precursor results in three noncovalently bound subunits: the stable signal peptide (SSP, aa 1–58), the surface exposed GP1 (aa 59–265), and the transmembrane GP2 (266–498) to make a (SSP/GP1/GP2)3 trimeric complex. SSP is myristoylated (9) and rearranges after signalase cleavage to translocate its C-terminal end back to the cytosolic side of the membrane, thus traversing the membrane twice (10). Its interactions with the cytosolic domain of GP2 are responsible for SSP retention as part of the mature GPC, as shown for Junin virus (11). GP1 and GP2 are proteolytically derived from the GPC ectodomain by subtilase SKI-1/S1P (Fig. 1A) in the early Golgi compartments. This cleavage event is essential for productive infection and viral spread (12–15). GP1 is responsible for interactions with cellular receptors for entry. Old World arenaviruses such as LCMV or Lassa fever virus use α-dystroglycan as entry receptor (16, 17), whereas the pathogenic subgroup of New World arenaviruses uses the transferrin receptor-1 (TfR1) (18, 19). The interaction of GP1 with the receptor results in virion uptake by the cell into an endosomal compartment, the acidic environment of which triggers a fusogenic conformational change in GP2 (20), the membrane fusion subunit. GP2 contains a bipartite fusion peptide, with one of the segments located at the very N terminus of the protein, as observed in many class I viral fusion proteins. GP2 also has a disulfide-bond-stabilized internal fusion loop located close to the N terminus (21), similar to the Ebola virus (22) and the avian sarcoma/leucosis retrovirus fusion proteins (23). For Junin virus—and by extension, for all arenaviruses—the acid sensitivity was shown to be controlled, at least in part, by interactions between the GP2 ectodomain and the luminal loop connecting the two transmembrane (TM) segments of SSP (24).
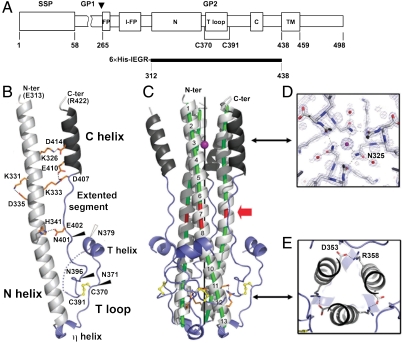
Fig. 1.
Overall structure of the LCMV GP2 trimer. (A) Diagram of the arenavirus GPC glycoprotein precursor (top) and the GP2 construct used in this study (bottom, full black line), numbered according to their position in the LCMV (WE-HPI strain) GPC sequence (Genbank accession no. CAC01231.1). An arrowhead marks the GP1/GP2 cleavage site. The boxes highlight the two fusion loops (N-FP and internal I-FP), the N and C helices, the T loop and the TM helix. IEGR indicates the Factor Xa cleavage site. (B) Ribbon diagram of the LCMV GP2 protomer. The conserved disulfide is displayed in yellow ball-and-stick. Residues involved in intrachain (B) and interchain (C) salt bridges are shown in orange. Full and empty arrowheads, respectively, mark conserved and nonconserved potential N-glycosylation sites in arenavirus GP2. (C) LCMV GP2 trimer. A vertical, central black line marks the coiled coil axis, with a magenta sphere indicating the location of a central Cl- ion. Green tubes mark the curved N-helix axes, with the stutter in red (highlighted with a full red arrow). (D) Blow up of the Cl- ion (magenta sphere) binding site, viewed down the coiled coil axis. The ion is chelated by Asn325 which also hydrogen bonds a structural water molecule (red spheres). (E) Interchain salt bridge stabilizing the base of the coiled coil mentioned in the text.
Amino acid sequence analyses and biochemical studies with recombinant fragments have indicated that, despite the presence of a bipartite fusion loop, GP2 is a typical class I fusion protein, forming an alpha helix-rich trimer in its postfusion state (25). These studies identified two α-helical regions (N- and C-terminal) with a heptad repeat pattern in the N-terminal helix (25, 26). A loop containing a conserved disulfide bond connects the two helices. A model of the structure has been proposed by comparison with the GP2 counterpart from Ebola virus (26), for which the structure is known (27, 28). Ebola virus, however, belongs to a different viral family (the Filoviridae), and other than the presence of the characteristic heptad repeat pattern typical of coiled coils—as well as a similar location of cysteine residues—shares poor sequence identity (less than 20%) with the arenavirus GP2.
We report here the structure of the recombinant LCMV GP2 ectodomain at 1.8-Å resolution. As expected, it forms an alpha-helical coiled coil similar to that of other class I viral fusion proteins in their postfusion conformations. The structure also shows a number of intrachain salt bridges stabilizing the postfusion hairpin, one of them involving an aspartic acid that is believed to interact with SSP in the prefusion conformation. Furthermore, comparison with the available structures of other viral fusion proteins reveals the striking conservation of a central stutter present in the alpha-helical coiled coil of GP2. The stutter provides a register allowing for unambiguous structural alignment of these proteins along the coiled coil. We show that the shared central location of the stutter in fusion proteins from many unrelated viruses—including those from class III, which also exhibit a central parallel 3-helix bundle—permits a direct comparison of the relative location of their functional elements, revealing similarities and clear distinctions among them that have not been discussed previously.
Results and Discussion
Structure Determination.
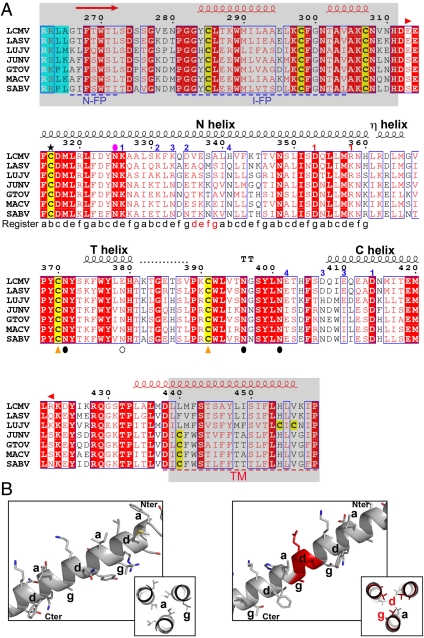
The details of the construct used for crystallization, the boundaries of which were delineated earlier (25), are outlined in Fig. 1A. The details of the crystallization and the structure determination procedures are described in SI Text. In brief, we crystallized the fragment spanning amino acids 312–438 of the LCMV GPC precursor, lacking the bipartite fusion peptide/loop region of GP2, which was omitted to avoid aggregation of the protein. In addition, to prevent artificial interchain disulfide bond formation, we mutated Cys316 to serine because this residue is believed to make a disulfide bond with one of the cysteines upstream (Cys285, Cys298, or Cys307; see Fig. 2A), which are in the fusion loop region. The experimental electron density map was interpretable for residues 313–422 (red arrowheads in Fig. 2A), with a break between amino acids 382–388 (dots above the sequence in Fig. 2A), which are disordered in the crystal, as are the C-terminal residues 423–438.
Fig. 2.
Amino acid sequence alignment from representative arenaviruses. (A) Highly conserved residues are displayed in white font on red background, cysteines in yellow background, with two full orange triangles below the alignment marking the cysteines involved in the disulfide bond in the T loop. A black star above the alignment marks the cysteine mutated to serine in the construct. The N-terminal light-blue background highlights the consensus motif for the SKI-1/S1P cleavage site. A light-gray background marks segments of the GP2 sequence not included in the crystallized construct. The observed secondary structure is indicated above the sequence in black, while predicted secondary structures in the segments missing in the structure are in red. Red arrowheads on the sequence mark the limits of the atomic model built from this crystallographic study, with a dotted line indicating a disordered region in the T loop. Intra- and interchain salt bridges are numbered above the alignment in red and blue, respectively. A full red circle above the alignment marks Asn325, which chelates the central chloride ion. Conserved and nonconserved predicted N-glycosylation sites are indicated by full and open black circles, respectively, below the alignment. The “register” line provides the abcdefg heptad repeat assignment, with the x layer type stutter in red. Dashed lines under the sequences indicate the fusion peptide and loop (in blue at the N-terminal end) and the TM region (in red, at the C-terminal end). The sequences are truncated before the respective cytoplasmic tails. (B) Left: Central coiled coil region, showing residues with regular “knobs-into-holes” packing. Right: Region of the x layer type stutter (in red), with Val336 (position d, knob-into-knob packing) and Ala339 (position g) pointing toward the coiled coil axis.
Description of the Molecule.
The structure shows that recombinant LCMV GP2 is an elongated trimer about 75 Å long (Fig. 1C), in which each protomer is folded in the typical “hairpin” postfusion conformation of viral fusion proteins. The protomer begins and ends as an alpha helix: a long amino-terminal helix spanning amino acids 315–360 (N helix) and a carboxy-terminal helix comprising residues 408–420 (C helix), which interact with each other in the trimer. Amino acids 313 and 422, at the amino and carboxy termini, respectively, are thus located at the same level, at one end of the trimeric rod (Fig. 1B). This conformation insures proximity of the two membrane-interacting regions (C-terminal TM segment and N-terminal fusion peptide/loop) in the full-length GP2 protein after the fusogenic conformational change, as shown for many other viral fusion proteins. Likewise, the N helix forms a central trimeric coiled coil, while the C helices pack against the grooves formed in between N helices in the trimer. The C helix makes four full turns and spans only about one third of the length of the N helix, which makes 13 turns (Fig. 1C). An extended polypeptide segment, spanning residues 391–407 and running “backwards” along the coiled coil grooves, connects the C helix to the “turn” of the molecule—i.e., the region where the chain reverts orientation at the C terminus of the N helix. Several highly conserved bulky aromatic or aliphatic side chains (Trp392, Leu400, Phe405; see Fig. 2A) anchor this segment in an extended conformation into the interhelical grooves of the coiled coil. At the end of the N helix, the chain reversal begins by a one-turn 3/10 helix (η helix; Fig. 1B) running roughly at 90 degrees from the N helix. Then the polypeptide makes an excursion to form a loop (the “T loop”), locked at its base by a highly conserved disulfide bond between Cys370 and Cys391. A third alpha helix (“T helix,” two turns) is present at the apex of the T loop (Fig. 1 B and C). There is a break in the electron density between the T helix and the disulfide at the base (dotted lines in the ribbon diagram, Fig. 1B).
As depicted in Fig. 1C, the GP2 trimer can be divided roughly in two halves, a membrane proximal “top” half and a “bottom” half (i.e., in the orientation of Fig. 1C, above and below the red arrow, respectively). The top half is a six-helix bundle with a central ion, presumably chloride as in other viral coiled coils, coordinated by the strictly conserved Asn325 (Fig. 1D). The bottom half has a three-layered structure, with an inner core formed by the C-terminal half of the central triple-stranded coiled coil, a middle layer formed by the extended segment between Cys391 and the C helix, and a third layer made by the T loop packing as a “flap” against the two inner layers. The fact that part of the flap is disordered in the crystal indicates that it packs more loosely than the rest of the molecule and is held in place in part by the disulfide connecting it to the end of the extended segment in the second layer (Fig. 1C). An interchain salt bridge, formed by residues Asp353 and Arg358 under the flap, stabilizes the end of the inner core (Fig. 1E). This salt bridge is strictly conserved in all arenavirus family members (Fig. 2A), suggesting a structural role. Although the recombinant protein used for crystallization was produced in Escherichia coli and is therefore not glycosylated, the structure shows that the four predicted N-linked glycosylation sites in arenavirus GP2 (at positions 371, 379, 396, and 401, marked with circles under the sequence in Fig. 2A) cluster in the bottom half of the trimer (arrowheads in Fig. 1B). Removal of glycosylation sites was shown to affect proper folding of the arenavirus envelope protein in the prefusion state, but the GP2 ectodomain can obviously fold into its postfusion hairpin conformation when produced in E. Coli, in the absence of glycans and/or ER chaperones, as was shown first for the influenza virus hemagglutinin HA2 protein (29). LCMV GP2 lacks the second arenavirus predicted glycosylation site (at position 379, empty circle in Fig. 2A), which lies in the second turn of the T helix (empty arrowhead in Fig. 1B). Of note, the first turn of this helix contains the epitope (374-Lys-Phe-Trp-Tyr-Leu-378) of an arenavirus broadly cross-reactive monoclonal antibody (mAb 83.6) (30). This antibody was shown to be more reactive after the acid pH-induced fusogenic conformational change of GP2 (20), in line with its location in the flap, at the membrane-distal end of the molecule. The presence of a glycan immediately downstream, at position 379, is likely to partially mask this epitope in many of the virulent arenaviruses.
The GP2 residues strictly conserved in the alignment of Fig. 2A form two clusters in the GP2 structure, one at the top and the other at the bottom of the molecule (Fig. S1). Inspection of Fig. 2A indicates that this pattern is maintained in the region of the top of the molecule that is missing from the structure (at the N- and C-terminal ends of the ectodomain). The bottom cluster includes residues of all three layers of the molecule, including the flap. The conservation of these residues highlights their importance in maintaining a very stable postfusion conformation.
Intrachain Salt Bridges Stabilize the GP2 Postfusion Hairpin.
In addition to the hydrophobic interactions of the C helix and the extended segment packing against the central coiled coil, a number of intraprotomer polar/electrostatic interactions are seen stabilizing the hairpin conformation, especially in the top half the trimer (Fig. 1B). There are two salt bridges between lysine side chains in the N helix and aspartate and/or glutamate residues in the C helix (Lys326 with Asp414, and Lys333 with Asp407 and Glu410). The first salt bridge is highly conserved (Fig. 2A). In particular, a mutation to alanine of the Asp414 counterpart in Junin virus (Asp400 in Junin GPC numbering) was shown to restore the wild-type pH sensitivity of a Lys33Gln mutant in the SSP subunit (24). This observation suggests that the polypeptide region that becomes the C helix in the postfusion hairpin conformation makes electrostatic interactions with SSP in the prefusion form, although we cannot rule out that a more global effect of these mutations on the protein affects the pH dependence of the conformational change.
A third salt bridge, this time LCMV-specific, connects His341 in the N helix with Glu402 in the extended segment. Finally, a fourth salt bridge connects two residues close in the sequence, Lys331 and Asp335, from two consecutive turns of the N helix (Fig. 1B). This interaction is part of a trimer contact, with the Asp335 side chain making van der Waals interactions with Ser406 from the adjacent protomer. This salt bridge is conserved only in Old World arenaviruses. Junin and Machupo viruses lack the equivalent to Lys331 (which becomes threonine), but at the same time the residue at position 406 is arginine, whereas aspartate or glutamate are maintained at position 335 (Fig. 2A). We therefore predict that in Machupo and Junin viruses there is an interchain salt bridge connecting the residues at positions 335 and 406 (LCMV numbering). Importantly, it was shown that mutation of Arg406 to alanine abolished membrane fusion induced by Junin virus (31). Taken together, these observations highlight the functional implication of salt bridges stabilizing the postfusion hairpin conformation of GP2 during the membrane fusion reaction.
A Stutter in the Coiled Coil Region.
At the center of the GP2 trimer, the coiled coil is markedly open due to the presence of an x layer type stutter at the seventh turn of the N helix (red in Fig. 1C). The stutter is a perturbation of the heptad repeat pattern of the coiled coil in the form of a four-residue insertion, with a “defg” motif intercalated in between two “abcdefg” repeats (32), as labeled in red under the corresponding sequence in Fig. 2A. Instead of the regular “knob-into-hole” packing of side chains normally observed at a and d positions in the heptad, the side chain of the residue at the stutter (Val336) packs as “knob-into-knob” against its counterparts from the other two helices, with the side chains pointing toward the central threefold axis, as illustrated in Fig. 2B. The presence of the stutter induces a slight curvature of the N helices, creating an internal cavity within the coiled coil delimited above and below by two aromatic layers formed by the side chains of Phe332 and Phe343, respectively.
Stutter-Based Alignment of Viral Coiled Coils.
We inspected the CC+ coiled coil database (33) and found the presence of an x layer type stutter in the coiled coil of the majority of class I and III viral fusion proteins, with the notable exception of those from lentiviruses (HIV-1 and SIV gp41). Further examination of the 51 trimeric parallel coiled coils currently present in the database indicated that the stutter is present only in the structure of postfusion viral fusogenic proteins, independent of the length of the coiled coil helices. It is absent in the other parallel, trimeric coiled coils from either cellular or viral origin.
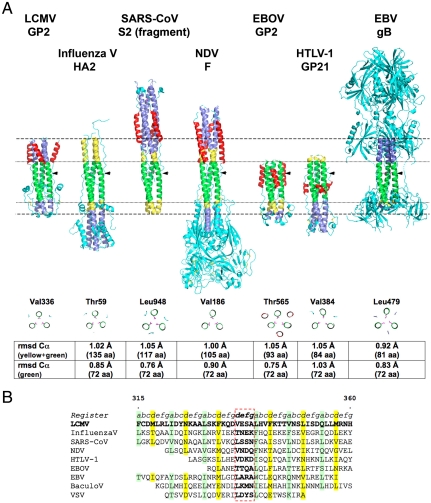
We found that the location of the stutter allows for an unambiguous alignment of the central coiled coils of the corresponding fusion proteins (Fig. 3). The side chain at the stutter (Val336 in LCMV) is branched in all the examples that we have analyzed (Fig. 3B). The corresponding superposition of the 3D structures revealed a common trimeric coiled coil core comprising 24 residues (i.e., GP2 residues 329–352 in the N helix, corresponding to roughly seven turns and to 72 residues in the trimer). The Cα atoms in this segment align with rmsd under 1 Å with all of the viral fusion proteins displayed in Fig. 3A. The matching segment is longer than this minimal core in the superposition with the GP2 counterparts from several viruses (green + yellow in Fig. 3A). For instance, influenza virus HA2 has 135 α-carbons (i.e., 45 per protomer, corresponding to 13 turns or the entire LCMV GP2 coiled coil) matching within 1 Å rmsd. This very good match indicates that the packing of the alpha helices is almost identical in spite of the poor amino acid sequence identity in the same region. The SARS coronavirus and the Newcastle disease virus (NDV, a paramyxovirus) also display matching segments longer than the minimal core (Fig. 3A). Superposition on the stutter also brings into coincidence the Cl- ion observed in LCMV GP2 with one of the two central Cl- ions identified in the SARS coronavirus S protein (34) (see SI Text). The list of viral proteins with a structurally very similar coiled coil region includes the Epstein Barr virus (EBV) glycoprotein B (gB), which is a class III fusion protein (Fig. 3A).
Fig. 3.
Stutter-based structural alignment of classes I and III viral fusion proteins. (A) Top: Side-to-side comparison of the postfusion structures of the indicated viral envelope proteins, after superposing the stutters (arrow). Horizontal dashed lines indicate the limits of the LCMV coiled coil, and horizontal dotted lines indicate the extent of the common core structure (in green). The remainder of the N helix is in blue and the C helices in red. Yellow indicates N-term and C-term extensions of the core, to highlight the region of each protein superposing within 1.0 Å rmsd onto LCMV GP2. Bottom: Cross-sections at the x layer type stutters (displaying “knob-into-knob” packing), indicated by black arrows in the alignment at the top. The table shows the corresponding rmsd values. Accession numbers for the structures used are provided in SI Text. (B) Stutter-based sequence alignment of viral fusion glycoproteins from classes I and III. The position of the x layer type stutter is indicated by the dashed red frame. Positions a and d of the central residues in the coiled coil are indicated in green and yellow, respectively.
Although the conserved coiled coil core of LCMV GP2 and the viral proteins represented in Fig. 3 align very well, we found that this is not the case with all members within the corresponding viral families, in spite of the conservation of the stutter. This is illustrated in Fig. S2, which shows that the fusion (F) proteins from some of the other members of the Paramyxoviridae family have a proline residue in the helical turn immediately downstream the stutter. This feature distorts the corresponding α-helices such that the register of the heptad repeats is altered, resulting in a second stutter. This is reflected in a higher rmsd obtained after superposition to the core coiled coil (around 2 Å; Fig. S2A). Despite this distortion, the side-to-side comparisons of the molecules presented in Fig. S2A shows that the conserved stutter provides the correct structural alignment of these homologous molecules, which in turn validates their comparison to LCMV GP2.
In the Retroviridae family, the stutter is absent altogether in the lentiviruses, and in that case the best alignment obtained when superposing the core segment of LCMV GP2 onto the coiled coil of SIV gp41 yields 1.8 Å rmsd, again a clearly poorer match than those reported in Fig. 3. Concerning other members of this viral family, the amino acid sequence alignment of HTLV-1 and MoMLV allows the prediction of the stutter in the MoMLV coiled coil (Fig. S3) in spite of its absence in the available structure, which appears truncated. Similarly, the class III fusion proteins, which are present in viruses belonging to unrelated families (gB from herpes viruses, gp64 from baculovirus, and G from the vesicular stomatitis virus) all display the stutter and can be aligned accordingly. However, these molecules align to EBV gB with a higher rmsd (between 1.3 Å and 2 Å; Fig. S4) than does EBV gB to LCMV GP2 (Fig. 3). This indicates that the conformation of the helices in the coiled coil can also be affected by their particular amino acid sequence and/or the local environment within the protein. Indeed, the central alpha helices in HSV-1 gB, baculovirus gp64 and VSV G appear more straight and less coiled around each other when compared to EBV gB or LCMV GP2 (Fig. S4). Yet it is clear from Fig. S4 that the stutter provides a useful means of aligning them, highlighting for instance the differences at the turn of the hairpin and showing that VSV G is clearly the most distant.
In summary, although the class I fusion proteins have been compared side to side before, the stutter provides a useful reference for superposing the trimeric molecules, with a single register along the heptad repeat. This alignment allows a comparison of the relative locations of the functional elements of the molecule (i.e., fusion loop and TM region) with respect to the conserved core. For instance, it is clear that there is an extension, in the form of a longer coiled coil, between the coronavirus core and the membrane-interacting elements. This feature is shared only with the paramyxovirus F protein, in addition to the presence of a bigger “head” of the molecule (absent form the structure of the coronaviruses S protein, which was truncated). These similarities between the two proteins could be an indication of homology between them, although more structural studies of the coronavirus S protein are needed to confirm this hypothesis. The alignment of Fig. 3 thus also indicates that the common core of the fusion protein is positioned at a different distance from the fused membrane for different viruses. Finally, it is clear from Fig. 3 that there are extensive variations in the region of the turn of the molecule, as well as toward the membrane interaction region (top) region, of the respective hairpins.
Concluding Remarks.
The structure of the recombinant LCMV GP2 ectodomain points to a number of salt-bridges stabilizing its postfusion hairpin conformation. The availability of mutagenesis data on some of the residues making these interactions, which display fusion deficient phenotypes, highlights their functional importance. The link in Junin virus between the Lys33Gln and Asp414Ala (LCMV numbering) mutants with the pH sensitivity of the fusion reaction (24) also suggest that the polypeptide segment that forms the C helix in the postfusion form may be involved in interactions with SSP in the prefusion conformation. The structure further reveals that the stutter provides a common register to align the trimeric coiled coils of many viral fusion proteins, a striking observation that had not been previously reported. For some viruses, the conservation of the central core appears to be the result of convergent evolution (for instance, when comparing GP2 with the class III proteins, and probably also with the paramyxovirus and coronavirus fusion proteins). In contrast, comparison with the Ebola and HTLV fusion proteins suggests divergent evolution from an ancestral gene—as pointed out before—which could have further diverged in the case of the lentiviruses to the point that the stutter is lost. It is striking, however, that the closest structural homolog of LCMV GP2 is the influenza virus HA2 protein. Finally, the broad conservation of the stutter in viral fusion proteins—and its absence in other proteins with a parallel, trimeric coiled coil—suggests a functional implication of this architectural feature, which may provide clear advantages to a protein that folds initially in one conformation and adopts later a different one in order to induce membrane fusion.
Materials and Methods
We used a synthetic DNA (Entelechon) corresponding to LCMV strain WE-HPI (35), codon optimized for E. coli production, inserted into the pET-19b vector (Novagen). The gene was expressed in the cytoplasm of E. coli Rosetta-gami (DE3) strain and cells were broken by two passages through an Emulsiflex-C5 homogenizer (Avestin) to recover the protein (25). The soluble fraction was separated by centrifugation, purified by Ni-nitrilotriacetic acid affinity chromatography (Amersham Biosciences) followed by gel filtration with a Superdex 75 16/60 column (GE Healthcare). The structure was determined to 1.8-Å resolution (Tables S1 and S2), as explained in the SI Text. All illustrations were prepared with the PyMOL Molecular Graphics System (36).
Supplementary Material
Acknowledgments.
We thank Ahmed Haouz and Patrick Weber from the robotized crystallization facility (PF6) at Institut Pasteur for help in crystal screening and the staff at the Swiss Light Source (SLS) for assistance during data collection. Diffraction data were collected with the Pilatus detector at the PXI beamline, SLS synchrotron (Villigen, Switzerland). F.A.R. acknowledges support from the European Union (VirApt consortium 018753), Institut Pasteur, Centre National de la Recherche Scientifique, and Merck Serono. H.H. and B.E. acknowledge support from the Swiss National Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3MKO).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108910108/-/DCSupplemental.
References
- 1.Delgado S, et al. Chapare virus, a newly discovered arenavirus isolated from a fatal hemorrhagic fever case in Bolivia. PLoS Pathog. 2008;4:e1000047. doi: 10.1371/journal.ppat.1000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briese T, et al. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 2009;5:e1000455. doi: 10.1371/journal.ppat.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oldstone MB. A suspenseful game of ‘hide and seek’ between virus and host. Nat Immunol. 2007;8:325–327. doi: 10.1038/ni0407-325. [DOI] [PubMed] [Google Scholar]
- 4.Jahrling PB, Peters CJ. Lymphocytic choriomeningitis virus. A neglected pathogen of man. Arch Pathol Lab Med. 1992;116:486–488. [PubMed] [Google Scholar]
- 5.Mets MB, Barton LL, Khan AS, Ksiazek TG. Lymphocytic choriomeningitis virus: An underdiagnosed cause of congenital chorioretinitis. Am J Ophthalmol. 2000;130:209–215. doi: 10.1016/s0002-9394(00)00570-5. [DOI] [PubMed] [Google Scholar]
- 6.Fischer SA, et al. Transmission of lymphocytic choriomeningitis virus by organ transplantation. N Engl J Med. 2006;354:2235–2249. doi: 10.1056/NEJMoa053240. [DOI] [PubMed] [Google Scholar]
- 7.Peters CJ. Lymphocytic choriomeningitis virus—an old enemy up to new tricks. N Engl J Med. 2006;354:2208–2211. doi: 10.1056/NEJMp068021. [DOI] [PubMed] [Google Scholar]
- 8.Palacios G, et al. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med. 2008;358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- 9.York J, Romanowski V, Lu M, Nunberg JH. The signal peptide of the Junin arenavirus envelope glycoprotein is myristoylated and forms an essential subunit of the mature G1-G2 complex. J Virol. 2004;78:10783–10792. doi: 10.1128/JVI.78.19.10783-10792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agnihothram SS, York J, Trahey M, Nunberg JH. Bitopic membrane topology of the stable signal peptide in the tripartite Junin virus GP-C envelope glycoprotein complex. J Virol. 2007;81:4331–4337. doi: 10.1128/JVI.02779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briknarova K, Thomas CJ, York J, Nunberg JH. Structure of a zinc-binding domain in the Junin virus envelope glycoprotein. J Biol Chem. 2011;286:1528–1536. doi: 10.1074/jbc.M110.166025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lenz O, ter Meulen J, Klenk HD, Seidah NG, Garten W. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc Natl Acad Sci USA. 2001;98:12701–12705. doi: 10.1073/pnas.221447598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beyer WR, Popplau D, Garten W, von Laer D, Lenz O. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J Virol. 2003;77:2866–2872. doi: 10.1128/JVI.77.5.2866-2872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunz S, Edelmann KH, de la Torre JC, Gorney R, Oldstone MB. Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions. Virology. 2003;314:168–178. doi: 10.1016/s0042-6822(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 15.Rojek JM, Lee AM, Nguyen N, Spiropoulou CF, Kunz S. Site 1 protease is required for proteolytic processing of the glycoproteins of the South American hemorrhagic fever viruses Junin, Machupo, and Guanarito. J Virol. 2008;82:6045–6051. doi: 10.1128/JVI.02392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao W, et al. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science. 1998;282:2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- 17.Spiropoulou CF, Kunz S, Rollin PE, Campbell KP, Oldstone MB. New World arenavirus clade C, but not clade A and B viruses, utilizes alpha-dystroglycan as its major receptor. J Virol. 2002;76:5140–5146. doi: 10.1128/JVI.76.10.5140-5146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radoshitzky SR, et al. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature. 2007;446:92–96. doi: 10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abraham J, et al. Host-species transferrin receptor 1 orthologs are cellular receptors for nonpathogenic new world clade B arenaviruses. PLoS Pathog. 2009;5:e1000358. doi: 10.1371/journal.ppat.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Simone C, Zandonatti MA, Buchmeier MJ. Acidic pH triggers LCMV membrane fusion activity and conformational change in the glycoprotein spike. Virology. 1994;198:455–465. doi: 10.1006/viro.1994.1057. [DOI] [PubMed] [Google Scholar]
- 21.Klewitz C, Klenk HD, ter Meulen J. Amino acids from both N-terminal hydrophobic regions of the Lassa virus envelope glycoprotein GP-2 are critical for pH-dependent membrane fusion and infectivity. J Gen Virol. 2007;88:2320–2328. doi: 10.1099/vir.0.82950-0. [DOI] [PubMed] [Google Scholar]
- 22.Ito H, Watanabe S, Sanchez A, Whitt MA, Kawaoka Y. Mutational analysis of the putative fusion domain of Ebola virus glycoprotein. J Virol. 1999;73:8907–8912. doi: 10.1128/jvi.73.10.8907-8912.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delos SE, White JM. Critical role for the cysteines flanking the internal fusion peptide of avian sarcoma/leukosis virus envelope glycoprotein. J Virol. 2000;74:9738–9741. doi: 10.1128/jvi.74.20.9738-9741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.York J, Nunberg JH. Intersubunit interactions modulate pH-induced activation of membrane fusion by the Junin virus envelope glycoprotein GPC. J Virol. 2009;83:4121–4126. doi: 10.1128/JVI.02410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eschli B, et al. Identification of an N-terminal trimeric coiled-coil core within arenavirus glycoprotein 2 permits assignment to class I viral fusion proteins. J Virol. 2006;80:5897–5907. doi: 10.1128/JVI.00008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallaher WR, DiSimone C, Buchmeier MJ. The viral transmembrane superfamily: Possible divergence of arenavirus and Filovirus glycoproteins from a common RNA virus ancestor. BMC Microbiol. 2001;1:1. doi: 10.1186/1471-2180-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weissenhorn W, Carfi A, Lee KH, Skehel JJ, Wiley DC. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 28.Malashkevich VN, et al. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc Natl Acad Sci USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Skehel JJ, Wiley DC. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc Natl Acad Sci USA. 1999;96:8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber EL, Buchmeier MJ. Fine mapping of a peptide sequence containing an antigenic site conserved among arenaviruses. Virology. 1988;164:30–38. doi: 10.1016/0042-6822(88)90616-2. [DOI] [PubMed] [Google Scholar]
- 31.York J, Agnihothram SS, Romanowski V, Nunberg JH. Genetic analysis of heptad-repeat regions in the G2 fusion subunit of the Junin arenavirus envelope glycoprotein. Virology. 2005;343:267–274. doi: 10.1016/j.virol.2005.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lupas A, et al. Model structure of the Omp alpha rod, a parallel four-stranded coiled coil from the hyperthermophilic eubacterium Thermotoga maritima. J Mol Biol. 1995;248:180–189. doi: 10.1006/jmbi.1995.0210. [DOI] [PubMed] [Google Scholar]
- 33.Testa OD, Moutevelis E, Woolfson DN. CC+: A relational database of coiled-coil structures. Nucleic Acids Res. 2009;37:D315–322. doi: 10.1093/nar/gkn675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duquerroy S, Vigouroux A, Rottier PJ, Rey FA, Bosch BJ. Central ions and lateral asparagine/glutamine zippers stabilize the post-fusion hairpin conformation of the SARS coronavirus spike glycoprotein. Virology. 2005;335:276–285. doi: 10.1016/j.virol.2005.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beyer WR, Miletic H, Ostertag W, von Laer D. Recombinant expression of lymphocytic choriomeningitis virus strain WE glycoproteins: a single amino acid makes the difference. J Virol. 2001;75:1061–1064. doi: 10.1128/JVI.75.2.1061-1064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.